Abstract
The identification of novel, synthetic targeting ligands to endothelial receptors has led to the rapid development of targeted nanoparticles for drug, gene and imaging probe delivery. Central to development and optimization are effective models for assessing particle binding in vitro. Here, we developed a simple and cost effective method to quantitatively assess nanoparticle accumulation under physiologically-relevant laminar flow. We designed reversibly vacuum–sealed PDMS microfluidic chambers compatible with 35 mm petri dishes, which deliver uniform or gradient shear stress. These chambers have sufficient surface area for facile cell collection for particle accumulation quantitation through FACS. We tested this model by synthesizing and flowing liposomes coated with APN (KD ~ 300 µM) and VCAM-1-targeting (KD ~ 30 µM) peptides over HUVEC. Particle binding significantly increased with ligand concentration (up to 6 mol%) and decreased with excess PEG. While the accumulation of particles with the lower affinity ligand decreased with shear, accumulation of those with the higher affinity ligand was highest in a low shear environment (2.4 dyne/cm2), as compared with greater shear or the absence of shear. We describe here a robust flow chamber model that is applied to optimize the properties of 100 nm liposomes targeted to inflamed endothelium.
Key Terms: Liposomes, HUVEC, VCAM-1, Aminopeptidase N, lipo-PEG-peptide
Introduction
Drug delivery using targeted nanoparticles is an attractive alternative to traditional molecular therapeutics. Nanoparticles are well suited for chemotherapeutic delivery, as they can increase the drug’s circulation half-life9, reduce cytotoxic side-effects16, and passively accumulate on the tumor interstitium via the enhanced permeability and retention (EPR) effect13. Selective screening methods such as in vivo phage display have led to the identification of numerous ligand-receptor pairs20. By coating nanoparticles with these targeting moieties, their use can be expanded to the localized delivery of genes, drugs and imaging probes for cardiovascular disease27, diabetes7 and other chronic conditions14.
Central to the development and optimization of targeted nanoparticles is an effective model for quantifying particle binding under near in vivo conditions. Inflammatory cell responses such as leukocyte tethering to endothelium take place under a certain range of shear stress (1 to 6 dyne/cm2 )22, and in vitro particle delivery can be significantly enhanced7 or reduced5 by the addition of shear. In order to predict the in vivo properties of targeted nanoparticles, shear stress must be included in its in vitro characterization.
Previous studies of targeted particles under shear stress have utilized functionalized microparticles flowing through microfluidic chambers seeded with cells or receptors and manual counts of the bound particles2, 5, 19. Such quantification is time consuming and a suboptimal strategy for nanoparticles, whose dimensions are below the optical threshold. Flow cytometry is an attractive alternative, as it is a fast and sensitive method for quantifying fluorescent nanoparticle delivery per cell, provided that a large number of cells are collected. We propose a microfluidic chamber model that allows for the facile collection of ample cells for flow cytometric analysis post-shear treatment. For this purpose, we employ reversibly vacuum-sealed polydimethylsiloxane (PDMS) microfluidic chambers. Vacuum sealing allows PDMS to bind to many surfaces with well characterized vacuum to fluid-pressure tolerance3. The device has been designed to fit into a 35 mm petri dish, but the chamber surface treatment area has been scaled up to allow for adequate cell collection. By employing microfluidic chambers, physiological shear stress can be reproduced with fluid flow rates on the order of tens of microliters per minute, conserving precious treatment materials. The vacuum sealable chamber allows for cells to be grown in standard 35 mm petri dishes, facilitates cell collection post-treatment and allows for chamber reuse. Collected cells can then be analyzed via flow cytometry.
Using this system, we characterized the effects of the targeting ligand, ligand density, and polyethylene-glycol (PEG) density on endothelial accumulation of particles under static and dynamic conditions. Fluorescently-labeled liposomal nanoparticles were synthesized and coated with NGR (cyclic CNGRC targeting aminopeptidase N (APN)17) or VHP (linear VHPKQHR targeting VCAM-18), two peptides with KD values of ~300 µM and ~30 µM, respectively15, 18. As APN expression is up-regulated at angiogenic sites and VCAM-1 at inflammatory sites, particles targeting these proteins can be used to selectively treat or image diseases such as cancer or atherosclerosis, respectively. As liposome binding strength increases with multivalency25, we expect particle accumulation under flow to increase with increasing concentrations of ligand and then plateau as binding is maximized. Liposomes of 0 to 6 mol% ligand density were synthesized by varying lipid-PEG-peptide complex (LPP, lipo-peg-peptide) content, and their binding to endothelial cells under flow was compared. PEG is a hydrophilic polymer that plays a key role in in vivo drug delivery, inhibiting opsonization by forming a steric barrier. Though the effect on particle accumulation of the PEG brush length relative to the ligand linker length has been studied27, the effect of PEG concentration (in addition to PEG within the LPP) on particle accumulation is unclear. Liposomes consisting of 6 mol% LPP and 0 to 6 mol% lipid-PEG were synthesized and optimized for particle accumulation. Flow cytometry results were corroborated with in situ post-treatment fluorescent microscopy images. Finally, to better understand the relationship between shear stress and particle binding, a second chamber model with a gradient shear stress was designed and particle delivery was compared to the shear stress experienced.
Materials and Methods
Peptide, FAM-labeled peptide, and lipo-PEG-peptide (LPP) synthesis
Cyclized NGR, linear VHP and the appropriate scrambled peptide (sVHP) were synthesized. Their full sequences with linker domains are as follows; NGR = cCNGRC, VHP = Boc-VHPKQHR-GGSK(ivDde)GC, and sVHP = Boc-QRHPHVK-GGSK(ivDde)GC. Peptides were synthesized on Pal resin (Applied Biosystems, Foster City, CA) or Rink amide MBHA resin (NovaBiochem, La Jolla, CA) using solid phase peptide synthesis with standard Fmoc chemistry. Fmoc-amino acids and peptide coupling reagents were purchased from NovaBiochem. Solvents and other reagents of analytical purity were obtained from Sigma-Aldrich (Milwaukee, WI) and VWR (Brisbane, CA).
Carboxyfluorescein (FAM) labeled VHP and sVHP peptides (FAM-VHP and FAM-sVHP) were synthesized by removing the ivDde protecting group with 2 % Hydrazene in dimethyl formamide (DMF), and then reacting the exposed amine with a FAM using peptide coupling reagents.
Peptides were coupled to form LPP conjugates using the method previously described by Zhang et al27. In brief, after synthesis of the desired peptide sequence, the peptide was coupled (at the cysteine’s free amine for NGR and an amine exposed at the lysine following ivDde removal for VHP) with three Fmoc-PEG(1300) groups, an Fmoc-Lys(Fmoc) and Stearic acid. Products were cleaved off resin, purified using Prostar HPLC (Varian, Palo Alto, CA) and the molecular mass was confirmed using the matrix assisted laser desorption ionization time of flight (MALDI-ToF) 4700 (Applied Biosystems, Carlsbad, CA).
