In this perspective, Marzluff considers Wilusz et al.'s report in the November 1, 2012, issue that a conserved AU-rich triple-helical structure at the 3′ end of two long noncoding RNAs (MALAT1 and MEN β) serves to stabilize and regulate these nonpolyadenylated transcripts.
Keywords: MALAT1, MEN β, NEAT1, translation, uridylation, RNA decay, RNA stability
Abstract
The 3′ ends of two large noncoding RNAs, MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) and MEN β, are formed by cleavage by RNase P and are capped but not polyadenylated. In the November 1, 2012, issue of Genes & Development, Wilusz and colleagues (pp. 2392–2407) show that when these 3′ ends are formed on a GFP reporter, the resulting mRNA is exported to the cytoplasm and translated. The 3′ end forms a novel triple-helical structure that supports export and translation as well as a poly(A) tail does.
The great majority of eukaryotic mRNAs end in a poly(A) tail, which in mammals typically is ∼200 nucleotides (nt) long when it is formed on the nuclear pre-mRNA. The poly(A) tail is essential for efficient translation of an mRNA in vivo. The only cellular mRNAs described thus far in eukaryotes that do not end in a poly(A) tail are the replication-dependent histone mRNAs in metazoans. In the November 1, 2012, issue of Genes & Development, Wilusz et al. (2012) reported the surprising finding that the 3′ ends of two largely nuclear noncoding RNAs—MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) and MEN β, which are transcribed by RNA polymerase II (Pol II) and whose 3′ ends are formed by cleavage by RNase P (Wilusz et al. 2008; Sunwoo et al. 2009)—support translation of a GFP reporter gene as efficiently as the poly(A) tail. They showed that these 3′ ends form a novel triple-helical structure that is essential for stimulating translation and stability of the RNA as well as supporting export to the cytoplasm. Although it is not clear whether these two larger nuclear noncoding RNAs are ever in the cytoplasm or encode peptides or proteins, it certainly is possible that there are other Pol II transcripts, currently classified as “noncoding” RNAs, that end in similar 3′ ends that encode peptides or proteins, since many of these noncoding RNAs are primarily cytoplasmic.
Wilusz, Spector, and coworkers (Wilusz et al. 2008; Sunwoo et al. 2009) had previously reported that the 3′ end of the MALAT1 transcript was formed by RNase P cleavage, resulting in two RNAs: the noncoding MALAT1 RNA and a small tRNA-like RNA, termed mascRNA (MALAT1-associated small cytoplasmic RNA), both of which accumulate in the cell. Subsequently, the same mechanism was found to form the 3′ end of the MEN β noncoding RNA (Sunwoo et al. 2009). Using a reporter construct that contains a GFP ORF and ends with the 3′ end of MALAT1, they found that the RNA was processed at the 3′ end identical to authentic MALAT1 RNA, but surprisingly, the resulting RNA was cytoplasmic and was translated to produce GFP in amounts similar to a polyadenylated GFP reporter driven by the same promoter. Thus, the 3′ end of the MALAT1 (and MEN β) RNA supports both nuclear export and translation, resulting in a relatively stable “mRNA.” There is a nuclear retention signal in the body of the MALAT1 RNA that prevents export, resulting in essentially all of the MALAT1 RNA remaining in the nucleus (Wilusz et al. 2012). However, they noted that in the ribosome profiling study of Ingolia et al. (2011), some MALAT1 RNA was found bound to ribosomes at sites near the 5′ end, consistent with potential translation initiation sites. Since MALAT1 is as abundant as GADPH mRNA, a small fraction of the RNA in the cytoplasm would have escaped detection and could potentially be translated. There is no conservation of potential peptide sequences, suggesting that MALAT1 translation does not play any role in its function, which remains unknown.
