Abstract
CD46 is a complement regulator with important immune-related roles. CD46 functions as a pathogen receptor and is a potent co-stimulator for the induction of interferon-γ (IFN-γ)-secreting T helper 1 (TH1) effector T cells and their subsequent switch into interleukin-10 (IL-10)-producing regulatory T cells. Here, we identify the Notch protein family member Jagged1 as a new physiological ligand for CD46. Further, CD46 regulates Notch receptors and ligands expression during T cell activation and disturbance of the CD46-Notch crosstalk impedes IFN-γ induction and IL-10 switching. Importantly, CD4+ T cells from CD46-deficient patients and patients with hypomorphic Jagged1 mutations (Alagille Syndrome) fail to mount appropriate TH1 responses in vitro and in vivo suggesting that CD46-Jagged1 crosstalk is responsible for the recurrent infections in subpopulations of these patients.
CD46 (membrane cofactor protein, MCP) was initially discovered as complement regulatory protein1, then as a cell entry receptor hijacked by several viruses to promote infection2 and is now emerging as an immunomodulatory molecule with vital functions in human T helper (TH1) T cell co-stimulation and regulation3-5. The ligands and their binding sites within CD46 are defined for the first two activities; complement activation fragments C3b and C4b bind to complement control protein (CCP) domains CCP2-41, whereas viral ligands such as adenovirus knob proteins or the measles virus hemagglutinin commonly interact with domains CCP1-26,7. However, not all activities of CD46 can be explained by its interaction with the currently known ligands; the egg-sperm fusion event mediated by CD46 requires CCP18 and although intrinsic T cell-derived C3b generation is required for CD46 stimulation on CD4+ T cells4, the molecular basis of CD46-mediated co-stimulatory activity is unknown. Similarly, while the importance of the Notch system in TH1 and TH2 lineage induction is undisputed and key signalling events mediated by Notch activation on CD4+ T cells have been identified9, many functional aspects of this system in T cell biology remain puzzling and cannot be explained solely by Notch and Notch ligand interactions. Given the intriguing similarities between these two evolutionarily old systems in regards to TH1 biology, we investigated if there may be a functional connection between CD46 and Notch proteins and if this potential complement and Notch system crosstalk is required for TH1 effector function.
We here demonstrate that CD46 activation on CD4+ T cells not only regulates the expression of Notch and Notch ligands but also identify Jagged1 as a novel physiological ligand for CD46. The Jagged1 binding site within CD46 is located in the N-terminal CCP domains 1 and 2 and interference with the CD46 and Jagged1 interaction greatly reduces TH1 induction in vitro. We provide support for the in vivo importance of this novel protein interaction by establishing that patients with either CD4610,11 or Jagged1 mutations (Alagille Syndrome patients12) share key features: These patients suffer from recurrent infections and though T cell proliferation and TH2 effector function is unaffected, in vitro TH1 induction (or regulation) was absent or severely compromised. Mechanistically, faulty TH1 induction seems to involve altered responsiveness to interleukin-2 (IL-2) cytokine family members as all patients presented with marked deviations in the expression of either the IL-7 receptor α-chain (CD127) or the common γ-chain (CD132) or both. Importantly, patient-derived T cells with defective TH1 induction in vitro were also unable to undergo TH1 induction in vivo when assessed in a humanized mouse graft versus host disease (GvHD) model. These data uncover a functional connection between the complement and Notch systems that is critical for human TH1 induction and regulation in infection and immune homeostasis.
