Abstract
Background
Acne inversa is a chronic, suppurative relapsing inflammatory skin disease that primarily affects the axillae, perineum and inframammary regions. Evidence suggests that the innate immune system is involved in the pathogenesis of acne inversa.
Objective
To investigate the role of the innate immune system in acne inversa.
Methods
Skin biopsies were obtained from inflammatory skin lesions (n=17) and from non-lesional skin (intraindividual control, n=17) of patients with acne inversa. Additional skin lesions were taken from patients with chronic venous leg ulcers (interindividual control, n=5). Quantitative real-time reverse transcription-polymerase chain reaction was used to determine the mRNA levels of antimicrobial peptides and proteins (AMPs), including human β-defensin (hBD)-1, hBD-2 and hBD-3, LL-37 (cathelicidin) and Ribonuclease 7 (RNase 7). mRNA levels were also determined for inflammatory and anti-inflammatory cytokines, including tumor necrosis factor-alpha (TNF-α), matrix metalloproteinase-1 (MMP1), interleukin (IL)-1β, IL-6, IL-8 and IL-10.
Results
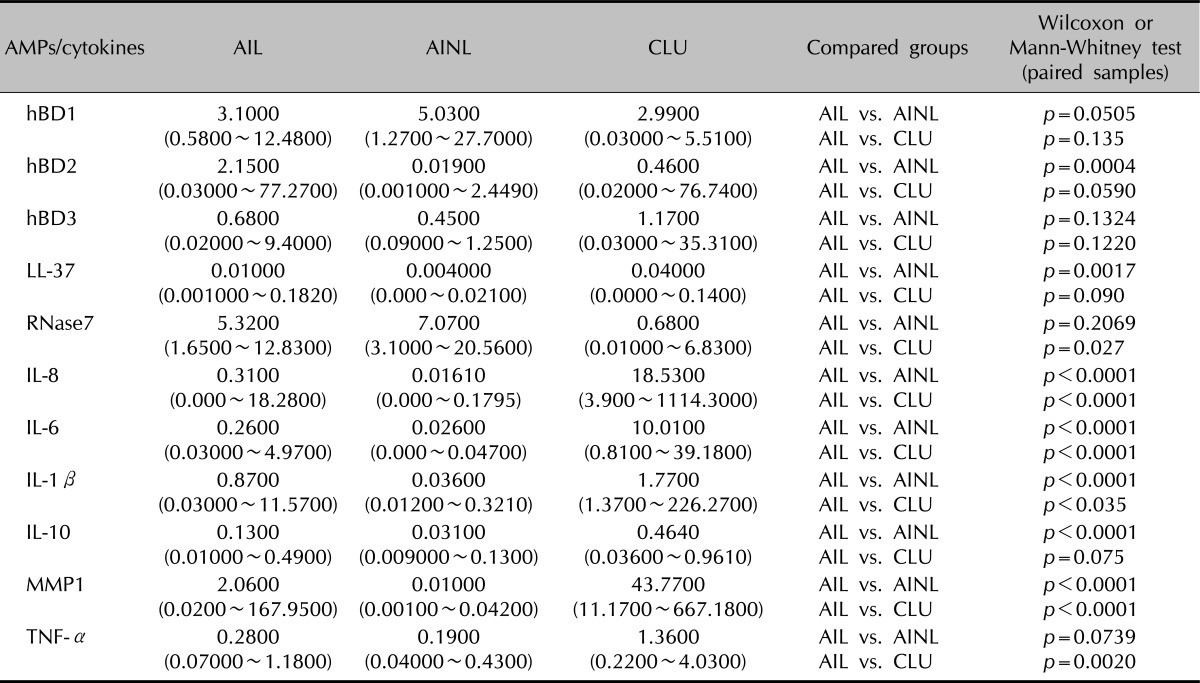
The mRNA levels of hBD-2, LL-37, IL-1β, IL-6, IL-8, IL-10 and MMP1 were significantly higher in acne inversa lesions compared to non-lesional skin (p<0.05). A significant positive correlation expression was observed between hBD-2 mRNA expression and LL-37 (ρ=0.53, p=0.03), and between hBD-2 and RNAse 7 (ρ=0.68, p=0.006). When compared to the chronic venous leg ulcer lesions, acne inversa lesions showed a significantly higher expression of RNase 7 mRNA, while IL-1 β, IL-6, IL-8, TNF-α and MMP1 mRNA expression was significantly higher in the chronic venous leg ulcer lesions (p<0.05).