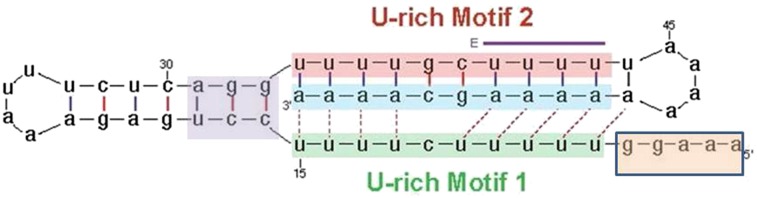
Wilusz et al. (2012) went on to determine the structure of the 3′ end of the mRNA as well as the regions essential for translation and stability. The 3′ end of the MALAT1 RNA is a 15-nt oligo(A) tract interrupted by a GC 4 nt from the 3′ end (Fig. 1). Upstream of the 3′ end are two U-rich sequences: The one closest to the 3′ end, U-rich motif 2, is interrupted by a GC and could form a hairpin with the 3′ end. Two nucleotide mutations in this sequence abolished accumulation of the GFP-MALAT1 mRNA, and accumulation was restored by compensatory mutations. However, larger mutations in U-rich motif 2 were not rescued by compensatory mutations, leading to the proposal that the other U-rich region, U-rich motif 1, could potentially form a triple helix at the 3′ end of the mRNA (Fig. 1). Mutagenesis and structural prediction studies confirmed that this is almost certainly the case. A triple-helical structure had been previously reported by Steitz and coworkers (Mitton-Fry et al. 2010) on the 3′ end of the PAN (polyadenylated nuclear) noncoding RNA, an abundant nuclear RNA synthesized by Kaposi's sarcoma-associated herpesvirus. The 3′ end of the poly(A) tail forms a triple helix with an upstream region, resulting in stabilization of the PAN RNA, protecting it from deadenylation (Mitton-Fry et al. 2010). This structure was elucidated by crystallography. The PAN RNA sequesters PABP [poly(A)-binding protein] in the nucleus, helping the virus shut down host protein synthesis (Borah et al. 2011). Thus, complex tertiary structures at the 3′ end of RNAs can clearly stabilize the RNAs and, in the case of the MALAT1 and MEN β RNA, efficiently support translation.
Figure 1.
Structure at the 3′ end of MALAT1 RNA. The triple-helix structure formed by the two U-rich motifs (shaded in pink and green) and the A-rich 3′ end of the RNA (shaded in cyan) is indicated. Mutation of the nucleotides shaded in purple blocked translation of the RNA, and mutation of the nucleotides shaded in orange destabilized the RNA. (Adapted from Fig. 4A in Wilusz et al. 2012.)
Further mutagenesis of the MALAT1 3′ end allowed Wilusz et al. (2012) to identify RNAs ending in MALAT1 that were stable but not translated well and ones that were much less stable but still translated, and these properties mapped to two distinct regions flanking residues in the triple helix but likely do not disrupt the basic triple-helix structure (Fig. 1). How is the Malat1 RNA degraded? Cloning sequences at the 3′ end of the unstable MALAT1 mRNA resulted in identification of RNAs that had either nontemplated Us or nontemplated As at the 3′ end. The nontemplated A tails were added to the 3′ end of the RNA, but many of the oligo(U) tails were added onto intermediates that had been partially degraded into the 3′ end structure. Degradation of replication-dependent histone mRNAs, the only known cellular nonpolyadenylated mRNAs, proceeds by oligouridylation of the stem–loop at the 3′ end of the histone mRNA, allowing recruitment of Lsm1–7 to the mRNA to activate degradation. Degradation intermediates partially degraded into the stem–loop are also uridylated, suggesting that an exonuclease activity initiates degradation after addition of the oligo(U) tail and, when it stalls, must be reprimed by a new oligouridylation (Mullen and Marzluff 2008). A similar mechanism may be degrading the MALAT1 mRNA.
The poly(A) tail: a dynamic regulator of translation and stability
For “standard” polyadenylated mRNAs, the poly(A) tail is bound to PABP in the cytoplasm, with one molecule of PABP binding to ∼15 adenosines. An actively translated mRNA is thought to have multiple PABP molecules bound to its 3′ end. One function of the poly(A) tail is to stimulate translation of the mRNA. The PABP interacts directly with eIF4G, which, together with the interaction of eIF4G with the cap-binding protein eIF4e, results in a complex that promotes initiation of translation. There are proteins—PAIP1 (Craig et al. 1998) and PAIP2 (Khaleghpour et al. 2001)—that interact with PABP and either enhance (PAIP1) or reduce (PAIP2) the efficiency of translation. There is compelling evidence that regulation of the poly(A) tail length is an important mechanism for regulating translation during early development (Sheets et al. 1995; Groisman et al. 2002) and likely also at synapses in metazoans (Costa-Mattioli et al. 2009). In Xenopus oocytes, mRNAs encoding critical proteins for development, such as c-mos, are stored with a short A tail, and lengthening of the poly(A) tail after progesterone treatment results in activation of translation (Sheets et al. 1995). Some mRNAs undergo a round of adenylation and deadenylation in each cell cycle in early Xenopus development (Groisman et al. 2002). There is also excellent evidence that yeast mRNAs are deadenylated [actually, the poly(A) tail is shortened to ∼10 nt] and stored when yeast are starved for glucose, and the poly(A) tail is lengthened and the mRNA is translated again when the yeast are refed (Coller and Parker 2005). Mammalian mRNAs are stored in stress granules when translation is blocked due to a variety of environmental stresses (Anderson and Kedersha 2008). Finally, binding of microRNAs to the 3′ untranslated region (UTR) leads to deadenylation and inhibition of translation (Wu et al. 2006). Thus, the poly(A) tail is a dynamic component of most mRNAs that plays an important role in modulating translation during the lifetime of those mRNAs.