RESULTS
Jagged1 binds to CCPs 1 and 2 of CD46
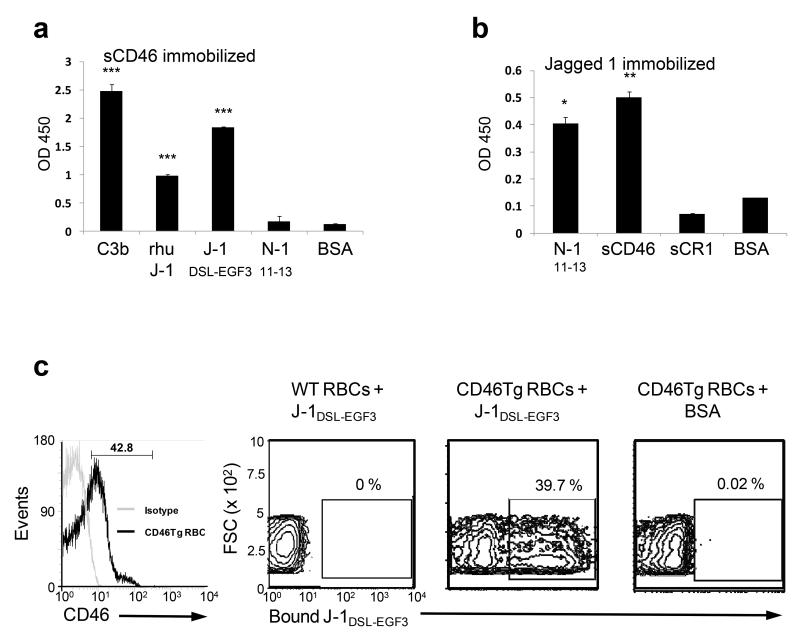
Because several of CD46’s biological activities cannot be explained by its interaction with C3b or C4b, the existence of another physiological ligand has long been suspected. Using an initial ELISA-based screen with recombinant (r) soluble CD46 (Supplementary Fig. 1a) and available rNotch receptor and rNotch ligand family proteins (Supplementary Fig. 1b and c), we identified Jagged1 as a novel CD46-binding protein. CD46 bound to C3b (positive control), full-length Jagged1 and a Jagged1 construct containing the Notch1 binding site (DSL and first three epidermal growth factor (EGF)-like domains; J-1DSL-EGF3 (Reference 13), Supplementary Fig. 1c) but not to Notch1 (or a recombinant soluble construct comprising Notch1 EGF-like domains 11-13 containing the Jagged1 binding site, N-111-13, Supplementary Fig. 1b) or other Notch family proteins (Fig. 1a and data not shown). Conversely, Jagged1 bound CD46 but not soluble rComplement Receptor 1 (sCR1), which shares CD46’s ability to bind C3b or C4b (Fig. 1b). We also did not observe binding of Jagged1 to factor H or C4bp, two fluid phase complement regulators containing C3b or C4b binding sites (data not shown). The interaction between CD46 and Jagged1 is therefore specific. Further, our data using either immobilized CD46 or J-1DSL-EGF3 indicate that both the CD46 and Notch1 binding sites within Jagged1 reside in this same region (Fig. 1a and b). Jagged1 binding to CD46 was confirmed with cell-surface expressed CD46 using red blood cells (RBCs) from mice transgenic for human CD46 (CD46Tg). CD46Tg animals express CD46 on all cells14 while wild-type (WT) mice lack CD46 expression on somatic cells15. Importantly, RBCs also lack Notch receptor and Notch ligand expression. Using this system, we observed that J-1DSL-EGF3 bound to RBCs from CD46Tg mice but not from WT mice (Fig. 1c), confirming that physiological, cell-expressed CD46 is competent for Jagged1-binding.
Figure. 1. Jagged1 is a ligand for CD46.
a and b, Analysis of the CD46 and Jagged1 interaction by ELISA. ELISA assays were performed utilizing either (a) immobilized soluble CD46 (CCP1-4) incubated with indicated recombinant soluble proteins (rhu J-1, recombinant human Jagged1; J-1DSL-EGF3, a recombinant protein containing the Notch1 binding site DSL and the first three EGF-like domains; N-111-13, a recombinant protein containing Notch1 EGF-like domains 11-13) or bovine serum albumin (BSA) or (b) immobilized J-1DSL-EGF3 incubated with indicated soluble proteins (sCR1, soluble recombinant complement receptor 1) in Ca2+ buffer (for further details on utilized proteins, see Supplementary Fig. 1). Mean ± SD (n = 5). (c) Analysis of Jagged1 binding to cell surface-expressed CD46. Red blood cells (RBCs) from CD46Tg mice express CD46 on the surface (left panel). RBCs from CD46Tg and from WT mice were incubated with biotinylated J-1DSL-EGF3 (middle panels) or BSA (right panel) and J-1DSL-EGF3-binding to RBCs measured using fluorochrome-labeled streptavidin. Shown is one representative data set of three independently performed experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.001 when compared with BSA binding to immobilized protein.
