Abstract
Immune effector cells recruited into tissues after stress integrate multiple signals to help restore the epithelial barrier homeostasis. Experimental measures for immune cell-mediated lysis of tumors or virally infected targets rely on average responses of permeability or apoptotic changes within a population of ‘targets’. Here we examined individual target cells following interaction with lymphoid effectors. We found that human peripheral blood lymphocytes (PBLs) not only provide lytic signals, but also promoted autophagy in the remaining cells. At high effector-to-target ratios, autophagy was induced in several human tumors, as assessed by induction of LC3 puncta and diminished p62. NK cells are a primary mediator of this process. In addition, target cell autophagy was enhanced by provision of IL-2, while IL-10 attenuated this effect, and cell-to-cell contact strongly enhanced lymphocyte-mediated autophagy. Although IFNγ can induce autophagy in target cells, IFNα acted directly on the targets or in concert with lymphocytes to diminish target autophagy in some cell types. Importantly, cell-mediated autophagy (C-MA) promoted resistance from treatment modalities designed to eradicate tumor cells. Our findings therefore demonstrate the lymphocyte-induced cell-mediated autophagy promotes cancer cell survival and may represent an important target for development of novel therapies.
Introduction
Cancer in adults arises in the setting of chronic inflammation associated with perpetual cell damage and cell death (1–3). In trauma, infection, autoimmunity, and cancer, damage to cells results in release of Damage Associated Molecular Pattern Molecules (DAMPs) (4, 5), and the up-regulation of stress ligands/pattern recognition receptors on the remaining cells (6, 7). Enhanced survival of cancer cells following lymphocyte interaction has been observed (8, 9). Those remaining cells not lysed by anti-tumor effectors have previously not been carefully examined. Here, we examine how epithelial cancer cells respond to short-term interactions with human peripheral blood lymphocytes.
Natural killer (NK) cells are rapidly recruited to wounds (10, 11) and tumor sites (12) and can release cytokines including interferons which rapidly modify target-cell biology, up-regulate MHC-I and diminish proliferation. Recently, the NK-family of cell types has been expanded to include several diverse innate lymphoid cells (ILCs) with unique capabilities and functions, not just the cytolytic function attributed to these cells (13). NK cells clearly perform both regulatory and “helper” roles in several contexts (14, 15). Interestingly, the IL-22-producing NK cell subset (NK22/ILC22) is involved in tissue repair and epithelial proliferation (16, 17), expanding on the classic repertoire of NK cell functions. Here, we suggest that peripheral NK cells, and possibly other immune effectors, can induce autophagy and enhance cell and/or tissue survival.
Classic cytotoxicity assays use release of a dye or radionuclide (chromium-51), that is loaded into cells to determine the extent that lymphocytes lyse targets (18, 19). Several paradoxes have been observed with these assays, including plateaus that do not reach the 100% lysis level in spite of increasing effectors (20), and frequent observations of negative measures of cytotoxicity, with enhanced survival in the presence of lymphocytes. Here we show for the first time that classic “cytolytic cells,” including NK cells, can often promote survival and autophagy in target cells, perhaps providing an explanation for these discordant findings.
Materials and Methods
Cell Culture
Human pancreatic cancer lines Panc2.03 and Panc3.27 from ATCC (Manassas, VA). Colon cancer HCT116s were a gift from Dr. Bert Vogelstein (Baltimore, MD), 786-0 from Dr. Jodie Maranchie and WSS1 from Lisa Butterfield (Pittsburgh, Pa). Human bladder cancer cell lines (T-24) stably transfected with GFP-LC3 were a gift from Umamaheswar Duvvuri (Pittsburgh, Pa). 786-0 and WSS1 cells were authenticated using genetic profiling (IDEXX Radil, Columbia, Mo). Cells were cultured in “complete media”; RPMI 1640 (Thermo/Hyclone, Waltham, Ma) supplemented with 10% heat-inactivated fetal bovine serum (Mediatech, Manassas, VA), and antibiotic–antifungal mixture (PenStrep, Gibco/Invitrogen, Ca) in a humidified incubator 37°C with 5% CO2. Cell lines were passaged, split, counted and seeded 4,000–5,000 cells/well in 55 μl of complete media on 384-well collagen-I coated plates (BD Labware 35-6832, Bedford, Ma) for 24h.
Treatment of Cells
We used recombinant human IFNγ (R&D Systems, Minneapolis, MN) and recombinant IFNα from Intron-A (Schering-Plough, Kenilworth, NJ). In cell-cell contact and diffusible factor experiments, supernatant was removed from coculture wells at 2 and 6 hours, spun down, and the top fraction was applied to naive culture wells for the remaining 22 and 18 hours (respectively). Transwell experiments were performed in 24-well plates using 0.4 μm polycarbonate transwell supports (Corning/Costar, Lowell, MA).
Human PBMC
PBMCs were isolated as previously described (21). Briefly, PBMC were collected from the buffy coat using Ficoll-Paque Plus (Amersham Biosciences, Uppsala, Sweden), rinsed and stored in 20% FBS with 10% DMSO at −80°C. Cells were thawed and incubated for 2h in T-75 flasks to remove adherent monocytes. Lymphocytes (82% of cells in PBLs) were rinsed and cultured +/− recombinant human IL-2 (Proleukin; Prometheus, San Diego, Ca) in RPMI+10% FBS. These peripheral blood lymphocytes (PBLs), were harvested and mixed with tumor cells at various E:T ratios in complete media. NK cells were negatively selected with the EasySep Human NK cell enrichment kit (19055, Stem Cell Technologies, Vancouver, Canada) resulting in 74% purity.
Cancer cell lines pre-plated for 24h on 384-well plates were challenged with lymphocytes or purified NK cells. Experimental conditions were prepared in round bottom 96-well plates (BD Falcon 35-3077; Franklin Lakes, NK). 100 μl complete media were used to setup dilution series of lymphocytes (4:1 dilution factor). IL-2 and other dilutions were executed in a similar manner. 20 μl from each of the setup wells was added directly to the existing media on the assay plate with the cancer cells. Experimental plates were incubated in a 37°C incubator with 5% CO2.
Staining, Microscopy and Analysis
Cells were stained for immune-fluorescent imaging, and microscopy was performed with Cellomics Arrayscan VTi (see Supplementary Methods). Autophagy was measured by LC3 morphology (22).
Results
Increased Autophagy in Surviving Tumor Cells Following Lymphocyte Coculture
We have developed a coculture system where dividing human cancer cell lines adhered onto collagen substrate were overlain with primary human lymphocytes. Cytolysis was measured directly by automated adherent cell counting, and was equivalent to the standard 51Cr release assay and the CCK8 cell survival assay (Supplementary Figure 1).