Peptide targeted liposome fabrication
Liposomes decorated with various molar percentages of NGR or VHP peptides were prepared for the comparison of binding avidity between static and dynamic conditions. 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt) (DSPE-PEG2000), mini extruders, membranes, and filters were purchased from Avanti Polar Lipids (Alabaster, AL), Calcein was obtained from Applied Biosystems, and Sephadex G-75 was obtained from GE Healthcare (Little Front, UK). For our studies 0, 1, 2, 4, 6I, 6II and 6III % NGR and VHP liposome were fabricated, where 0 % = (DPPC:DSPE-PEG2000: LPP-XXX = 94:6:0), 1 % = (94:5:1), 2 % = (94:4:2), 4 % = (94:2:4), 6I % = (94:0:6), 6II % = (92:2:6) and 6III % = (88:6:6). The liposomes were prepared using methods described previously27. Briefly, lipids dissolved in chloroform were dried under a stream of nitrogen gas, lyophilized overnight and then re-solubilized in 5 or 10 mM Calcein (pH 7.4, 300 mOs) suspended in DPBS +/+ by incubating and sonicating at 60°C. The lipid solution was then extruded through 100 nm pores 21 times at 60°C, and the liposomes were isolated via size exclusion chromatography using Sephadex G-75. The size and absorbance of the collected liposomes were verified using a Nicomp 380ZLS particle sizer (Nicomp PSS, Santa Barbara, CA) and a Nanodrop-1000 spectrophotometer (Thermo Scientific, Weltham, MA), respectively. Particle diameters were 110.6 ± 4.0 nm for control liposomes, and 131.3 ± 8.3 nm for peptide targeted liposomes. The difference in size is due to the longer PEG brush (3900 Da) on the LPP, compared to the DSPE-PEG2k (2000 Da) brush layer. Liposome concentration was normalized by its 495 nm absorbance (Calcein’s maximal absorbance).
Cell culture and protein expression
Human umbilical vein endothelial cells (HUVEC) were purchased from Clonetics (San Diego, CA) and were cultured in endothelial growth media (EGM) supplemented with 0.1 % (v/v) human endothelial growth factor (hEGF), 0.1 % (v/v) hydrocortisone, 0.1 % (v/v) GA-1000, 0.4 % (v/v) bovine brain extract (BBE) and 10 % (v/v) fetal bovine serum (FBS), all from Lonza (Portsmouth, NH) inside a 95 % air/ 5 % CO2 atmosphere incubator maintained at 37°C. HUVECs from passages 5 to 7 were used for the experiments.
VCAM-1 expression was up-regulated by incubating HUVEC with tumor necrosis factor alpha (TNFα) (R&D Systems, Minneapolis, MN) in EGM for 4 hours. To minimize the effect of VCAM-1 up-regulation induced by shear stress23 (as shear stress will be applied in later studies), a range of TNFα concentration was explored, and the concentration with maximal up-regulation without visible cytotoxic effects was employed. VCAM-1 and APN protein expression on HUVEC was verified by immunolabeling and flow cytometry analysis using fluorescence activated cell sorting (FACS, Becton Dickinson, Franklin Lakes, NJ). Fluorescein-labeled antibodies and the corresponding isotype controls were purchased from R&D Systems and eBiosciences (San Diego, CA). For the anti-APN mAb, the WM15 clone was used.
To assess NGR-liposome specificity, HUVEC were pre-treated with 5 µg/ml of anti-CD13 mAb in 0.2 % BSA DPBS+/+ for 15 minutes, followed by a 10 minute treatment with 1 % NGR liposomes at 4°C. Cells were collected and their fluorescence was analyzed using FACS. NGR-liposome binding was also tested by pre-blocking with 10× (free peptide to LPP-NGR ratio of 10 to 1) and 20× free peptide. VHP peptide specificity has been previously investigated by Nahrendorf et al15. To confirm this, we compared VHP and sVHP monomer binding to cells. HUVEC were treated with 0.8 µM FAM-VHP or FAM-sVHP for 60 minutes at 37°C in media, rinsed, collected and analyzed using FACS.
Microfluidic chamber construction
Two vacuum sealable microfluidic PDMS chambers were designed. The first chamber was designed for treating and analyzing cells using FACS, requiring a large cell treatment area with uniform shear stress throughout the majority of the chamber (uniform shear chamber, or USC). The second chamber was designed for optically analyzing particle binding to cells under a spatially dependent shear stress profile (gradient shear chamber, or GSC). The designs were based upon those fabricated by Simon et al21, 23. The wall shear stress at the center of a microfluidic chamber can be estimated using , where μ is the viscosity [0.015 dyn*s/cm2 for water at 4°C], Q is the volumetric flow rate, h is the height [50 µm], and w is the width of the chamber. For this equation, the fluid must be Newtonian, and the width must be significantly greater than the height. With all other factors kept constant, the theoretical shear stress can be manipulated by altering the width. For the USC, a rectangular chamber (4 mm × 19 mm) with constant width throughout, save for the tapered inlet and outlet (Figure 1a) was designed. For the GSC, the Hele-Shaw design based on Usami et al 24 was employed. In this model, the first 16.5 mm from the inlet, the chamber width can be expressed by , where w0 is the entrance width, L is the total chamber length, and x is the distance from the inlet along L (Figure 1b). The wall shear stress for this chamber is expressed as , and linearly decreases with distance from the inlet. Values of w0 equal to 2 mm and L equal to 20 mm were used. Both chambers were designed over a spider web of vacuum networks.
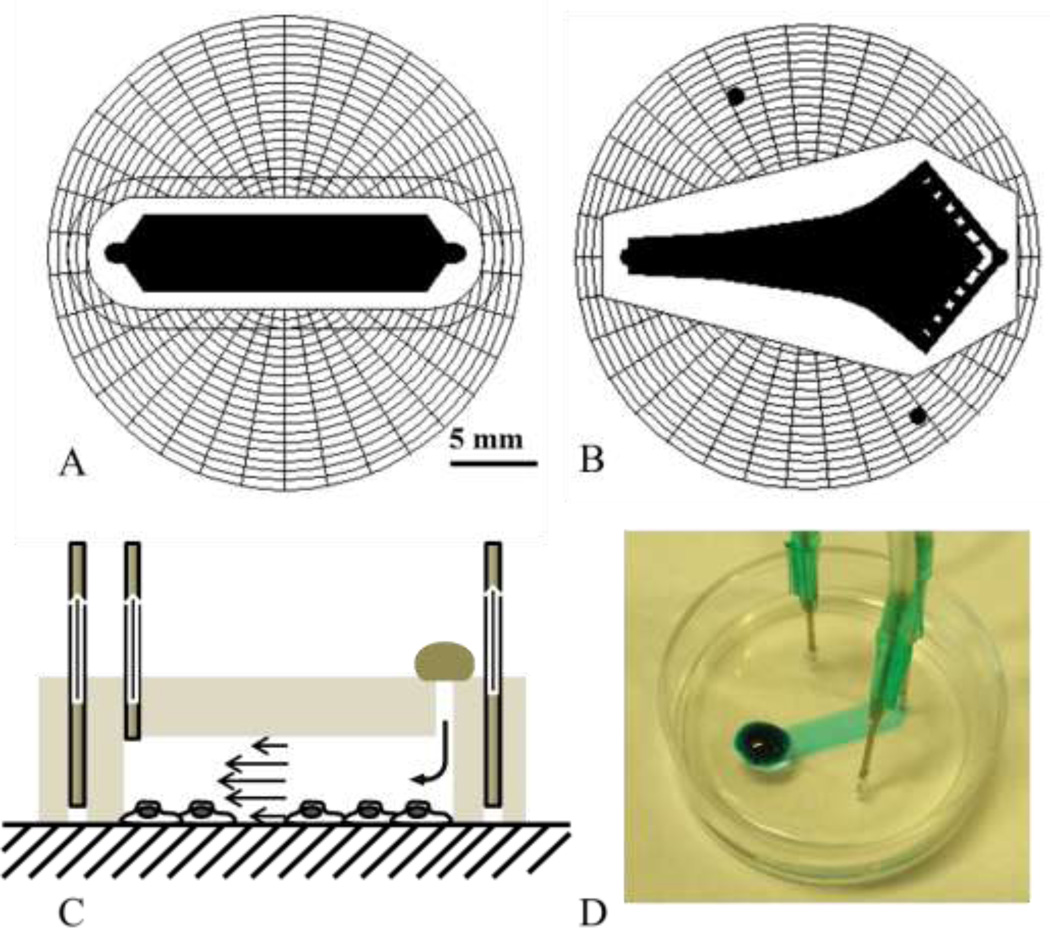
Figure 1.