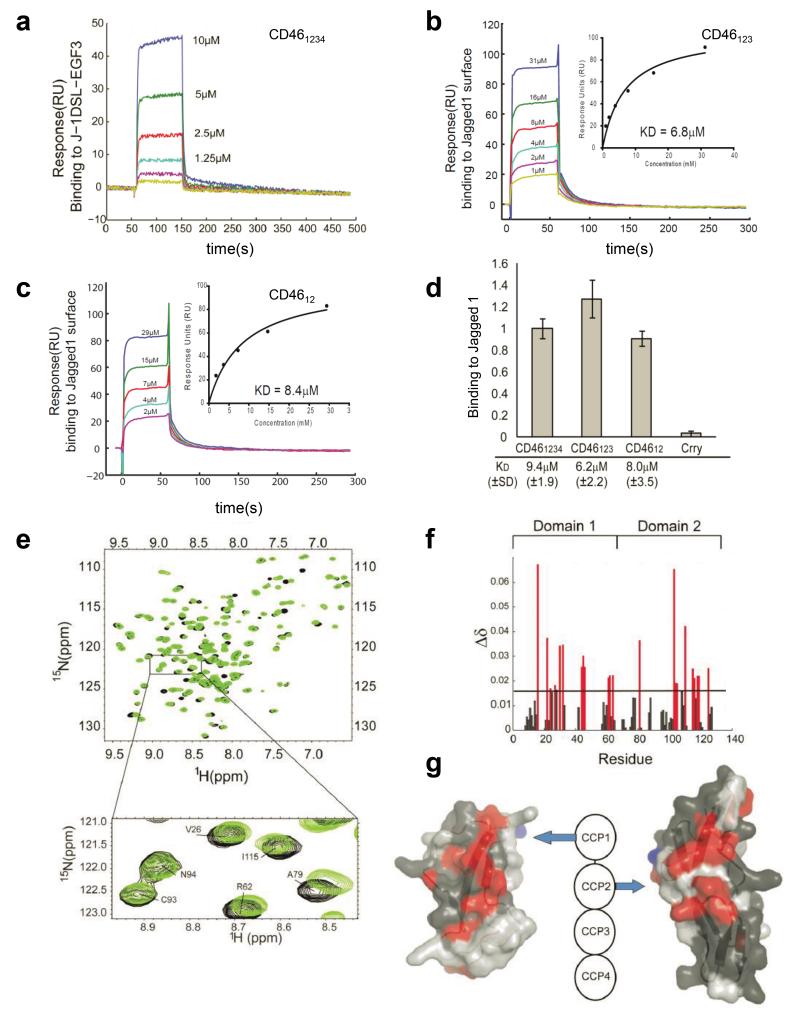
To further characterize the CD46-Jagged1 interaction, we measured binding affinities between J-1DSL-EGF3 and truncated CD46 constructs using surface plasmon resonance (SPR) and mapped the Jagged1 binding site on CD46 using nuclear magnetic resonance (NMR) spectroscopy. J-1DSL-EGF3 binding to CD46 constructs comprising domains 1-4, 1-3 and 1-2 all showed similar interaction affinities and kinetics, but did not bind to the closely related murine complement regulatory molecule Crry16 (Fig. 2a-d). These experiments established that the two N-terminal domains of CD46 (CD4612) are fully competent for Jagged1-binding. The dissociation constant (KD) for the CD46-J-1DSL-EGF3 interaction is about 8μM and with this in a less tight affinity range as the interaction between CD46 and C3b (1 μM, Dr. Claire Harris, University of Cardiff, UK; personal communication) but tighter than the assumed KD for interactions between sN-111-13 and J-1DSL-EGF3, where protein concentrations in excess of 50μM have previously been required to observe an interaction and the interaction was found not to be saturated with protein concentrations up to 160μM13. Using purified, soluble, proteins to characterize interactions has demonstrated that the majority of cell-surface proteins interact with one another with KDs in the μM-range and concomitant fast on and off rates. This has often been interpreted as facilitating fine-tuning of interactions by the avidity effects either due to many weak interactions being shared across two interacting cells or increases in avidity driven by the fact that cell-surface molecules on the same cell need to diffuse only in 2D rather than 3D to find interacting partners. This means that even apparently transient interactions will occur in vivo more frequently and for a longer duration than the solution studies will imply and suggest that similar ideas will be important in the case of the interaction between CD46 and Jagged1.
Figure. 2. Jagged1 binds to CD46 CCPs 1 and 2.