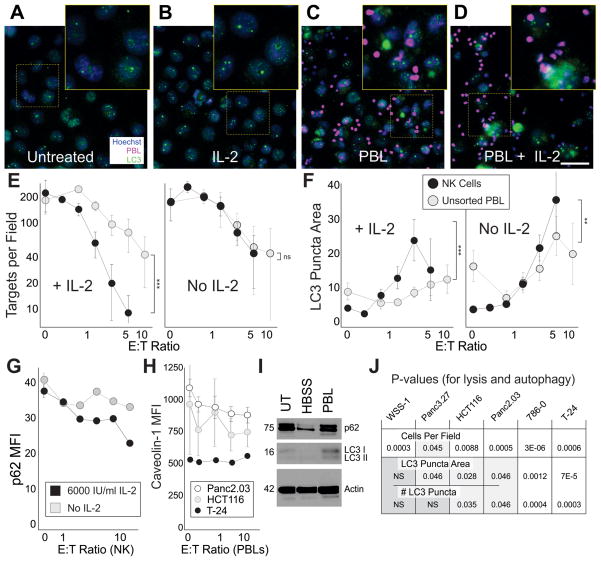
With the coculture model, not only could we determine the rate of lysis in a dose-dependent manner, we also found that many target cells remained adherent even with the highest lymphocyte number delivered, for extended periods (8). Autophagy is a common cellular response to stress and we hypothesized that autophagy could sustain the surviving cells and potentially protect them from lysis. Unlike controls (Figure 1A,B), high effector to target (E:T) ratio cocultures have lymphocyte clusters around the remaining tumor cells (Figure 1C), especially in the presence of IL-2 (Figure 1D). The high lymphocyte/IL-2 condition (Figure 1D) has fewer adherent target cells remaining due to lysis. Importantly, the remaining cells have heightened autophagy, as demonstrated by increases in LC3 puncta area, density and number of puncta. Not only did the fraction of highly autophagic cells increase in the high E:T conditions, but the overall number of autophagic cells increased, confirming induction of heightened autophagy, and not solely a selection of autophagic cells. We next performed a dose-response assessment for unsorted lymphocytes and negatively selected human primary NK cells. NK cells are able to kill targets more effectively than unsorted cells (Figure 1E, ANCOVA comparing regressions of NK vs. PBL p=4.1x10−5) in the presence of IL-2. Both unsorted lymphocytes and NK cells promoted autophagy as assessed by LC3 puncta area (NK No IL2 p=1x10−15, IL2 p=0.035, PBL No IL2 p=1x10−4, IL2 p=4x10−5 regression analysis). Compared with baseline, autophagy was doubled with 2:1 E:T ratios of NKs, and was increased four-fold with 5:1 E:T NK cells. Higher E:T ratios (~40:1) of unsorted lymphocytes (Figure 1F, also Figures 3–7) were required to produce comparable levels of autophagy, suggesting that a major role of NK cells is to promote autophagy (ANCOVA comparing regression of NK vs. PBL for LC3 puncta area, p<0.001). Additionally, CD56+CD3-CD19− cells promoted autophagy to a greater extent than the CD56− lymphocyte population (not shown). Interestingly, other immune cells including macrophages and T cells (Supplemental Figure 2) mediate C-MA as well. Consistent with previous findings, the autophagy markers p62 (23) and caveolin-1 (24) diminish with increasing autophagy (Figure 1G,H, regressions for p62 IL-2 6000 p=3.6x10−8, No IL-2 p=0.00063) as LC3 levels increased (Figure 1I). Induction of target autophagy required the autophagy gene ATG5 (Supplemental Figure 3). Lymphocytes and macrophages can promote “cell-mediated autophagy” (C-MA) in human epithelial cancer cell lines.
Figure 1. Human Peripheral Lymphocytes and NK cells Induce Autophagy in Human Tumors.
A–D Fluorescence microscope images of CAKI-1 renal cancer cells cultured for 24h; untreated (A), treated with 500 IU/ml IL-2 (B), 100:1 effector:target (E:T) ratio of peripheral lymphocytes (C), or 100:1 lymphocytes concurrently treated with 500 IU/ml IL-2 (D) for 16h. 100:1 ratio was used to allow identification of lymphocyte-target interactions. Cancer cell nuclei (blue), PBL nuclei (false-color magenta), LC3 (green). E–F Line charts representing the mean +/−SD for the number of remaining cancer cells (targets) per field (E) and LC3 puncta area (F) of T-24 bladder cancer cells exposed to varying E:T ratios (on a log scale) of negatively selected NK cells (black markers) or unsorted lymphocytes (gray markers) for 24h (ANCOVA comparing NK vs. PBLs ** p<0.01, ***p<0.001). The effector series was in the presence of 6000 IU/ml IL-2 (left panels), or without IL-2 (right panels). All effector series were significant. G, Cytoplasmic p62 mean fluorescence intensity (MFI) in T-24 cells. H, Membrane-associated Caveolin-1 intensity. I, Western blot for LC3 I and II, p62, and actin in HCT116 cells untreated (UT), treated by 4 hour starvation (HBSS) or 30:1 E:T ratio of lymphocytes (PBL). 30:1 was used to limit the effector number required for protein analysis. J, P-values (Chi squared) for cell mediated lysis (remaining cells per field), and two measures of autophagy (LC3 puncta area and #LC3 puncta). Scale bar 50 μm.
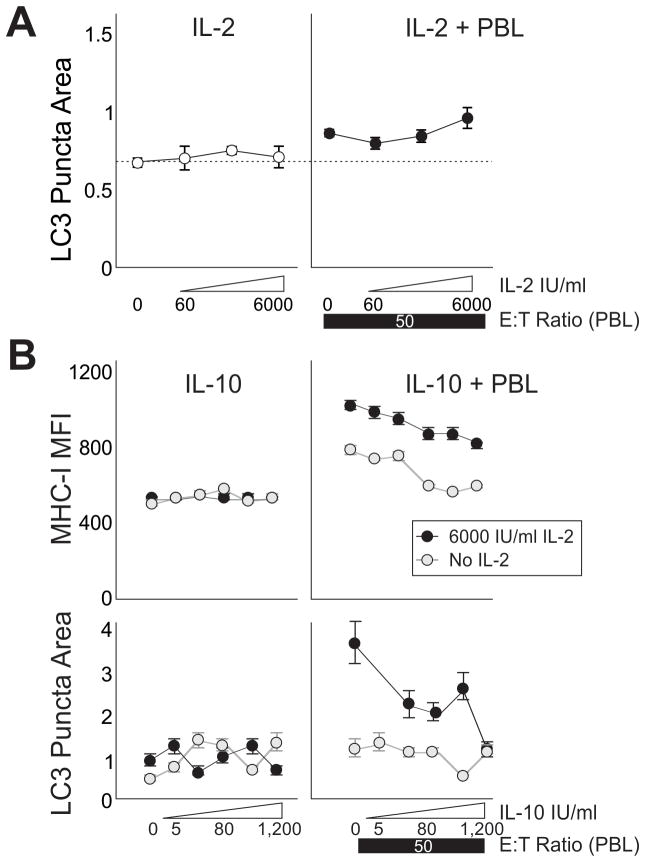
Figure 3. IL-2 Promotes and IL-10 Inhibits PBL Induced Autophagy in Cocultures.