PDMS microfluidic chambers. A) Uniform shear chamber (USC) with 4 mm width and 19 mm total length (15 mm without taper). B) Gradient shear chamber (GSC) based upon the Hele-Shaw design, where shear stress along the center line decreases linearly with distance from the inlet (narrower end), with inlet width of 2 mm, and chamber length of 20 mm. C) Schematics of the flow chamber in use – the chamber is affixed onto the surface over cells by applying a vacuum through the spider web network via the outer two holes. Treatment solution is directly placed over the inlet, and the outlet is connected to a microinjector that withdraws the solution at a controlled rate. D) USC demonstrated with a blue dye. The chamber is small enough to be affixed directly over cells on a standard 35 mm petri dish.
The prototype chamber molds were designed in Powerpoint (Microsoft) and fabricated by photopatterning dry-film28, 29, a method that is fast and requires neither a photomask nor a clean room. Later generation chambers were designed using AutoCAD-LT2000 (Autodesk) and photomasks were ordered from Output City (Bandon, OR). Standard soft lithography techniques were used to create PDMS (Sylgard 184, Dow Corning) chambers with 50 µm tall features. Once cured, an inlet, outlet, and two vacuum holes were punched through the chamber using a sharpened 16 gauge blunt needle. During experiments vacuum tubes are inserted into the vacuum holes, evacuating the air inside the spider web, creating a seal and affixing the chamber onto the surface. A tube connected to a PHD2000 microinjector (Harvard Apparatus, Holiston MA) is inserted into the outlet, withdrawing solution through the chamber at a specified flow rate. The treatment solution is deposited directly over the inlet hole, onto the PDMS surface. (Figure 1c)
Microfluidic chamber flow characterization
The spatial distribution of shear stress throughout the USC was obtained experimentally for a flow rate of 10 µl/min through velocity and height profile measurements (Supplementary Methods). Shear stress measurements through relevant portions of the GSC were conducted in a similar manner for 10 and 20 µl/min and are detailed in the supplementary methods.
Chamber performance with nanoparticles was tested by flowing liposomal particles over cells and optically analyzing the plate for spatial dependencies in particle-cell binding. HUVEC were grown on petri dishes pretreated with rat tail collagen (BD Biosciences, Franklin Lakes, NJ) to prevent non-specific liposomal binding to the petri dish surface. After reaching confluency, cells were treated with TNFα, chilled to 4°C, and rinsed with 0.2 % BSA DPBS+/+. The chamber was then affixed on the top of the cell plate, and cells underwent a treatment of 2 min rinse, 5 min treatment with 1 or 2 % VHP liposomes (10 mM Calcein, normalized to λ495 = 0.1 in 0.2 % BSA DPBS+/+) and 3 min rinse. The USC was tested using a flow rate of 10 µl/min, and the GSC was tested at 10 and 20 µl/min. The chambers were gently removed, and cells were rinsed and incubated in media at 37°C for 4 hours. Cells were then fixed with 3 % paraformaldehyde (PFA) in DPBS−/− and stained with 60 ng/ml DAPI in DPBS−/−. Cells were imaged using a fluorescence microscope (Y-IDP, Nikon, Japan) with a 5× objective, and a 20×6 or 20×10 frame mosaic was collected for each USC and GSC treated dish, respectively. Background images were subtracted from the mosaic images using ImageJ. Areas inside the USC were segmented into cross-sectional areas along an axis (1 mm by 4 mm along the x-axis and 1 mm by 16 mm along the y-axis), and a profile along the axis was calculated by taking the average fluorescence of each area. For the GSC, mean fluorescence along the x axis was measured and plotted against the shear stress. In order to identify spatial binding patterns, each fluorescence profile was normalized by subtracting its mean value before comparing with other samples of the same treatment group.
Particle binding studies - Static binding studies
To assess the relative binding avidity of each liposome formulation, particles were statically incubated with the target cells, which were then analyzed using FACS. HUVEC were grown to confluence in 24-well tissue culture plates and chilled to 4°C. Cells were rinsed and treated with 150 µl of chilled liposome solution (0, 1, 2, 4, 6I, 6II or 6III % of NGR or VHP solution normalized to λ495 = 0.05) for 5 minutes at 4°C, then rinsed 3×, treated with cold media and incubated at 37°C for 4 hours. Cells were then rinsed with DPBS−/−, collected and analyzed using FACS.