(a) Surface plasmon resonance demonstrates that J-1DSL-EGF3, binds to CD461-4 coupled on the surface of the chip with a KD of ~8μM (normalised by subtraction of mock-coupled channel). (b and c) Binding of (b) CD461-3 and (c) CD461-2 to immobilized J-1DSL-EGF3 normalized by subtraction of a mock-coupled channel. Insert chart shows equilibrium values of binding and KD fit using SigmaPlot. All data processed using the BIAevaluation software. (d) Injections of CD461-4 (21 μM), CD461-3 (25 μM) CD461-2 (19 μM) over immobilized J-1DSL-EGF3 in triplicate. The rodent complement regulator Crry (CCP1-4) was used as a negative control (20μM). Data are normalised for construct molecular weight and represented as mean ± SD. The KD values reported below the chart are derived from a minimum of three repeated dilution series ± SD. (e) NMR spectroscopy overlay of the 1H,15N-heteronuclear single quantum coherence (HSQC) of CD461-2 (black) demonstrates chemical shift perturbation with the addition of unlabelled J-1DSL-EGF3 (green) at a molar ratio of 0.6. (f) Chemical shift perturbation by residue for those unambiguously assigned and baseline resolved in the 15N,1H-HSQC for CD461-2. (g) Surface structure of CCP1 and CCP2 (3O8E.pdb) showing residues with unambiguous assignment (dark grey) and chemical shift perturbation > 0.15ppm (red) upon J-1DSL-EGF3 addition. N-linked glycosylation sites are shown in blue.
CD46 interaction site mapping was performed using NMR chemical shift perturbation. A backbone assignment of the CD461-2 construct was performed using standard triple resonance methods17. The Jagged1 binding site mapped to an interaction surface comprised of CCP1-2 (Fig. 2e-g). The Jagged1 interaction surface is clearly distinct from that for C3b and C4b (which utilise CD46 domains CCP2-4) but in the same two domains used by measles virus and adenoviruses 11, 21, and 35 (References 6 and 7) (Supplementary Fig. 1d).
These data identify Jagged1 as a physiological, and not pathogen-derived ligand for CD46 and map the binding site of Jagged1 to CCP1-2 within CD46.
CD46 regulates Notch and Notch ligand expression
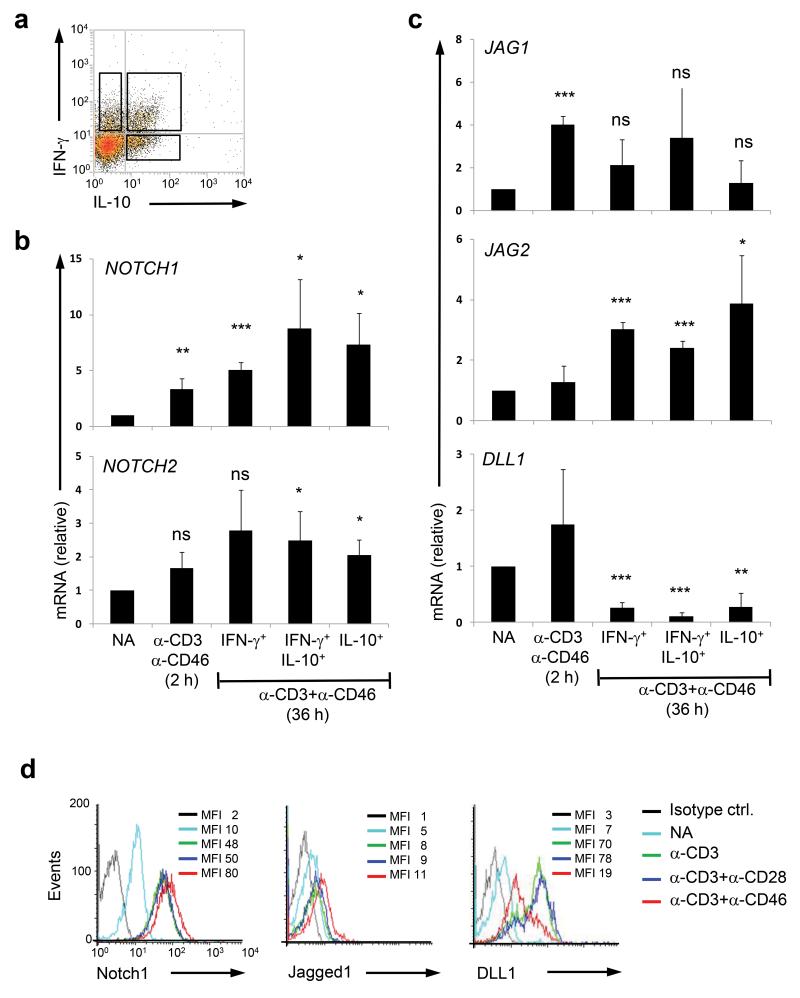
Although CD46 was initially discovered as a complement regulatory molecule, it transmits intracellular signals upon cell surface engagement and can modulate the function of several immune competent cell types18. For example, CD46 is a T cell co-stimulator3 and regulates IFN-γ vs. IL-10 production by human TH1 cells; co-engagement of CD3 and CD46 in the presence of low IL-2 induces proinflammatory TH1 cells (IFN-γ+IL-10−), while the presence of high environmental IL-2 initiates co-expression of IL-10, and a switch to a regulatory phenotype (IFN-γ+IL-10+) and finally IFN-γ shut down (IFN-γ−IL-10+)4,18,19. This CD46 and IL-2 driven (self) regulatory pathway is defective in T cells from rheumatoid arthritis patients, which fail to switch4. Similarly, signalling events initiated by the human Notch receptor and ligand family members (Notch1-4, Jagged1 and Jagged2 and Delta-like (DLL)1, 3 and 4, Supplementary Fig. 1b and c) play a crucial role in TH1/TH2 lineage induction9,20 and specifically in IL-10 co-expression in TH1 cells21. Consequently, we next assessed if the CD46 and Jagged1 interaction is important in TH1 IFN-γ and IL-10 regulation, and first measured the expression of CD46, Jagged1 and other Notch family members on resting T cells and then on T cells that had been activated with