Conclusion
The AMP, cytokine milieu and tissue proteases in acne inversa lesions differ significantly from non-lesional skin and chronic venous leg ulcers. The positively correlating up-regulation of AMPs in acne inversa indicates an important role of the innate immune system in the pathogenesis of this disorder.
Keywords: Antimicrobial peptides, Hidradenitis suppurativa, Human β-defensin, LL-37, Ribonuclease 7
INTRODUCTION
Acne inversa (AI, hidradenitis suppurativa) is a chronic, recurrent suppurative inflammatory skin disease that primarily affects the axillae, perineum and inframammary regions1. Although it has historically been described as a disorder of apocrine origin, there is now evidence for a follicular-based pathogenesis with a secondary inflammation of apocrine glands2,3. Based on the chronic relapsing nature of the disease, alteration of host defense factors has recently been considered to be its etiopathogenesis. Schlapbach et al.4 were the first to show that the mRNA expression of the antimicrobial peptides (AMPs) human β-defensin (hBD)-1 and psoriasin are significantly increased in AI lesions compared to non-lesional skin. Additionally, several other epithelial chronic inflammatory conditions of the skin, intestines and airways have shown an altered expression of AMPs5-8.
Based on these initial findings, we investigated a variety of AMPs including hBD1, hBD2, hBD3, Ribonuclease 7 (RNase 7) and LL-37, and inflammatory cytokines including interleukin (IL)-1β, IL-6, IL-8, IL-10, tumor necrosis factor-alpha (TNF-α) and matrix metalloproteinase-1 (MMP1) in AI lesions, non-lesional skin and chronic inflammatory skin to further elucidate a possible role of AMPs in AI.
MATERIALS AND METHODS
Patients
In this prospective study, we recruited 17 patients (10 females, 7 males, 22~50 years of age) diagnosed with AI. The diagnosis was based on clinical criteria. Patients had a longstanding history (at least 18 months) of chronic AI. Tissue samples were obtained from all patients from the affected skin area and from non-affected skin that was at least 10 cm from the clinically visible AI lesions. In addition to the intraindividual control, skin samples of five patients (4 females, 1 male, 49~70 years of age) with chronic leg ulcers served as a control. The study was conducted in light of the declaration of Helsinki. All subjects who participated in the investigation signed an informed consent.
Real-time reverse transcription-polymerase chain reaction (RT-PCR)
The mRNA levels of hBD1, hBD2, hBD3, RNase7, LL-37, TNF-α, IL-1β, IL-6, IL-8, IL-10 and MMP1 were analyzed. Quantitative analysis of the real-time RT-PCR was performed as previously suggested9. The total cellular RNA was isolated from the skin tissue samples using RNeasy® Lipid Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. Prior to cDNA synthesis, RNA was digested with RNase-free DNase I (Roche Diagnostics, Indianapolis, IN, USA). The cDNA was synthesized by reverse transcription from DNase I-treated RNA using MultiScribe™ reverse transcriptase enzyme and random hexamer primers (TagMan®, Reverse transcription reagents; Applied Biosystems, Foster City, CA, USA). Real-time PCR was performed using a Power SYBR® Green PCR Master Mix and GeneAmp® Sequence Detection System (Applied Biosystems). The PCR primers for AMPs, cytokines and the housekeeping gene (RPL-38) were designed using the computer program Primer Express (Applied Biosystems) and produced by the custom oligonucleotide synthesis service TIB Molbiol (Berlin, Germany) (Table 1). PCR amplifications were performed in a total volume of 25µl, which contained 5µl of cDNA sample, 5µM of each primer and 12.5µl of Power SYBR® Green PCR Master Mix. The PCR was started for 2 minutes at 50℃ and an initial 10 minutes at the denaturing temperature of 95℃, followed by a total of 40 cycles of 15 seconds of denaturing and 1 minute of annealing and elongation at 60℃. The reaction products were separated by 2% agarose gel electrophoresis. Relative mRNA expression levels were calculated using the comparative Δ-ΔCt method as previously suggested by Livak and Schmittgen10. Quantities of all targets in the test samples were normalized to the corresponding RPL-38 RNA transcript in the skin samples. The quantities of mRNA levels were determined using the median log transform of the gene expression.