FACS analyzed dynamic binding studies
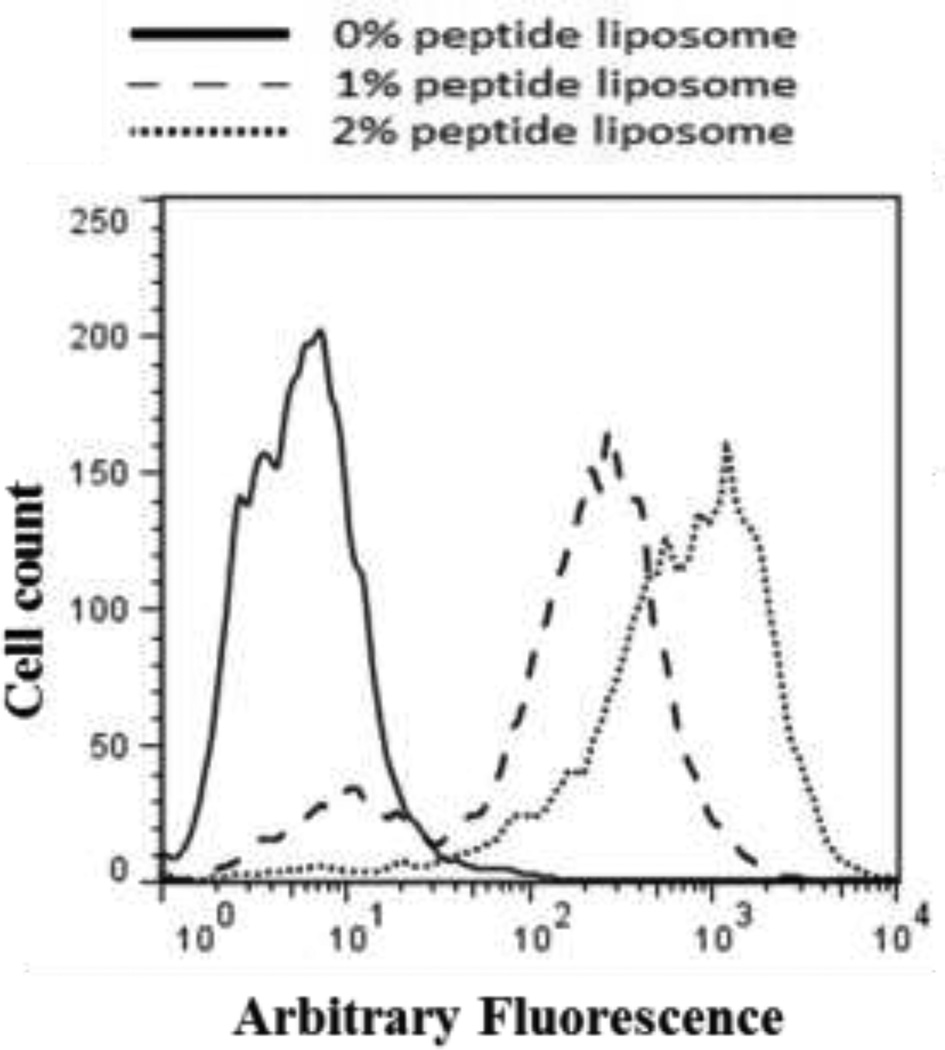
Dynamic binding of liposomes was assessed by flowing nanoparticles through the USC over cells and analyzing their fluorescence via FACS. Results were then compared with those obtained via static binding. Like static studies, treatments were conducted at 4°C to prevent internalization during treatment. HUVEC were grown to confluence on 35 mm petri dishes, rinsed, and the USC was vacuum affixed over the cells onto the petri dish. Rinsing solution was pulled through the chamber at 10 µl/min for 2 minutes, followed by treatment solution (liposome solution normalized to λ495 = 0.05) for 5 minutes, and rinsed again for 3 minutes. The vacuum was turned off, the flow chamber carefully removed, and the area outside the chamber was cleaned to remove untreated cells. Cells were rinsed 3 additional times, incubated in media at 37°C for 4 hours, and collected for FACS analysis. FACS data of dynamic studies resulted in two distinct peaks; one peak corresponding to cell’s auto-fluorescence, and a higher fluorescence peak which varied with liposome formulation (Figure 2). Fluorescence microscopy of in situ cells (Figure 5a) revealed that cells were labeled throughout the entirety of the inner chamber surface. By thoroughly cleaning the channel exterior, we reduced the non-fluorescent population of cells collected, from 81.4 ± 6.9 % to 5.9 ± 4.4 %. We concluded that the lower peak belonged to untreated cells collected from outside the chamber, and therefore report the median value of the higher peak.
Figure 2.
Representative example of FACS data for 0% (solid line), 1% (dashed line) and 2% (dotted line) VHP-conjugated liposome binding to HUVEC under flow. When double peaks were present, the lower peak was coincident with the control treatment.
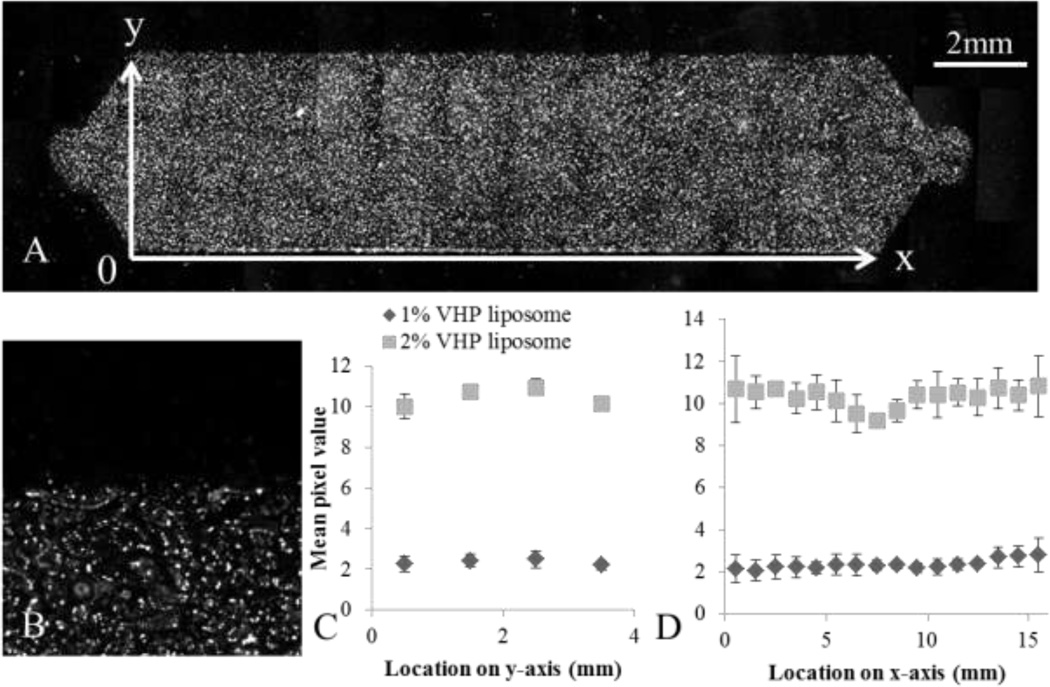
Figure 5.