A, Human pancreatic cancer cells (Panc2.03) were cultured alone (left panel) or with lymphocytes (right panel) and treated with IL-2. Markers show LC3 puncta area. B, T-24 bladder cancer cells were treated with a dose series of IL-10 (left panels), or a dose series of IL-10 over a constant E:T ratio of 50:1 PBLs (right panels), with and without IL-2 (black or gray markers). Markers show MHC-I mean fluorescent intensity (upper panels) and LC3 puncta area (lower panels). Error bars are SEM of field replicates.
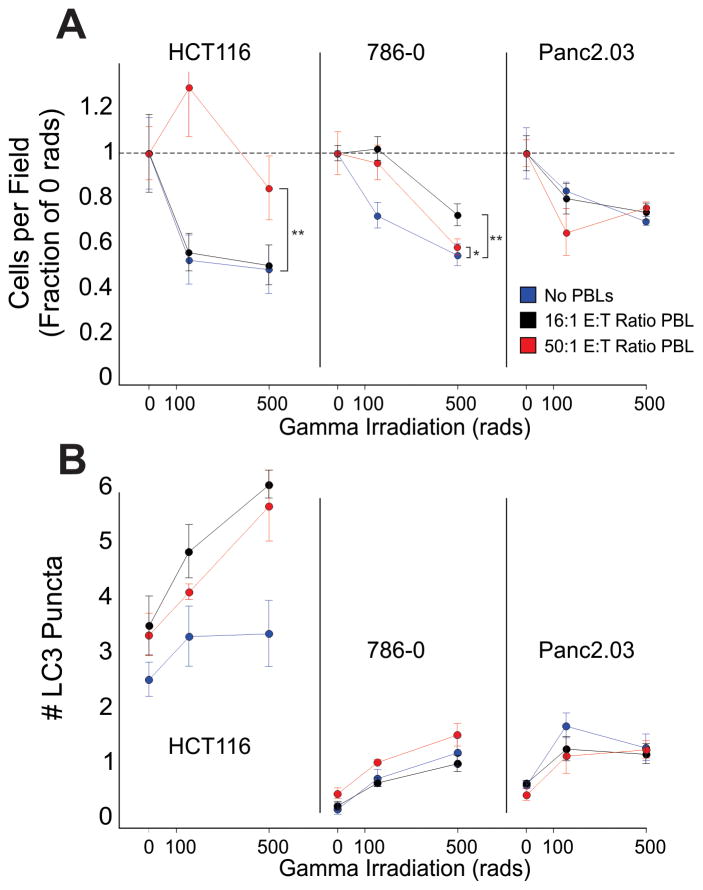
Figure 7. Coculture with Lymphocytes protects HCT116 and 786-0 against subsequent Gamma Irradiation.
HCT116, 786-0 and Panc2.03 cells were cultured with human PBLs for 24h. Following the culture, the lymphocytes were rinsed off, and the cells were harvested and exposed to gamma irradiation. A, Remaining cell numbers, displayed as a fraction of the control (no gamma irradiation) for cultures previously exposed to 16:1 E:T ratio of PBL (black) and 50:1 E:T ratio of PBL, or no PBLs (red), all with 6000 IU/ml IL-2, are plotted as the mean+/−SD of well replicates (** p < 0.01, ** p < 0.05). B, Number of cytoplasmic LC3 puncta are plotted (mean+/− SD of well replicates), indicative of sustained autophagy at 24h post irradiation.
Lymphocytes Effectively Lyse and Induce Autophagy in Multiple Epithelial Cancer Lines
We examined whether lymphocytes had the ability to induce autophagy in several human epithelial cancer cell lines including those derived from the colorectum, pancreas, kidney, and bladder. All cell types were sensitive to lysis with increasing PBL E:T ratios (Figure 1J, first row, chi-square analysis, independent experiments with statistically decreasing slopes). Autophagy was consistently induced and the measurement of LC3 by “LC3 puncta area” (average size of the autophagosomes), was repeatable across cell types and experiments (Figure 1J, second row). WSS-1 did not share this phenotype, possibly due to more variable basal autophagy. Lymphocyte-induced changes in the number of LC3 puncta (Figure 1J, third row) and the individual punctum LC3 density were more cell-type specific. These results indicate significant and reliable autophagic changes induced by lymphocytes for pancreatic, colorectal, renal, and bladder cancer cell lines.
To verify that the autophagic increase was not simply a precursor to cell death, cultures were established for various time periods (Supplemental Figure 4A–C). Autophagic cells could be identified for at least five days, and both the number and percent of autophagic cells increased at high E:T conditions when compared with vehicle controls (Supplemental Figure 4D). Additional studies using caspase inhibitors did not prevent C-MA (Supplemental Figure 5). Therefore, induction of autophagy by lymphocytes isn’t caused directly by induction of cell death pathways, and it likely relates to extended cell survival. Bafilomycin, a late autophagy inhibitor, was used to halt progression of so-called ‘autophagic flux’, which results in un-cleared autophagosomes otherwise targeted for lysosomal degradation. In the presence of PBLs, bafilomycin-treated targets had both enhanced LC3 puncta as well as the lysosome marker lysosomal-associated membrane protein, LAMP1 (Supplemental Figure 6), consistent with a block in autophagosome fusion and degradation.