Table 1.
Primers of antimicrobial peptides and proteins, cytokines and the housekeeping gene used in the real-time reverse transcription-polymerase chain reaction study
hBD: human β-defensin, RNase7: ribonuclease 7, IL: interleukin, MMP1: matrix metalloproteinase-1, TNF-α: tumor necrosis factor-alpha.
Statistical analyses
Data analysis was performed using the statistical package MedCalc software (MedCalc, Mariakerke, Belgium). Non-parametric analyses of the real-time RT-PCR results were used because a non-normal distribution was observed in subsets of data (D'Agostino-Pearson test). The Wilcoxon rank-sum test was applied to test for overall significant differences of data between groups (AI vs. non-lesional skin, paired data). A Mann-Whitney test was used to test for significant differences of data between AI vs. chronic venous ulcer lesions (unpaired data). Correlation studies were performed using Spearman's coefficient of rank correlation (mRNA data). Corrected values of p <0.05 were considered significant.
RESULTS
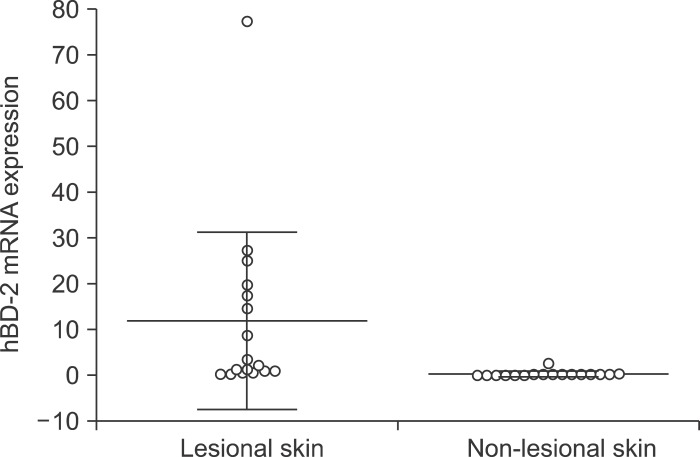
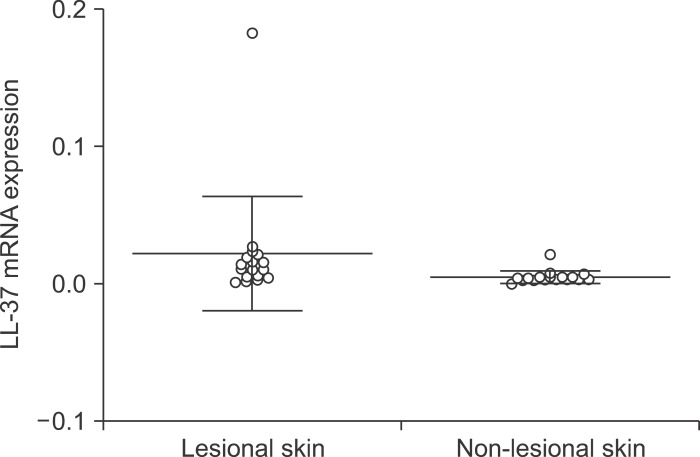
In total, we studied 17 patients (10 females, seven males; 22~50 years of age, mean age 35.1±8.8 years) diagnosed with AI and 5 patients (4 females, 1 male; 49~74 years of age, mean age 65.2±9.6 years) diagnosed with chronic venous ulcers. Non-lesional skin of AI patients served as the control. The mRNA expression of hBD-2, LL-37, IL-1β, IL-6, IL-8, IL-10 and MMP1 was significantly higher in AI compared to non-lesional skin (p<0.05) (Fig. 1, 2, Table 2). The mRNA expression levels of hBD1, hBD3, RNase7 and TNF-α showed no significant differences (p>0.05). The expression of hBD-2, LL-37, hBD-2 and RNase7 showed a significant degree of correlation (Spearman's rank correlation coefficient ρ=0.53, p=0.03 and ρ=0.68, p=0.006). The mRNA expression of RNase7 was significantly higher in AI when compared to CVU patients. For IL-1β, IL-6, IL-8, TNF-α and MMP1 mRNA, the expression was found to be significantly higher in CVU compared to AI.