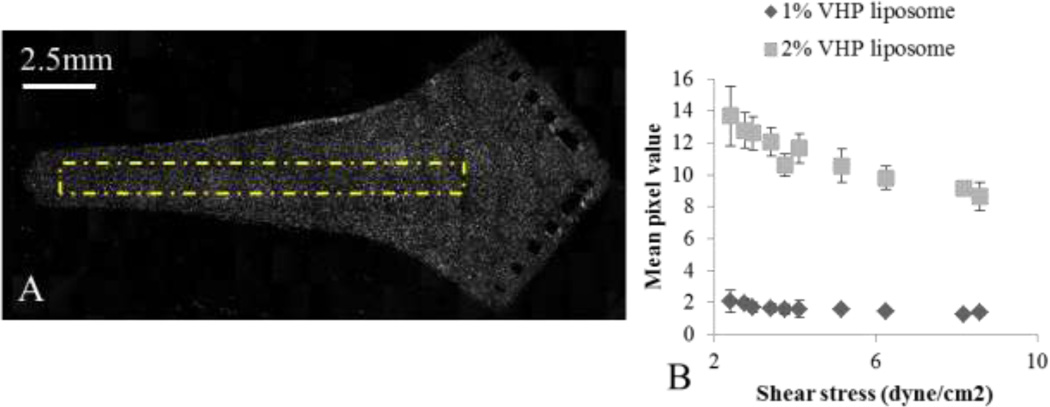
Spatial binding pattern of liposomal nanoparticles throughout the USC was characterized by flowing 1 (N = 8) and 2 % (N = 6) VHP-conjugated liposomes over HUVEC and measuring fluorescence through microscopy. A) 20×6 frame mosaic image composed of fluorescence images acquired using a 5× objective, with background subtraction. B) An enlarged image of the edge of the chamber, showing the individual cells with punctate fluorescence, seen at 5×. C) The average fluorescence along the y-axis. D) The average fluorescence along the x-axis. One way ANOVA results showed significant spatial dependence of 2% VHP-conjugated liposomes capture along the y-axis (p < 0.01), but not along the x-axis. Capture of 1% VHP-conjugated liposomes was not spatially dependent along either axis (p > 0.01).
Results
Cell protein expression and peptide particle specificity
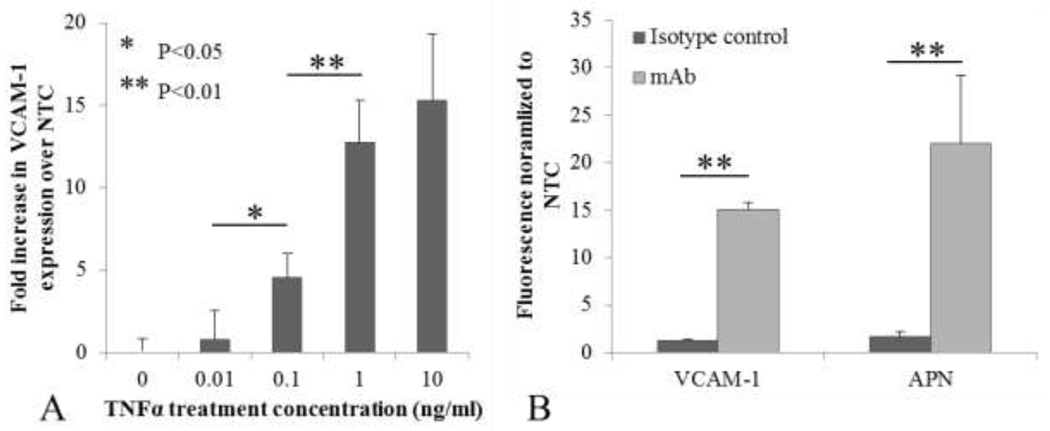
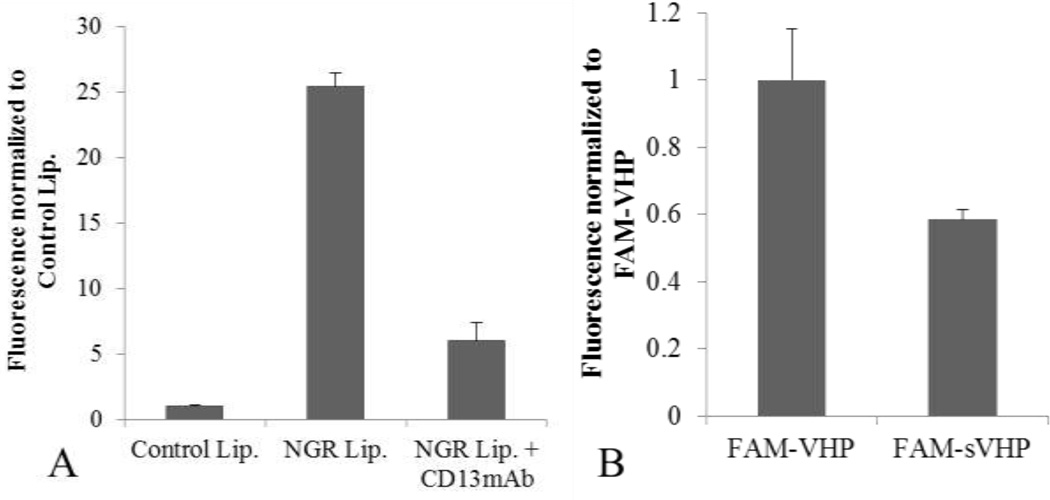
VCAM-1 expression levels were characterized through pretreatment with 0, 0.01, 0.1, 1.0 or 10 ng/ml of TNFα followed by immunolabeling. VCAM-1 expression increased with TNFα concentration in a non-linear manner (Figure 3a) with 10 ng/ml resulting in the highest expression level, however the increase between 1 and 10ng/ml TNFα was not significant (P > 0.05). We thus chose the 10 ng/ml level to maximize accumulation and minimize the dependence of expression on shear stress. For all subsequent VHP-liposome related studies HUVEC were pre-treated with 10 ng/ml of TNFα. Both APN and VCAM-1 mAb binding was significantly greater than that of their respective IgG isotype controls, confirming APN and VCAM-1 presence on HUVEC (Figure 3b). NGR-liposome’s specificity to APN was tested by pre-treating cells with anti-APN mAb or free NGR peptide. Pre-treatment with the mAb reduced NGR liposome binding five-fold (Figure 4a), and pre-treatment with 10× free peptide reduced binding ten-fold (data not shown). Increasing free peptide concentration from 10× to 20× did not further inhibit binding. To assess VHP specificity, FAM-VHP monomer binding was compared against FAM-sVHP monomer (Figure 4b). A reduction in binding of up to 42% was observed when cells were incubated with FAM-sVHP instead of FAM-VHP.
Figure 3.
Verifying protein expression on HUVEC. A) VCAM-1 up-regulation characterized post incubation with varying concentrations of TNFα. VCAM-1 expression increased non-linearly with TNFα concentration. Differences between 1 and 10 ng/ml were not found to be significant. N = 3 for each group. Error bars represent standard deviations. B) Comparing VCAM-1 and APN expression on HUVEC. Both proteins were confirmed to be expressed on the HUVEC surface. Iso-VCAM-1 and Iso-APN are isotype control mAbs IgG1 kappa and IgG2A, respectively. N = 3 for each group.
Figure 4.
Assessing particle specificity. A) NGR-conjugated liposome binding to HUVEC was reduced five-fold by pre-treating with 5 µg/ml free anti-APN/CD13 mAb for 15 minutes. All differences were found to be significant (P < 0.01, N = 3). B) 0.8 µM FAM-VHP and FAM-sVHP monomer were incubated with HUVEC for 1 hour and fluorescence was measured using FACS. Fluorescence was background subtracted and normalized to the FAM-VHP signal. With sVHP, a 42% reduction in binding (P < 0.01, N = 3) was observed compared to VHP.