Autophagic Flux Measured by Imaging LC3
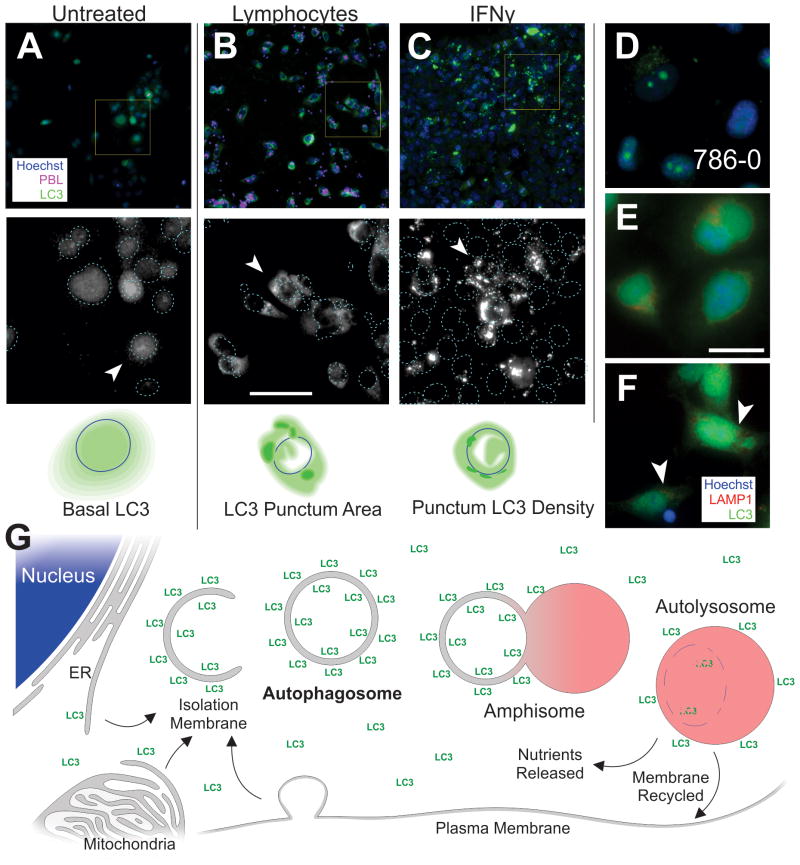
Some autophagic characteristics of epithelial cancer cells are shown in Figure 2. These patterns were context dependent and associated only with particular cell lines or treatments. Cells in control conditions (Figure 2A) had diffuse LC3 with difficult-to-discern puncta. In the pancreatic, renal, and bladder cancer cell lines there was a pool of nuclear LC3 and less cytoplasmic LC3. Several patterns emerged that were mirrored across most cell lines tested. The addition of lymphocytes produced autophagy with more puncta appearing in the cytoplasm. These puncta were quite large and possessed indistinct borders (Figure 2B). The overall amount of LC3 in the cytoplasm and the number of puncta were somewhat higher than the control, but the largest difference was in the area (size) of the puncta. Other treatments, such as IFNγ, produced a different pattern of autophagy. Although there was an increase in the size and number of LC3 puncta, the largest increase was the LC3 density within individual puncta (i.e. the intensity of LC3, Figure 2C). These bright puncta were also easier to identify because of more distinct borders. The localization of the puncta within the cell was also distinct, depending on the context (cell type and treatment). The renal cancer cell line 786-0 (Figure 2D), had a high level of juxta-nuclear LC3 at basal conditions, which was often observed as a diffuse perinuclear signal with defined borders and one or two bright perinuclear puncta. Occasionally a small “mist” of cytoplasmic LC3 was also observed. The classic autophagy inducer, serum starvation and chloroquine inhibition also produced distinct LC3 patterns (Supplemental Figure 7). Staining with the lysosomal marker LAMP1 indicated puncta of both LC3 and LAMP1 in the peri-nuclear region (Figure 2E), which were enhanced after lymphocyte coculture (Figure 2F). Classic progression of the macro-autophagic pathway is illustrated in Figure 2G, indicating the source, progress, and recycling of the recruited membranes associated with autophagy. Imaging LC3 morphology can reveal changes in the density of LC3 on the autophagosomes, amphisome or autolysosome and denote the size of these membranous organelles. The nature and quality of the puncta is variable depending on the type and source of lymphocytes, cytokine types, or cancer cell lines, reflecting the variability often observed within cytolysis assays.
Figure 2. Autophagic Flux and Phenotype in Human Cancer Cell Lines.
Epi-fluorescent imaging of human pancreatic and renal cell lines plated for 24h under various conditions. A–C, Panc2.03, top row; 20x images, Hoechst (blue), LC3 (green), and PBL (false-color magenta), yellow box is magnified in micrographs below. Second row, magnified fields, LC3 channel in grayscale for enhanced contrast; nuclei are outlined with dotted lines. Arrowheads indicate an illustrative cell which is cartooned below. Third row, cartoon of cell showing nuclear outline, diffuse LC3 (light green), and LC3 puncta (dark green). D, A renal cancer cell line (786-0) with resting LC3 staining. E–F, Panc2.03 cells stained for Hoechst (blue), LC3 (green) and LAMP1 (red). Perinuclear puncta from both LC3 and LAMP1 are visible in treated condition (arrow heads). Cells in control conditions (A,D,E), lymphocyte coculture (B,G) and IFNγ (C). G, Overview of classical macroautophagy process. Cytoplasmic LC3 is microtubule associated while membrane LC3 is lipidated. Scale bars 50 μm.
Cytokines Regulate Induction of Autophagy
Unsorted PBLs were used henceforth since they can readily be induced to express lymphokine-activated killer (LAK) activity (25) and to establish the response for the mixture of cells that would exist in a tumor microenvironment in individuals treated with IL-2. We sought to determine the dose response for IL-2 in regards to lymphocyte-induced target autophagy. With no lymphocytes, cancer cells maintained the same level of autophagy through increasing IL-2 treatment (as expected, no significant effect of IL-2). With peripheral lymphocytes present, IL-2 enhanced autophagy in the target cells (Figure 3A right panel, linear regression, p = 0.035). Denatured (boiled) IL-2 was unable to increase autophagy, indicating that IL-2’s mannitol and SDS excipient did not contribute to induction (data not shown). IL-10 was expected to act on the lymphocytes to regulate autophagy in the targets (26). MHC-I expression was used as a positive control. In Figure 3B (upper right), MHC-I was increased in cancer cells by lymphocyte coculture, especially with the addition of IL-2. MHC-I was unchanged by direct treatment of cancer targets with dose escalating IL-10. IL-10 steadily decreased the expression of MHC-I otherwise induced by high E:T ratio of lymphocytes both with (p<0.001) and without IL-2 (p<0.001). The effect on autophagy was similar (Figure 3B, right panels). LC3 puncta area was diminished with IL-10 treatment in coculture, particularly with concurrent IL-2 treatment (No IL-2 p<0.005, with IL-2 p<0.0005). Therefore IL-2 potentiated cell-mediated autophagy, while IL-10 attenuated it.
TGF-β can act on NK cells to diminish target lysis and IFNγ production (8). We have found that blocking TGF-β in the coculture diminishes target-cell autophagy in a dose-dependent manner (Supplemental Figure 8), indicating that TGF-β may be one of the molecular signals responsible for autophagy induction.