Fig. 1.
Human β-defensin (hBD)-2 mRNA expression in acne inversa lesional skin and non-lesional skin. Significantly higher expression in lesional skin (p<0.01).
Fig. 2.
LL-37 mRNA expression in acne inversa lesional skin and non-lesional skin. Significantly higher expression in lesional skin (p<0.01).
Table 2.
Antimicrobial peptides and proteins (AMPs) and cytokine mRNA expression in acne inversa lesional skin (AIL) acne inversa non-lesional skin (AINL) and chronic leg ulcers (CLU)
Quantities of analyzed genes were determined using the median log-transformation of the gene expression. Relative mRNA expression levels were calculated using the comparative Δ-ΔCt method. Gene expression was normalized to the homogenously expressed housekeeping gene glyceraldehyde-3-phosphate dehydrogenase. hBD: human β-defensin, RNase7: Ribonuclease 7, IL: interleukin, MMP1: matrix metalloproteinase-1, TNF-α: tumor necrosis factor-alpha.
DISCUSSION
Antimicrobial peptides hBD1, hBD2, hBD3, LL37, RNase7 in AI
Various studies have shown an altered expression of antimicrobial peptides AMPs in chronic inflammatory conditions of different epithelial tissues. Psoriasis vulgaris, lichen sclerosus, atopic dermatitis and AI are among those skin conditions that have been investigated4,11-14.
In conjunction with the findings of Schlapbach et al.10, we were able to show a significant increase in mRNA expression of hBD2 in patients with AI. Interestingly, the present data revealed that LL37, another potent AMP with a critical role in mammalian innate immune defense against bacterial and fungal infections, was likewise significantly over-expressed. LL37 is a broad spectrum AMP that inhibits the growth of a variety of gram-negative bacteria, gram-positive bacteria and Candida species. Produced predominantly by neutrophils and epithelial cells, it is particularly known to be expressed in skin keratinocytes in vivo under inflammatory conditions15. Because we were able to show a significant correlation of hBD2 and LL37 mRNA expression (ρ=0.53, p=0.03), a combined up-regulation of both AMPs has to be considered.
In contrast to the increase of hBD2 mRNA expression, which is preferentially a gram-negative bactericidal AMP, the broad-spectrum AMP, hBD3, showed no significant difference in AI patients. The regulation of hBD3 expression differed from hBD2 expression regulation. While hBD2 expression is highly dependent on IL-1 and direct bacterial induction, interferon-γ, insulin-like growth factor-I and transforming growth factor-α are strong hBD3-inducing cytokines16. In addition to pro-inflammatory cytokines, such as IL-1 and IL-6, interferon-γ, insulin-like growth factor-I and transforming growth factor-α induce the transactivation of the epithelial growth factor receptor, which leads to an increased expression of hBD3. However, an up-regulation of hBD3 expression was absent in our group of AI patients. The same was true for hBD1 mRNA expression, which was not up-regulated in the AI patients. Because hBD1 mRNA expression has previously been shown not to be up-regulated under inflammatory conditions in mucosa or skin, this finding was expected for AI lesions, as previously shown17,18.