Microfluidic chamber characterization
Shear stress experienced inside the chambers was characterized by measuring flow velocity and chamber height throughout the chamber. Cross sectional height measurements revealed that the USC height varied in a parabolic fashion across the width due to sagging of the upper wall. A decrease in chamber height and mean flow velocity of up to 42% and 30% (relative to the edge), respectively, was observed in the chamber center (Supplementary Figure 2). Following normalization of the velocity by height, the average shear stress of the USC was 4.4 ± 0.4 dyne/cm2 over the distance from 4.5 to 12.5 mm from the inlet (2 to 4 in Supplementary Figure 1). Due to the designed increase in width with distance from the inlet, shear stress in the GSC decreased linearly from 8.6 to 3.8 dyne/cm2 and 4.1 to 2.4 dyne/cm2 for 20 and 10 µl/min flow rates, respectively, over a distance of 2.7 to 13.5 mm from the inlet (Supplementary Figure 3). For the GSC, the chamber height decreased by 5% at most over the distance from 2.7 to 13.5 mm from the inlet.
Optical analysis of cells in situ
Optically-labeled liposomes were distributed throughout the USC (Figure 5a). After 4 hours of post treatment incubation and fixation, punctate fluorescence was observed in the cells (Figure 5b). One way ANOVA analysis of the 2% peptide-conjugated liposome treatment revealed a spatial dependence on binding along the y-axis (p < 0.01) with a decrease along the chamber edge, but not along the x-axis (p > 0.05) (Figure 5c and 5d). Increasing the surface concentration of peptide from 1 % to 2 % VHP increased the mean fluorescence intensity by approximately 4 fold.
To systematically examine the change in accumulation between 1 % and 2 %-conjugated VHP liposomes, we examined liposome capture under shear in the GSC at 10 and 20 µl/min, corresponding to a range in measured shear stress from 2.4 to 8.6 dyne/cm2. Figure 6a provides a representative image of endothelium sheared with 2 %-conjugated VHP liposomes at 10 µl/min. For both 1% and 2%-conjugated VHP liposomes, the greatest accumulation occurred at the lowest shear measured (2.4 dyne/cm2), and decreased steadily with increased shear stress (Figure 6b). As observed in earlier studies, the 2 %-conjugated ligand liposome accumulation was significantly greater than for 1 %-conjugated ligand liposomes.
Figure 6.
1% and 2% VHP-conjugated liposome binding to HUVEC, flown through the GSC at 10 or 20 ul/min. A) A representative image of 2% VHP-conjugated liposomes flown through at 10 ul/min. The yellow dashed box indicates the area from which signal profiles were analyzed. B) The signal from various locations plotted against the experimentally characterized shear stress. For both 1 and 2% VHP-conjugated liposomes, binding decreased with increasing shear stress.
Optimization of surface architecture
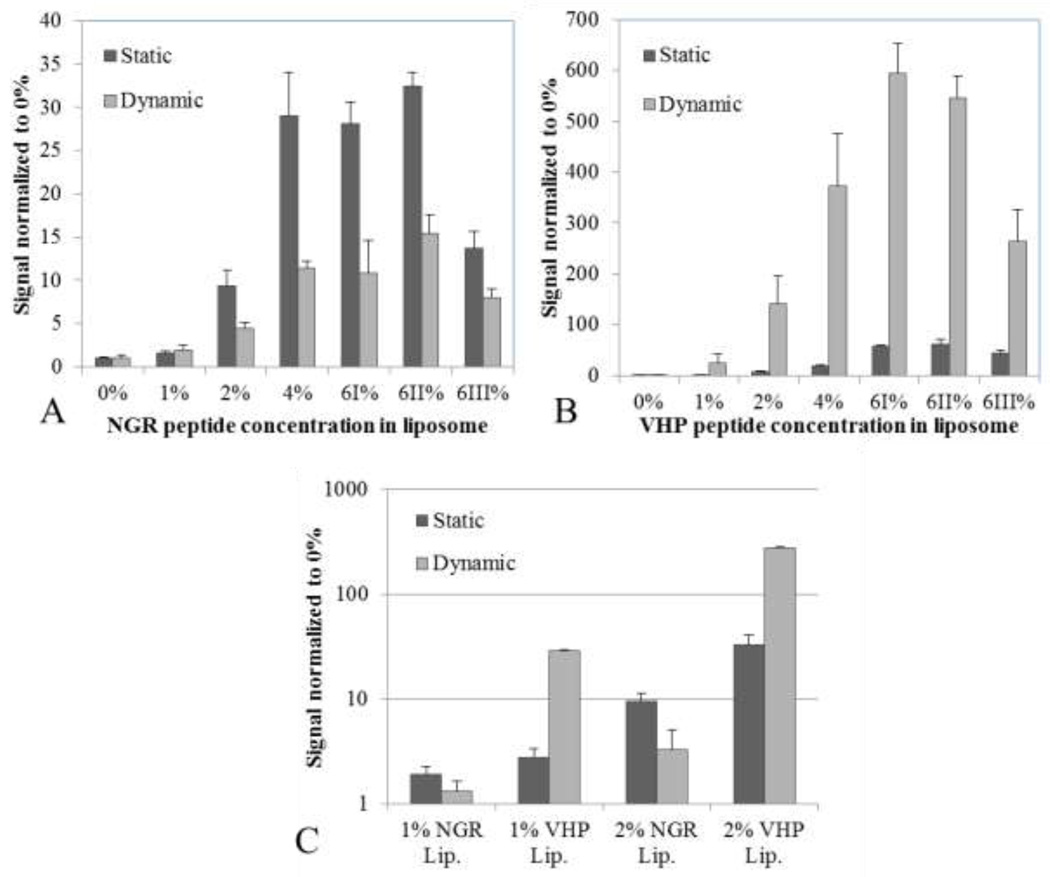
Particle binding under static and dynamic conditions was compared for two peptide-conjugated liposomes, with substantial increases in avidity as the peptide molar percentage increased from 0 to 6 % (Figures 7a and 7b). Further, increasing the PEG brush layer decreased accumulation in static and dynamic assays. For both peptides, increasing the molar percentage of DSPE-PEG2k from 0% to 6% decreased liposome accumulation by 50%. For maximal binding, we found 6% LPP without additional PEG to be optimal.
Figure 7.
Comparison of binding under static vs dynamic conditions for varying peptide molar concentrations for NGR and VHP-conjugated liposomes. A) NGR-conjugated liposome binding under static and dynamic (4.4 dyne/cm2 shear stress with the USC) conditions. Particle binding increased with peptide concentration (p < 0.01 except from 4 to 6%), but consistently decreased under shear (p < 0.01 except for 1%) (N ≥ 3). B) VHP-conjugated liposome binding under static and shear conditions. Particle binding consistently increased with peptide concentration (p < 0.01), and also increased under shear (p < 0.01 except for 1%, up to 13-fold) (N ≥ 3). C) Binding of NGR and VHP-conjugated liposome in side by side comparison.