Cell-Cell Contact Enhances Lymphocyte Mediated Autophagy
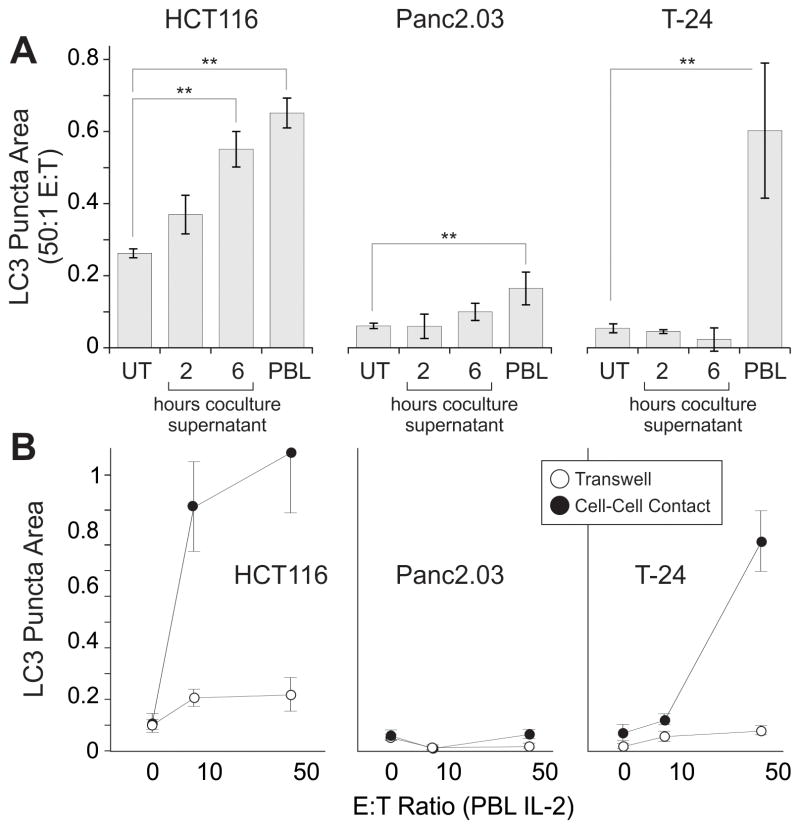
To evaluate mechanisms promoting cell-mediated autophagy, we asked whether soluble factors released from lymphocytes were sufficient for induction. Supernatants from lymphocyte/cancer cocultures were applied to adherent cancer cell cultures (Figure 4A). With supernatant treatment, autophagy was slightly increased when compared with untreated cancer cells (in HCT116 and Panc2.03) but less than with cell:cell contact (50:1 E:T ratio cocultures, ANOVAs HCT116 p = 0.0014, Panc2.03 p = 0.0013, T-24 p = 0.012, Dunnet’s posttests displayed in figure). In a parallel experiment, autophagy was promoted by direct lymphocyte coculture, with only minimal increases observed when cells were separated by a permeable transwell (Figure 4B, ANCOVAs comparing regressions between transwell and cell-cell contract for HCT116 p=0.022, Panc2.03 ns, T-24 p<0.001). Interestingly, overall LC3 intensity in the cancer cell lines was increased by both direct coculture and transwell conditions (data not shown), consistent with the presence of a diffusible factor that is sufficient to up-regulate LC3 expression, but not to markedly increase autophagic flux. Cell-cell contact was required for full increases of autophagy in the cancer cell lines tested.
Figure 4. Cell-Cell Contact Enhances Autophagy.
Lymphocytes were cocultured with hepatocellular, pancreatic, renal and bladder cancer cell lines to examine the extent of cell-mediated autophagy. A, induction of autophagy is measured in targets; untreated (UT), supernatant from 2 and 6 hour coculture, and direct coculture with lymphocytes (PBL). Bars show average with SD, ** p<0.01 by Dunnet’s posttests after ANOVA. B, Cancer cell lines were analyzed with lymphocytes in coculture (filled circles) or separated by a 0.4 μm transwell (open circles) for 24h. IL-2 was added concurrently at 6000 IU/ml under all conditions. Line chart shows averages with SEM, E:T ratio is plotted on a log scale, ANCOVAs were significant for HCT116 p=0.022, T-24 p<0.001.
NK cells express specific receptors which either activate or inhibit effector function. We have found that the activating receptor NKG2D is not responsible for triggering cell mediated autophagy (Supplemental Figure 9). On the other hand, the family of Killer Cell Ig-like Receptors (KIRs); do appear to play a role, at least in some cell types. KIR inhibition both by a pan-KIR blocking antibody and by a SHP2 phosphatase (downstream of the ITIM domain in KIR-L forms) inhibition produce increases in effector IFNγ production (27). Importantly, the inhibitors partially reduce target autophagy (Supplemental Figure 10); supporting the hypothesis that KIR engagement can both inhibit classic (lytic) effector function but induce other effector functions, including C-MA.
IFNγ Induces Autophagy
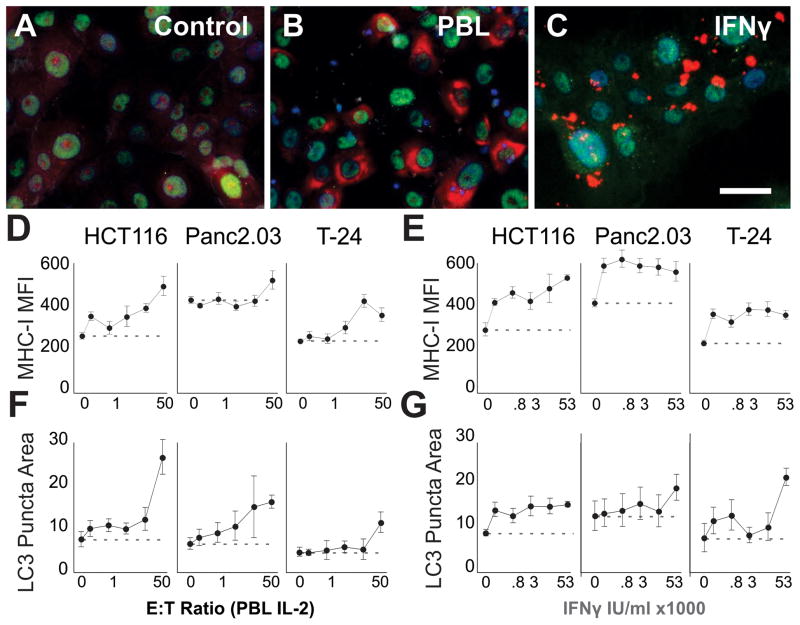
IFNγ is sufficient to induce target autophagy (28–30). Whereas resting cells had diffuse LC3 staining (Figure 5A), addition of lymphocytes to Panc2.03 markedly increased autophagy (Figure 5B). High doses of IFNγ led to substantial increases in the density of LC3 in autophagic puncta (Figure 5C). Using three cell lines, we tested the effects of lymphocyte coculture (up to 50 E:T ratio) in parallel with IFNγ on autophagy. We used MHC class-I expression as a control to demonstrate IFNγ efficacy. IFNγ rapidly and consistently leads to the up-regulation of MHC-I on the surface and in the cytoplasm of target cells (Figure 5E, linear regressions with first two concentrations, all cell types p<0.0003). As expected, the addition of lymphocytes in the coculture also leads to increases in MHC-I expression both on the surface and within the cytoplasm of target cells (Figure 5D, linear regressions all cell types, p<0.0001).
Figure 5. IFNγ Is Sufficient To Induce Autophagy In Epithelial Cancer Cell Lines.