Immunohistochemistry indicated a predominant epidermal expression of hBD2 and hBD3. Similarly to Schlapbach et al.4, we were able to observe a significant increase of hBD2 expression on the mRNA level, while the protein expression was significantly lower when compared to non-lesional skin by immunohistochemistry4.
Inflammatory and anti-inflammatory cytokines IL-1β, IL-6, IL-8, IL-10, MMP1 and TNF-α
The inflammatory and anti-inflammatory cytokines of the IL family are important regulators of the inflammatory response.
IL-1β is a potent proinflammatory cytokine expressed in activated macrophages and has been implicated in acute and chronic inflammatory conditions19. Its synthesis is induced by bacterial lipopolysaccharide and by LL37, linking the innate immune system to the adaptive immune system. LL37 activates the purinergic receptor P2X7, a nucleotide-gated channel that is a potent inducer of IL-1β synthesis20. Inflammatory pain hypersensitivity by induction of cyclooxygenase 2 and regulation of epidermal Langerhans cell migration are shown to be effects of IL-1β21,22.
IL-6 acts as both an inflammatory and anti-inflammatory cytokine and is expressed in T cells and macrophages as an acute phase response protein. It is involved in the differentiation of B-cells, lymphocytes and monocytes. Its anti-inflammatory effect is caused by inhibition of TNF-α and IL-1. This is consistent with our results, which showed that TNF-α was not expressed significantly higher (p>0.05) in the skin of AI patients.
IL-8 produced in macrophages and epithelial cells is a chemotactic factor that attracts basophils, neutrophils and T cells. It induces chemotaxis of neutrophils in response to inflammation and has an important role in the innate immune system response. Clinically, it is shown to be increased in localized inflammation, such as psoriasis and gingivitis. The same was observed in the skin of AI patients.
IL-10 is an anti-inflammatory cytokine produced by monocytes, lymphocytes and mast cells. It counteracts the inflammatory response via the inhibition of pro-inflammatory cytokines, such as interferon-γ, IL-2, IL-3, TNF-α or GM-CSF.
MMP1 is an enzyme also known as collagenase 1, and it degrades fibrillar collagens23. It is involved in physiological (embryonic development and tissue remodeling) and pathological processes (arthritis and metastasis). Because AI is an inflammatory process that potentially involves extensive tissue destruction of the skin, MMP1 up-regulation in inflammatory tissue was expected.
Other than TNF-α, all of the inflammatory and anti-inflammatory cytokines that were investigated (IL-1β, IL-6, IL-8, IL-10 and MMP1) were expressed significantly higher in AI, compared to non-lesional skin (p<0.05).
AMP, cytokine milieu and tissue proteases in AI lesions differ significantly from non-lesional skin and chronic venous leg ulcers. The positive correlation of up-regulation of AMPs in AI indicates an important role of the innate immune system in the pathogenesis of this disorder.
References
- 1.Bechara FG, Hartschuh W. Acne inversa. Hautarzt. 2010;61:39–46. doi: 10.1007/s00105-009-1813-2. [DOI] [PubMed] [Google Scholar]
- 2.ANderson MJ, Jr, Dockerty MB. Perianal hidradenitis suppurativa; a clinical and pathologic study. Dis Colon Rectum. 1958;1:23–31. doi: 10.1007/BF02616510. [DOI] [PubMed] [Google Scholar]
- 3.Kurzen H, Kurokawa I, Jemec GB, Emtestam L, Sellheyer K, Giamarellos-Bourboulis EJ, et al. What causes hidradenitis suppurativa? Exp Dermatol. 2008;17:455–456. doi: 10.1111/j.1600-0625.2008.00712_1.x. [DOI] [PubMed] [Google Scholar]
- 4.Schlapbach C, Yawalkar N, Hunger RE. Human beta-defensin-2 and psoriasin are overexpressed in lesions of acne inversa. J Am Acad Dermatol. 2009;61:58–65. doi: 10.1016/j.jaad.2008.12.033. [DOI] [PubMed] [Google Scholar]