NGR and VHP-targeted particles respond differently to shear stress
Under shear stress of 4.4 dyne/cm2, NGR-conjugated liposome binding significantly decreased while VHP-conjugated liposome binding significantly increased. The increase in accumulation for the 2%-conjugated VHP liposomes under shear stress was verified using fluorescence microscopy. Finally, the accumulation of NGR- and VHP-conjugated liposomes was compared for 1 and 2% ligand concentrations (Figure 7c). On average, VHP-conjugated liposomes accumulated 1.6 and 33.4 times better than NGR-conjugated liposomes under static and dynamic conditions, respectively. VHP-conjugated liposome accumulation increased approximately 13 fold when treated under 4.4 dyne/cm2 of shear stress, while NGR-liposome accumulation decreased to half under identical conditions.
Discussion
In this work, we have designed a flow chamber that facilitates the characterization of targeted nanoparticle accumulation on a monolayer of cells under physiological shear stress. The design allows for cells to be cultured in standard 35 mm petri dishes, uses a small volume of reagents and cells are collected with ease for FACS analyses. The concept of analyzing particle binding under shear in vitro is not new, and studies involving more complex microfluidic chambers that replicate the vascular intricacies19 have been used to identify the vascular structural features that favor binding. The strength of our model lies in its ability to rapidly and quantitatively characterize nanoparticle binding under flow on a per cell basis with high sensitivity, facilitating particle optimization. For optimal in vivo particle-based delivery, continued research will be important regarding both the vascular/flow field characteristics, and the particle surface chemistry that maximize accumulation.
Experimental shear stress characterization via velocity and chamber height measurements revealed that the chamber height varied as a result of the PDMS material properties26 and the high aspect ratio of the chamber (1:80), which was chosen in order to obtain large numbers of cells for flow cytometry. For future studies, alternative materials should be evaluated, balancing the requirement for obtaining a vacuum seal with the stiffness required by the large chamber width. Despite deformation, the shear stress throughout the centermost half of the USC was 4.4 ± 0.4 dyne/cm2 under a volume flow of 10 µl/min. In situ fluorescence microscopy of the cells treated with 2 %-conjugated VHP liposomes using the USC exhibited relatively uniform fluorescence of 10.29 ± 0.98 throughout the treatment area. The magnitude of the decrease in fluorescence at the chamber edge (as compared with the center) was only 7% (Figure 5c), and the effect on FACS data was therefore small. Analyzing GSC treated cells in situ revealed that the average liposome accumulation decreased by 9 % over the USC relevant shear stress range of 3.6 to 5.1 dyne/cm2 and by 35 % when shear stress was increased from 2.4 to 8.6 dyne/cm2. The effect of ligand concentration was far greater, with an 85 % decrease in accumulation observed when liposomes included 1 %, as compared with 2 %, of the conjugated peptide (Figure 6b).
Under shear stress, the difference in accumulation of particles containing the VHP and NGR ligands likely results from both the differences in avidity15, 18 of the particles and the nature of the receptor. We anticipate that the introduction of shear stress will disrupt receptor ligand interaction for the low affinity (NGR-APN) receptor-ligand bond. In addition, a substantial literature has demonstrated that VCAM-1 bonds exhibit catch-bond characteristics that are enhanced by shear stress4.
Treating cells with VHP-conjugated liposomes under a shear stress of 4.4 dyne/cm2 significantly increased particle delivery as compared to treatment under static conditions. This is not surprising, as studies involving the transfection of endothelial and neuronal cells using cationic lipoplexes have also resulted in a significant increase in delivery when supplemented with flow6. However, convection-diffusion based modeling by Lee et al shows that spheroids do not migrate towards chamber walls in the absence of external forces (such as gravitational or electrostatic forces), and that the addition of flow forces parallel to the wall does not enhance particle margination12. Korn et al modeled the mean first passage time (MFPT) of targeted nano-spheres to receptor-coated walls and found MFPT to decrease with increasing Peclet number (proportional to shear rate), receptor density and receptor length10. Consistent with Lee et al’s work, margination/sedimentation was found to be independent of convection; however, once the particle was in binding proximity (via external forces or diffusion) greater flow resulted in faster particle rotation and an increasing ligand-receptor encounter rate. Modeling the binding rate of surface tethered reactants on two surfaces moving against each other, Chang et al found the binding rate to initially increase with relative velocity due to an increased rate of encounter, but to then plateau due to a decrease in encounter duration1. This may explain why particle delivery decreased when shear stress was increased from 2.4 to 8.6 dyne/cm2 in the GSC, and the large difference in binding between NGR- and VHP-conjugated liposomes under flow.
Under both static and dynamic conditions, an increase in ligand concentration (from 0, 1, 2, 4, to 6 mol%) resulted in a steady increase in particle accumulation. This is consistent with the work of others, where MFPT and probability of capture of a targeted nanoparticle to a surface under flow was enhanced with increased ligand concentration10, 25. The critical shear stress required to dislodge a particle from a receptor-coated wall has been found to increase linearly with ligand density11. Korn’s work predicts that at low shear MFPT is dominated by the ligand density, but as shear is increased, MFPT decreases for all ligand densities, eventually converging. Although the binding of particles with differing ligand densities was quantified under only two shear stresses in this study (0 and 4.4 dyne/cm2), it will be of interest to quantitatively test particle accumulation with a greater range of shear stress to test Korn’s predictions.
The addition of PEGylated lipids in excess of 6 total mol % did not enhance particle accumulation, and in the case of 6 % excess (6III %), particle accumulation was halved compared to 0 % excess (6I %).
Although not explored in this paper, the chamber can accommodate physiological treatment fluids. The introduction of erythrocytes in in vitro flow studies has shown to significantly increase the accumulation of targeted microparticles on to surfaces2. Future studies will evaluate targeted nanoparticles in the presence of erythrocytes.
In summary, we have designed a flow chamber that allows for facilitated characterization of targeted nanoparticle binding to a monolayer of cells under physiological shear stress. The data obtained through FACS has been verified with fluorescence microscopy images of in situ cells, and although the chamber shear is not completely uniform, both the FACS and in situ imaging results were in agreement that the effects of non-uniformity was minimal compared to the effects of particle composition. Most importantly, we find that even for 100 nm liposomes with dense concentrations of ligand (up to 3000 per nanoparticle) the effect of shear stress on particle accumulation varies with the affinity of the ligand. For the ligands evaluated here, accumulation was maximized at the maximum surface density tested (6 mol%) and without an additional brush layer. Accumulation of NGR-conjugated liposomes decreased with the introduction of shear stress, however, for VHP-conjugated liposomes targeted to VCAM-1, accumulation peaked with the shear stress of 2.4 dyne/cm2, similar to the adhesion of neutrophils in similar shear stress.
Supplementary Material
Acknowledgements
We thank Dr Shengping Qin for the CFD modeling used for preliminary analysis and Arnold Chan for the fabrication of prototype PDMS molds. This work was supported by the Bioengineering research partnership NIHR01CA103828, NIHT32EB003827 and National Heart, Lung, and Blood Institute Contract no. HHSN268201000043.