Human colorectal (HCT116), pancreatic (Panc2.03), and bladder (T-24) cancer lines were cultured for 24h alone before the addition of primary lymphocytes or recombinant IFNγ. A–C Micrographs of Panc2.03 in the control condition (A), mixed with lymphocytes at 50:1 E:T ratio (B), or with 53,000 IU/ml IFNγ (C). Note the varied localization and distribution of the LC3 (red) when comparing PBLs with IFNγ treatment (blue Hoechst, green HMGB1). D–G Line charts summarizing the dose response of primary human lymphocytes (PBLs+IL-2, D,F), or IFNγ (E,G) showing Mean+/−StDev of MHC-I expression (D,E) or LC3 puncta area (F,G) on target cancer cells (x-axes log scale). Scale bar 50 μm.
In the same cells, we concurrently examined autophagy. The addition of IFNγ induced autophagy in the cell lines (Figure 5G, linear regression Panc2.03 and T-24 p<0.0001, HCT116 p<0.001 using the lowest two concentrations). IFNγ had the largest effect on LC3 density (Figure 5C, Figure 2C,G,K) but also led to increases in the number and area of the puncta (not shown). Lymphocytes induced increases in LC3 puncta area (Figure 5F, linear regressions p<0.0001 for all cell types). STAT1/2 are downstream of IFN signaling, and we observed increases in the nuclear localization of STAT1 and STAT2 following lymphocyte coculture (Supplemental Figure 11A-B), indicating STAT activation. While both lymphocytes and IFNγ induced autophagy, the phenotype of the target cells was different; suggesting that molecular pathways driven by lymphocyte coculture other than IFNγ may be involved. Additionally, soluble IFNγ receptor (IFNgR1) reduced upregulation of MHC-I but was not able to block C-MA (Supplemental Figure 11C–F), confirming that additional factors or surface molecules can trigger the process.
IFNα Inhibits Cell-Mediated Autophagy in HCT116 and 786-0 Cell Lines
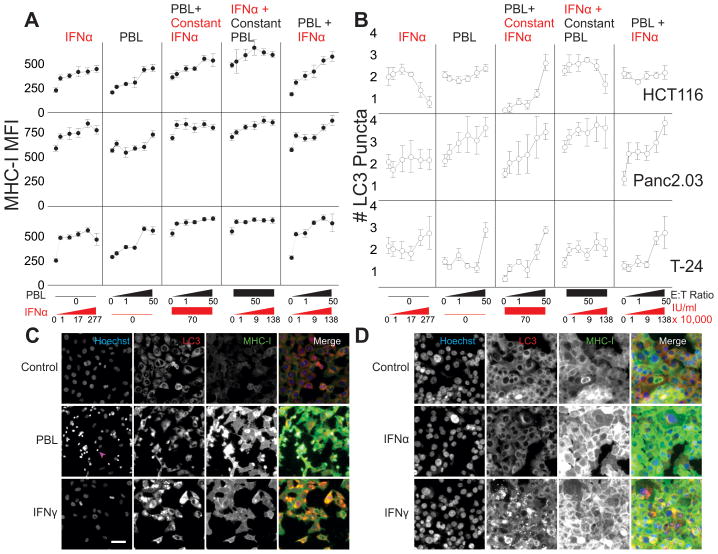
As expected, IFNα was sufficient to upregulate MHC-I on target cells (Figure 6A). Additive effects on MHC-I expression were observed with the combinations of lymphocytes and IFNα (Figure 6A, 2nd panel monotonic, 6A, 3rd panel additive, but no synergy). Increasing both cells and IFNα led to MHC increases with a two-fold greater rate of change (6A, 5th panel). Although IFNα produced the greatest MHC-I expression increase from 0 to 10,000 IU/ml then plateaued, the combination of lymphocytes and IFNα produced greater MHC-1 increases, suggesting that another cytokine (such as IFNγ) was also operative. All IFNα effects on MHC were significant across the concentration range tested (linear regression HCT116 p<0.0001, Panc2.03 p=0.0073, T-24 p=0.014), and for high E:T ratio or high doses of IFNα (t-tests, p<0.0001).
Figure 6. Cell-Mediated Autophagy is inhibited by Alpha Interferon.
Tumor cell lines were grown alone for 24h, then treated with combinations of IFNα and lymphocytes for 24h before measuring cytolysis, MHC-I expression, and autophagic puncta. Line charts showing MHC-I expression (A) and # LC3 puncta (B) with the five columns on each graph representing a dose response to IFNα, increasing E:T ratios of PBLs with no IFNα, a gradient of PBLs with a constant IFNα 70,000 IU/ml, Constant 50:1 E:T Ratio (PBLs) with increasing concentrations of IFNα, and a dual increase in both E:T ratio and IFNα. All x-axes are on a log scale. Micrographs of T-24 bladder cancer (C) and Panc2.03 (D) representing the various patterns of autophagy and MHC-I expression. C, Control growth of cells in Hoechst channel, LC3, MHC-I and merge (blue, red, green respectively). On the second row, 50:1 E:T ratio of lymphocyte coculture after 24h (arrowhead indicating lymphocyte nuclei). Bottom row showing IFN-gamma treatment for comparison. D, Panc2.03 control (top), 70,000 IU/ml IFNα (middle), and IFNγ (bottom) for comparison. Scale bar 50 μm.
Although both IFN γ and α produced similar target-cell changes in MHC-I expression, the effect on target cell autophagy was distinct. Depending on the cell type, IFNα either produced no significant changes in autophagy (Panc2.03, T-24), or reliably inhibited autophagy (HCT116 p=2x10−16). When IFNα was added to cells, autophagic LC3 puncta dropped in size and number in a dose-dependent manner (Figure 6B, 1st panel, p=0.0068). HCT116 autophagy was induced by increasing lymphocyte E:T ratios (Figure 6B, 2nd panel, p=0.0015). IFNα specifically inhibited autophagy induced by lymphocyte coculture (Figure 6B, panels 4, 5), except at the highest E:T ratio tested (panel 3). In all cases, cell-mediated autophagy was limited in the presence of IFNα (multiple regression’s interaction term in HCT116 and 786-0 p<0.001). Representative images are shown in Figure 6C,D. When combined with a late autophagy inhibitor, there is little expansion of autophagosome number with higher doses of IFNα, and when combined with an early autophagy inhibitor, there is little change in IFNα’s effect (Supplemental Figure 12). These results confirm that IFNα does not increase autophagy in the target cells we evaluated.