- 5.Beisswenger C, Kandler K, Hess C, Garn H, Felgentreff K, Wegmann M, et al. Allergic airway inflammation inhibits pulmonary antibacterial host defense. J Immunol. 2006;177:1833–1837. doi: 10.4049/jimmunol.177.3.1833. [DOI] [PubMed] [Google Scholar]
- 6.Dann SM, Eckmann L. Innate immune defenses in the intestinal tract. Curr Opin Gastroenterol. 2007;23:115–120. doi: 10.1097/MOG.0b013e32803cadf4. [DOI] [PubMed] [Google Scholar]
- 7.Menendez A, Brett Finlay B. Defensins in the immunology of bacterial infections. Curr Opin Immunol. 2007;19:385–391. doi: 10.1016/j.coi.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 8.de Jongh GJ, Zeeuwen PL, Kucharekova M, Pfundt R, van der Valk PG, Blokx W, et al. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J Invest Dermatol. 2005;125:1163–1173. doi: 10.1111/j.0022-202X.2005.23935.x. [DOI] [PubMed] [Google Scholar]
- 9.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods. 2001;25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 12.Gambichler T, Skrygan M, Tigges C, Kobus S, Gläser R, Kreuter A. Significant upregulation of antimicrobial peptides and proteins in lichen sclerosus. Br J Dermatol. 2009;161:1136–1142. doi: 10.1111/j.1365-2133.2009.09273.x. [DOI] [PubMed] [Google Scholar]
- 13.Gambichler T, Skrygan M, Tomi NS, Othlinghaus N, Brockmeyer NH, Altmeyer P, et al. Differential mRNA expression of antimicrobial peptides and proteins in atopic dermatitis as compared to psoriasis vulgaris and healthy skin. Int Arch Allergy Immunol. 2008;147:17–24. doi: 10.1159/000128582. [DOI] [PubMed] [Google Scholar]
- 14.Kreuter A, Jaouhar M, Skrygan M, Tigges C, Stücker M, Altmeyer P, et al. Expression of antimicrobial peptides in different subtypes of cutaneous lupus erythematosus. J Am Acad Dermatol. 2011;65:125–133. doi: 10.1016/j.jaad.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Frohm M, Agerberth B, Ahangari G, Stâhle-Bäckdahl M, Lidén S, Wigzell H, et al. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J Biol Chem. 1997;272:15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- 16.Schröder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci. 2006;63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, et al. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kesting MR, Stoeckelhuber M, Hölzle F, Mücke T, Neumann K, Woermann K, et al. Expression of antimicrobial peptides in cutaneous infections after skin surgery. Br J Dermatol. 2010;163:121–127. doi: 10.1111/j.1365-2133.2010.09781.x. [DOI] [PubMed] [Google Scholar]
- 19.Guarda G, So A. Regulation of inflammasome activity. Immunology. 2010;130:329–336. doi: 10.1111/j.1365-2567.2010.03283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elssner A, Duncan M, Gavrilin M, Wewers MD. A novel P2X7 receptor activator, the human cathelicidin-derived peptide LL37, induces IL-1 beta processing and release. J Immunol. 2004;172:4987–4994. doi: 10.4049/jimmunol.172.8.4987. [DOI] [PubMed] [Google Scholar]
- 21.Kuwano T, Nakao S, Yamamoto H, Tsuneyoshi M, Yamamoto T, Kuwano M, et al. Cyclooxygenase 2 is a key enzyme for inflammatory cytokine-induced angiogenesis. FASEB J. 2004;18:300–310. doi: 10.1096/fj.03-0473com. [DOI] [PubMed] [Google Scholar]
- 22.Cumberbatch M, Dearman RJ, Griffiths CE, Kimber I. Langerhans cell migration. Clin Exp Dermatol. 2000;25:413–418. doi: 10.1046/j.1365-2230.2000.00678.x. [DOI] [PubMed] [Google Scholar]
- 23.Sauter W, Rosenberger A, Beckmann L, Kropp S, Mittelstrass K, Timofeeva M, et al. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1127–1135. doi: 10.1158/1055-9965.EPI-07-2840. [DOI] [PubMed] [Google Scholar]