References
- 1.Chang KC, Hammer DA. The forward rate of binding of surface-tethered reactants: effect of relative motion between two surfaces. Biophys J. 1999;76:1280–1292. doi: 10.1016/S0006-3495(99)77291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charoenphol P, Huang RB, Eniola-Adefeso O. Potential role of size and hemodynamics in the efficacy of vascular-targeted spherical drug carriers. Biomaterials. 2010;31:1392–1402. doi: 10.1016/j.biomaterials.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Chen AN, Pan TR. Three-dimensional fit-to-flow microfluidic assembly. Biomicrofluidics. 2011;5 doi: 10.1063/1.3670368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans E, Kinoshita K. Using force to probe single-molecule receptor-cytoskeletal anchoring beneath the surface of a living cell. Methods Cell Biol. 2007;83:373–396. doi: 10.1016/S0091-679X(07)83016-0. [DOI] [PubMed] [Google Scholar]
- 5.Farokhzad OC, Khademhosseini A, Jon S, Hermmann A, Cheng J, Chin C, Kiselyuk A, Teply B, Eng G, Langer R. Microfluidic system for studying the interaction of nanoparticles and microparticles with cells. Anal Chem. 2005;77:5453–5459. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 6.Harris SS, Giorgio TD. Convective flow increases lipoplex delivery rate to in vitro cellular monolayers. Gene Ther. 2005;12:512–520. doi: 10.1038/sj.gt.3302397. [DOI] [PubMed] [Google Scholar]
- 7.Jeong JH, Lee M, Kim WJ, Yockman JW, Park TG, Kim YH, Kim SW. Anti-GAD antibody targeted non-viral gene delivery to islet beta cells. J Control Release. 2005;107:562–570. doi: 10.1016/j.jconrel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Kelly KA, Nahrendorf M, Yu AM, Reynolds F, Weissleder R. In vivo phage display selection yields atherosclerotic plaque targeted peptides for imaging. Mol Imaging Biol. 2006;8:201–207. doi: 10.1007/s11307-006-0043-6. [DOI] [PubMed] [Google Scholar]
- 9.Klibanov AL, Maruyama K, Torchilin VP, Huang L. Amphipathic polyethyleneglycols effectively prolong the circulation time of liposomes. FEBS Lett. 1990;268:235–237. doi: 10.1016/0014-5793(90)81016-h. [DOI] [PubMed] [Google Scholar]
- 10.Korn CB, Schwarz US. Mean first passage times for bond formation for a Brownian particle in linear shear flow above a wall. J Chem Phys. 2007;126:095103. doi: 10.1063/1.2464080. [DOI] [PubMed] [Google Scholar]
- 11.Kuo SC, Lauffenburger DA. Relationship between receptor/ligand binding affinity and adhesion strength. Biophys J. 1993;65:2191–2200. doi: 10.1016/S0006-3495(93)81277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SY, Ferrari M, Decuzzi P. Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 14.Nahrendorf M, Jaffer FA, Kelly KA, Sosnovik DE, Aikawa E, Libby P, Weissleder R. Noninvasive vascular cell adhesion molecule-1 imaging identifies inflammatory activation of cells in atherosclerosis. Circulation. 2006;114:1504–1511. doi: 10.1161/CIRCULATIONAHA.106.646380. [DOI] [PubMed] [Google Scholar]
- 15.Nahrendorf M, Keliher E, Panizzi P, Zhang H, Hembrador S, Figueiredo JL, Aikawa E, Kelly K, Libby P, Weissleder R. 18F-4V for PET-CT imaging of VCAM-1 expression in atherosclerosis. JACC Cardiovasc Imaging. 2009;2:1213–1222. doi: 10.1016/j.jcmg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papahadjopoulos D, Allen TM, Gabizon A, Mayhew E, Matthay K, Huang SK, Lee KD, Woodle MC, Lasic DD, Redemann C, Martin FJ. Sterically Stabilized Liposomes - Improvements in Pharmacokinetics and Antitumor Therapeutic Efficacy. P Natl Acad Sci USA. 1991;88:11460–11464. doi: 10.1073/pnas.88.24.11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumorhoming peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 18.Plesniak LA, Salzameda B, Hinderberger H, Regan E, Kahn J, Mills SA, Teriete P, Yao Y, Jennings P, Marassi F, Adams JA. Structure and activity of CPNGRC: a modified CD13/APN peptidic homing motif. Chem Biol Drug Des. 2010;75:551–562. doi: 10.1111/j.1747-0285.2010.00974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prabhakarpandian B, Wang Y, Rea-Ramsey A, Sundaram S, Kiani MF, Pant K. Bifurcations: focal points of particle adhesion in microvascular networks. Microcirculation. 2011;18:380–389. doi: 10.1111/j.1549-8719.2011.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruoslahti E. Vascular zip codes in angiogenesis and metastasis. Biochem Soc Trans. 2004;32:397–402. doi: 10.1042/BST0320397. [DOI] [PubMed] [Google Scholar]
- 21.Schaff UY, Xing MM, Lin KK, Pan N, Jeon NL, Simon SI. Vascular mimetics based on microfluidics for imaging the leukocyte--endothelial inflammatory response. Lab Chip. 2007;7:448–456. doi: 10.1039/b617915k. [DOI] [PubMed] [Google Scholar]
- 22.Smith ML, Smith MJ, Lawrence MB, Ley K. Viscosity-independent velocity of neutrophils rolling on p-selectin in vitro or in vivo. Microcirculation. 2002;9:523–536. doi: 10.1038/sj.mn.7800165. [DOI] [PubMed] [Google Scholar]
- 23.Tsou JK, Gower RM, Ting HJ, Schaff UY, Insana MF, Passerini AG, Simon SI. Spatial regulation of inflammation by human aortic endothelial cells in a linear gradient of shear stress. Microcirculation. 2008;15:311–323. doi: 10.1080/10739680701724359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Usami S, Chen HH, Zhao Y, Chien S, Skalak R. Design and construction of a linear shear stress flow chamber. Ann Biomed Eng. 1993;21:77–83. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- 25.Wattenbarger MR, Graves DJ, Lauffenburger DA. Specific adhesion of glycophorin liposomes to a lectin surface in shear flow. Biophys J. 1990;57:765–777. doi: 10.1016/S0006-3495(90)82597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia YN, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 27.Zhang H, Kusunose J, Kheirolomoom A, Seo JW, Qi J, Watson KD, Lindfors HA, Ruoslahti E, Sutcliffe JL, Ferrara KW. Dynamic imaging of arginine-rich heart-targeted vehicles in a mouse model. Biomaterials. 2008;29:1976–1988. doi: 10.1016/j.biomaterials.2007.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao S, Chen A, Revzin A, Pan T. Stereomask lithography (SML): a universal multi-object micro-patterning technique for biological applications. Lab Chip. 2011;11:224–230. doi: 10.1039/c0lc00275e. [DOI] [PubMed] [Google Scholar]
- 29.Zhao S, Cong H, Pan T. Direct projection on dry-film photoresist (DP(2)): do-ityourself three-dimensional polymer microfluidics. Lab Chip. 2009;9:1128–1132. doi: 10.1039/b817925e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.