Cell-Mediated Autophagy Protects Cells from Subsequent Gamma Radiation
We examined the remaining tumor cells following lymphocyte coculture to determine if the cancer cells were protected from radiation-induced cell death. The cancer cell lines were cocultured with human lymphocytes for 24 hours at individual E:T ratios, then harvested and exposed to gamma irradiation. The cancer cells were re-plated for an additional 24 hours before they were analyzed. Increasing doses of gamma irradiation eradicated almost half of the cancer cells in all three cell lines (Figure 7A, regressions for HCT116 p=0.0015, 786-0 p=2.8x10−10, Panc2.03 p=6.7x10−6). Interestingly, HCT116 and 786-0 cells which were previously cocultured with PBLs had a significantly higher fraction of cells remaining following gamma irradiation than those cells which were cultured alone (ANOVA p<0.0001, Dunnet’s post-tests HCT116 500 rads p = 1.2x10−5, 786-0 125 rads p = 0.055, 500 rads p = 0.001. Panc2.03 ns). Gamma irradiation itself induces a stress response in cells, increasing autophagy (Figure 7B). These results suggest that cell-mediated autophagy protects cells from subsequent stressors including radiation. Chemotherapeutic agents’ efficacy was only modestly diminished following previous coculture with lymphocytes (Supplemental Figure 13).
Discussion
Cell mediated lysis (CML) has been well documented since the pioneering studies of the Hellstroms and others (31). Multiple reasons for the failure of immune effectors to fully eradicate tumor have been advanced, including immune exhaustion, factors released within the tumor microenvironment to limit effector survival or efficacy (21) in vitro and in vivo (8, 9), etc. Relatively little attention has been given to the state of the remaining tumor cells. Here we suggest that cell-mediated autophagy (C-MA) may play a role in inducing resistance to immune effectors and also limit the ability of tumors to respond to subsequent chemotherapy or radiation therapy (32).
Target cells with high autophagy persisted in culture for at least five days following interaction with immune effectors. The induction of autophagy was demonstrated for six individual epithelial cancer cell lines and is ATG5 dependent. Measuring autophagic flux was achieved by quantifying LC3 puncta (33–35). IL-2 further activates lymphocytes to induce LAK activity (25), enhancing lysis but also enhancing autophagy induction in the non-lysed targets. Direct cell-cell contact produced the strongest C-MA. IFNγ was sufficient to induce autophagy in the target cells. However, the morphology of the autophagosomes was different than that observed with PBL-induced autophagy, with especially high-density LC3 puncta. The effect of IFNα depended on the specific target cell type used. IFNα was able to inhibit autophagy when combined with lymphocyte coculture. Both KIR and TGF-β inhibition diminished (but did not abolish) C-MA, making them candidates for the molecular mechanism.
Autophagy is a basal repair and stress response mechanism, and in many ways mediates “programmed cell survival”. Autophagy is important oncologically since it can both suppress tumorigenesis at early stages (36, 37), and enable cancer adaptation and recurrence after therapy in late stages (32, 38), thus suggesting an ‘autophagic switch’ arising during carcinogenesis. While IL-2 administration can be potentially curative, it creates a “systemic autophagic syndrome”, limiting vital processes within tissues at the expense of cell survival (39, 40). Combination with the autophagy inhibitor chloroquine significantly enhances IL-2’s effects (41). The balance between activating and inhibitory signals regulating NK-mediated lysis is now more nuanced, given that lymphocytes can also induce C-MA in targets. Classic activating signals for cytolysis (42) may also promote repair and survival in some circumstances.
We showed that IFNγ, but not IFNα, enhanced autophagy in cancer cell lines. IFNγ induces autophagy (43) both to enhance antigen processing and presentation through MHC-I (44, 45). Increased autophagy also enhances the process of viral/bacterial digestion through xenophagy (46). In the liver for example, IFNγ is likely responsible (along with IL-12/IL-18), for the clearance of hepatitis virus (47, 48). IFNα primarily enhances NK-cell cytotoxicity (49). The differential response produced from these type-I and type-II interferons provides a potential mechanism for lymphocytes to deliver differential repair/cytolytic instructions to target cells. Additional cytokines such as IL-10, which diminishes C-MA as well as IL-4 may play roles in negatively regulating C-MA.
Lymphocytes perform critical roles in the early detection and response to cellular/tissue stress. NK cells detect stress signals arrayed on the surface of cells (5, 6, 13). While it is well established that NK cells can lyse virus-infected, damaged and stressed cells, the role of innate lymphocytes in promoting survival and healing is only now being studied (50). Permitting epithelial cell survival, in some instances, could enhance barrier function and integrity.
Using a new image-based cytolysis assay, we show that lymphocytes can promote cell survival. The induction of autophagy, the precise signals and means for communication to induce this function, needs to be more fully evaluated to understand the complex multistep interactions between lymphoid cells and their targets. The potentially reparative or protective signal provided by lymphocytes to induce autophagy could promote wound healing or response to pathogens, but could also promote the development and persistence of cancer.
Supplementary Material
Acknowledgments
Fiscal support
UPCI Cancer Center Support Grant, NIH 5P30 CA47904
(Nancy E. Davidson) NIH 1 P01 CA 101944-04 (Michael T. Lotze)
We acknowledge Anne Geller, Nicole Schapiro, Jesse Payton, Jillian Bonaroti and Dr. D. Andy Clump for their assistance. We appreciate collaborators in the Pawel Kalinski laboratory for cell death inhibitors and Prometheus for the kind gift of IL-2. Dan Normolle in UPCI biostatistics was consulted on the model employed for performing multiple regressions of PBL/IFNα. We would like to give special thanks to our Cancer Center director, Nancy Davidson and the Department of Surgery Chairman, Timothy Billiar for supporting this work. Grant support includes an NKDC NIH P01 CA 101944-04 and NCI core support award P30CA047904.