Deadenylation is also the initial step in mRNA degradation for most mRNAs . These series of findings have led to the model that mRNAs can transition between an active and inactive state by modulating poly(A) tail length in the cytoplasm (Coller and Parker 2005), making the poly(A) tail a dynamic modification that serves to regulate translation. How the cell decides whether deadenylation results in translation inhibition and retention of the mRNA in an inactive form is not understood. Thus, the poly(A) tail provides a dynamic mechanism for regulating both the translation and half-life of mRNAs.
Unusual cellular 3′ ends: short poly(A) tails
There are cellular RNAs that do not end in long poly(A) tails. These include some highly abundant, stable mRNAs, such as serum albumin mRNA in frogs, which end in poly(A) tails of 20 nt, and long poly(A) tails are never formed on these mRNAs. Instead, there is a cis-element, termed the PLE, that directs the formation of a short poly(A) tail on these transcripts, and this sequence is also essential for their efficient translation (Peng and Schoenberg 2005). PABP does not appear to be essential for translation of these mRNAs.
Replication-dependent histone mRNAs
The only known cellular mRNAs that are not polyadenylated are the metazoan replication-dependent histone mRNAs. These mRNAs end in a simple six-base stem and four-base loop that must fulfill all of the essential functions of the poly(A) tail (Marzluff et al. 2008). The histone 3′ end interacts with a cellular protein—the stem–loop-binding protein (SLBP)—that appears to function only in histone mRNA metabolism. The 3′ ends of histone mRNA are essential for translation, synergizing with the cap like the poly(A) tail does for other mRNAs (Gallie et al. 1996), and SLBP binds the stem–loop and is also essential for translation (Sanchez and Marzluff 2002). SLBP homologs are only found in organisms that have histone stem–loops in their 3′ UTR, suggesting that there has been coevolution of this novel set of histone mRNAs and the SLBP protein (Lopez and Samuelsson 2008).
There is one other set of 3′ ends formed on the snRNAs transcribed by RNA Pol II. A processing complex dedicated to snRNA 3′ end formation cleaves the nascent transcripts (Baillat et al. 2005), a processing mechanism similar to the cleavage of either histone or polyadenylated pre-mRNAs, and the snRNAs are rapidly exported to the cytoplasm for assembly of snRNPs. mRNAs transcribed from genes with the histone 3′ end replaced with the snRNA 3′ end are exported but not translated (Sun et al. 1992), indicating that just having a transcript made by RNA Pol II with a secondary structure on the 3′ end is not sufficient for translation.