Footnotes
Conflict of Interest: No Conflict of Interest
References
- 1.Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol. 2004;4:641–8. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 2.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–45. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 4.Lotze MT, Deisseroth A, Rubartelli A. Damage associated molecular pattern molecules. Clin Immunol. 2007;124:1–4. doi: 10.1016/j.clim.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubartelli A, Lotze MT. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol. 2007;28:429–36. doi: 10.1016/j.it.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 7.Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 8.Wilson EB, El-Jawhari JJ, Neilson AL, Hall GD, Melcher AA, Meade JL, et al. Human Tumour Immune Evasion via TGF-beta Blocks NK Cell Activation but Not Survival Allowing Therapeutic Restoration of Anti-Tumour Activity. PLoS One. 2011;6:e22842. doi: 10.1371/journal.pone.0022842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlsten M, Norell H, Bryceson YT, Poschke I, Schedvins K, Ljunggren HG, et al. Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells. J Immunol. 2009;183:4921–30. doi: 10.4049/jimmunol.0901226. [DOI] [PubMed] [Google Scholar]
- 10.Agaiby AD, Dyson M. Immuno-inflammatory cell dynamics during cutaneous wound healing. J Anat. 1999;195 ( Pt 4):531–42. doi: 10.1046/j.1469-7580.1999.19540531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viswanathan K, Dhabhar FS. Stress-induced enhancement of leukocyte trafficking into sites of surgery or immune activation. Proc Natl Acad Sci U S A. 2005;102:5808–13. doi: 10.1073/pnas.0501650102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Y, Santoriello C, Mione M, Hurlstone A, Martin P. Live imaging of innate immune cell sensing of transformed cells in zebrafish larvae: parallels between tumor initiation and wound inflammation. PLoS Biol. 2010;8:e1000562. doi: 10.1371/journal.pbio.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–7. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 14.Koyasu S, Moro K. Type 2 innate immune responses and the natural helper cell. Immunology. 2011;132:475–81. doi: 10.1111/j.1365-2567.2011.03413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hepworth MR, Grencis RK. Disruption of Th2 immunity results in a gender-specific expansion of IL-13 producing accessory NK cells during helminth infection. J Immunol. 2009;183:3906–14. doi: 10.4049/jimmunol.0900577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–5. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: new players in mucosal immunity and tissue repair? Nat Rev Immunol. 2009;9:229–34. doi: 10.1038/nri2522. [DOI] [PubMed] [Google Scholar]
- 18.Perlmann P, Perlmann H, Muller-Eberhard HJ, Manni JA. Cytotoxic effects of leukocyres triggered by complement bound to target cells. Science. 1969;163:937–9. doi: 10.1126/science.163.3870.937. [DOI] [PubMed] [Google Scholar]
- 19.Kaliss N. Micromethod for assaying immune cytolysis by the release of 51Cr. Transplantation. 1969;8:526–30. doi: 10.1097/00007890-196910000-00047. [DOI] [PubMed] [Google Scholar]
- 20.Bol SJ, Rosdorff HJ, Ronteltap CP, Hennen LA. Cellular cytotoxicity assessed by the 51Cr release assay. Biological interpretation of mathematical parameters. J Immunol Methods. 1986;90:15–23. doi: 10.1016/0022-1759(86)90378-9. [DOI] [PubMed] [Google Scholar]
- 21.Ito N, DeMarco RA, Mailliard RB, Han J, Rabinowich H, Kalinski P, et al. Cytolytic cells induce HMGB1 release from melanoma cell lines. J Leukoc Biol. 2007;81:75–83. doi: 10.1189/jlb.0306169. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 24.Martinez-Outschoorn UE, Balliet RM, Rivadeneira DB, Chiavarina B, Pavlides S, Wang C, et al. Oxidative stress in cancer associated fibroblasts drives tumor-stroma co-evolution: A new paradigm for understanding tumor metabolism, the field effect and genomic instability in cancer cells. Cell Cycle. 2010;9:3256–76. doi: 10.4161/cc.9.16.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lotze MT, Grimm EA, Mazumder A, Strausser JL, Rosenberg SA. Lysis of fresh and cultured autologous tumor by human lymphocytes cultured in T-cell growth factor. Cancer Res. 1981;41:4420–5. [PubMed] [Google Scholar]
- 26.Harris J. Autophagy and cytokines. Cytokine. 2011 doi: 10.1016/j.cyto.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 27.Abeyweera TP, Merino E, Huse M. Inhibitory signaling blocks activating receptor clustering and induces cytoskeletal retraction in natural killer cells. J Cell Biol. 2011;192:675–90. doi: 10.1083/jcb.201009135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–7. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap GS, Ling Y, Zhao Y. Autophagic elimination of intracellular parasites: convergent induction by IFN-gamma and CD40 ligation? Autophagy. 2007;3:163–5. doi: 10.4161/auto.3655. [DOI] [PubMed] [Google Scholar]
- 30.Khalkhali-Ellis Z, Abbott DE, Bailey CM, Goossens W, Margaryan NV, Gluck SL, et al. IFN-gamma regulation of vacuolar pH, cathepsin D processing and autophagy in mammary epithelial cells. J Cell Biochem. 2008;105:208–18. doi: 10.1002/jcb.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hellstrom I. A colony inhibition (CI) technique for demonstration of tumor cell destruction by lymphoid cells in vitro. Int J Cancer. 1967;2:65–8. doi: 10.1002/ijc.2910020111. [DOI] [PubMed] [Google Scholar]
- 32.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–63. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 34.Tang D, Kang R, Xiao W, Zhang H, Lotze MT, Wang H, et al. Quercetin prevents LPS-induced high-mobility group box 1 release and proinflammatory function. Am J Respir Cell Mol Biol. 2009;41:651–60. doi: 10.1165/rcmb.2008-0119OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–98. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 36.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livesey KM, Tang D, Zeh HJ, Lotze MT. Not just nuclear proteins: ‘novel’ autophagy cancer treatment targets - p53 and HMGB1. Curr Opin Investig Drugs. 2008;9:1259–63. [PubMed] [Google Scholar]
- 38.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chavez AR, Buchser W, Basse PH, Liang X, Appleman LJ, Maranchie JK, et al. Pharmacologic administration of interleukin-2. Ann N Y Acad Sci. 2009;1182:14–27. doi: 10.1111/j.1749-6632.2009.05160.x. [DOI] [PubMed] [Google Scholar]
- 40.Lotze MT. Interleukin Therapy. In: Vincent T, DeVita SH, Steven A, Rosenberg, editors. Cancer: Principles and Practice of Oncology. 9. Philadelphia, Pa: Lippincott Williams and Wilkins; 2011. pp. 469–81. [Google Scholar]
- 41.Liang X, de Vera ME, Buchser WJ, Romo de Vivar Chavez A, Loughran P, Beer-Stolz D, et al. Inhibiting Autophagy During Interleukin 2 Immunotherapy Promotes Long Term Tumor Regression. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loux Tara J, MTL, Herbert J Zeh. NK cells as recipients of cytokine signals. In: Lotze Michael T AWT., editor. Natural Killer Cells Basic Science and Clinical Application. London, England: Academic Press; 2010. [Google Scholar]
- 43.Tu S, Quante M, Bhagat G, Takaishi S, Cui G, Yang XD, et al. Interferon-{gamma} inhibits gastric carcinogenesis by inducing epithelial cell autophagy and T cell apoptosis. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-10-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhl M, Kepp O, Jusforgues-Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 2009;16:991–1005. doi: 10.1038/cdd.2009.8. [DOI] [PubMed] [Google Scholar]
- 45.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–41. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cavanaugh VJ, Guidotti LG, Chisari FV. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71:3236–43. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura K, Kakimi K, Wieland S, Guidotti LG, Chisari FV. Activated intrahepatic antigen-presenting cells inhibit hepatitis B virus replication in the liver of transgenic mice. J Immunol. 2002;169:5188–95. doi: 10.4049/jimmunol.169.9.5188. [DOI] [PubMed] [Google Scholar]
- 49.Chijioke O, Munz C. Interactions of human myeloid cells with natural killer cell subsets in vitro and in vivo. J Biomed Biotechnol. 2011;2011:251679. doi: 10.1155/2011/251679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conigliaro P, Scrivo R, Valesini G, Perricone R. Emerging role for NK cells in the pathogenesis of inflammatory arthropathies. Autoimmun Rev. 2011 doi: 10.1016/j.autrev.2011.04.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.