Nonpolyadenylated viral RNAs often have 3′ ends essential for translation
There are a number of viral mRNAs that are translated efficiently without a poly(A) tail. Many plant RNA viruses are uncapped and have 3′ ends that end in complex structures. Six different classes of structures that are essential for translation of plant viral RNAs have been identified (for review, see Nicholson and White 2011); four of them interact with the translation initiation factor EIF4F, binding directly to either eIG4g or eIF4e, and a fifth interacts directly with the ribosome. A recent report demonstrated that the sixth class binds directly to PABP (Iwakawa et al. 2012), raising the possibility that RNA structures other than the poly(A) tail promote translation by interacting directly with PABP, which in turn could stimulate translation by binding to eIF4g. Some of these plant viral mRNAs are translated efficiently in animal cells (Gallie and Walbot 1990), in contrast to metazoan histone mRNAs that are not translated in plant cells (Gallie et al. 1996). In contrast, there are very few animal viral mRNAs that are not polyadenylated. A notable exception is the flaviviruses that cause West Nile disease, dengue fever, and yellow fever. These viruses have a +-strand genome that is not polyadenylated and is translated efficiently, even though these mRNAs lack the IRES present in other flavifiruses like hepatitis C and poliovirus. The flavirus RNAs end in an RNA structure whose secondary structure is predicted to consist of two stem–loops, preceded by a complex secondary structure, and the 3′ UTR promotes translation. The cellular trans-acting factors required for translation of the flaviviral RNAs are not known. There is a report that PABP interacts directly with the 3′ UTR dengue viral RNA and can affect translation in vitro (Polacek et al. 2009). However, the interacting region seems to be A-rich regions in the 3′ UTR rather than the 3′ stem–loop region that has also been directly implicated in translation (Holden et al. 2006), suggesting that other factors may be required as well.
How is MALAT1 RNA translated?
The 3′ end of the MALAT1 RNA likely promotes translation by circularizing the mRNA and promoting the recruitment of translation initiation factors. Whether there is a specific novel cellular trans-acting factor required or whether the 3′ end interacts directly with known initiation factors, including possibly PABP, is not known. Regardless of the mechanism, the fact that mRNAs ending in the MALAT1 3′ end are translated as efficiently as polyadenylated mRNAs suggests that there may be other Pol II transcripts that are not polyadenylated but that may be translated. Given that some estimates suggest that as many as 25% of the long noncoding RNAs may not be polyadenylated (Yang et al. 2011), there are likely to be more Pol II transcribed RNAs whose 3′ ends are formed by RNase P cleavage or other novel mechanisms, such as “partial splicing” (Box et al. 2008), and it is possible that there will be RNAs that are not polyadenylated that are translated to synthesize critical peptides or proteins under some conditions.
Acknowledgments
Supported by NIH grant GM29832 to W.F.M.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.207233.112.
References
- Anderson P, Kedersha N 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7: e1002300 doi: 10.1371/journal.ppat.1002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Tang W, Baumann P 2008. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature 456: 910–914 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sonenberg N, Richter JD 2009. Translational regulatory mechanisms in synaptic plasticity and memory storage. Prog Mol Biol Transl Sci 90: 293–311 [DOI] [PubMed] [Google Scholar]
- Craig AW, Haghighat A, Yu AT, Sonenberg N 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392: 520–523 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Walbot V 1990. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev 4: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Gallie DR, Lewis NJ, Marzluff WF 1996. The histone 3′-terminal stem–loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res 24: 1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Jung MY, Sarkissian M, Cao QP, Richter JD 2002. Translational control of the embryonic cell cycle. Cell 109: 473–483 [DOI] [PubMed] [Google Scholar]
- Holden KL, Stein DA, Pierson TC, Ahmed AA, Clyde K, Iversen PL, Harris E 2006. Inhibition of dengue virus translation and RNA synthesis by a morpholino oligomer targeted to the top of the terminal 3′ stem–loop structure. Virology 344: 439–452 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Tajima Y, Taniguchi T, Kaido M, Mise K, Tomari Y, Taniguchi H, Okuno T 2012. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3′ untranslated region. J Virol 86: 7836–7849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216 [DOI] [PubMed] [Google Scholar]
- Lopez MD, Samuelsson T 2008. Early evolution of histone mRNA 3′ end processing. RNA 14: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton-Fry RM, DeGregorio SJ, Wang J, Steitz TA, Steitz JA 2010. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 330: 1244–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, White KA 2011. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr Opin Virol 1: 373–380 [DOI] [PubMed] [Google Scholar]
- Peng J, Schoenberg DR 2005. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. RNA 11: 1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek C, Friebe P, Harris E 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol 90: 687–692 [DOI] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF 2002. The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol 22: 7093–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Wu M, Wickens M 1995. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature 374: 511–516 [DOI] [PubMed] [Google Scholar]
- Sun J-H, Pilch DR, Marzluff WF 1992. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res 20: 6057–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn C-D, Joshua-Tor L, Sharp PA 2012. A triple helix stabilizes the 3′ end of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL 2011. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 12: R16 doi: 10.1186/gb-2011-12-2-r16 [DOI] [PMC free article] [PubMed] [Google Scholar]