immobilized antibodies to CD3 and CD46. CD46 co-activation by antibody cross-linking rapidly increased notch 1 (gene symbol, NOTCH1) and notch 2 (NOTCH2), and jagged 1 (JAG1) and jagged 2 (JAG2) gene expression (similar to CD3 and CD3+CD28-activation, data not shown), which remained high in the CD46-induced TH1 and IL-10-switched TH1 cells (Fig. 3a-c). In contrast, delta-like 1 (DLL1) transcription was decreased at 36h in cells activated with the immobilized antibodies to CD3 and CD46. Similarly, CD46 cross-linking leads to rapid loss of CD46 itself from the cell surface (Supplementary Fig. 2a). CD46 down-regulation upon stimulation has been observed widely and its biological significance is unknown18. Gene transcripts for notch 3 and notch 4 (NOTCH3 and NOTCH4) as well as delta-like 3 and delta-like 4 (DLL3 and DLL4) were detected in resting cells but remained unaltered by any antibody-mediated activation conditions tested (data not shown). All changes were also observed at the level of cell surface-expressed proteins (Fig. 3d).
Figure. 3. CD46 regulates Notch receptors and ligand expression on human CD4+ T cells.
(a) CD3+CD46+IL-2 activation of human CD4+ T cells induces three distinct IFN-γ and IL-10-secreting subpopulations (36h post activation). (b and c) CD46 co-stimulation increases mRNA levels of NOTCH1, NOTCH2 and JAG1 and JAG2 genes but decreases DLL1 transcripts. mRNAs were assessed from non-activated T cells (NA), 2h α-CD3+CD46-activated T cells (with 50 U/ml rhIL-2) and from T cells corresponding to the three distinct cytokine-secreting populations. Expression of mRNA is presented relative to 18s mRNA expression in each sample. Results shown are mean ± SD (n = 4). mRNAs coding for NOTCH3 and NOTCH4 as well as DLL4 genes were present but unaltered by CD3 or CD3+CD46 activation at any time point measured (data not shown). (d) Protein expression of Notch1, Jagged1 and Delta-like1 (DLL1) on non-subsorted bulk CD4+ T cells after CD3+CD46 stimulation for 36h. Data represent mean ± SD (n=4). Note that increase in proteins also reached statistical significance when the respective mean fluorescence intensities (MFIs) of the histograms were compared. *, p < 0.05; **, p < 0 .005; ***, p < 0.001 when compared with non-activated cells. Ctrl, control; ns, statistically not significant.
These results demonstrate that CD46 activation induces a specific Notch receptor and Notch ligand expression pattern on CD4+ T cells which is typified by increased expression of Notch1 and Notch2 as well as Jagged1 and Jagged2 but by loss of DLL1 and CD46.
CD46-Notch crosstalk is vital for TH1 induction in vitro
Notch proteins must be processed by the metalloprotease (ADAM) and the presenilin/γ-secretase complex to become signalling competent and contributions of Notch-mediated signals are assessed utilizing ADAM and γ-secretase inhibitors9,20. This approach cannot be used to address if CD46-Notch crosstalk is a requisite for TH1 regulation since CD46 activation and signalling on CD4+ T cells also requires ADAM and γ-secretase processing 22 (Supplementary Fig. 2a and b). CD46 exists in four isoforms that arise through splicing of a glycosylated extracellular region and the two possible intracellular tails, CYT1 and CYT2 (Supplementary Fig. 1a). Jurkat T cells stably transfected with the CD46-CYT1 isoform (Jurkat-BC1) produce IL-10 upon CD3+CD46 activation whereas non-transfected or CD46-CYT2-transfected Jurkat cells are unable to express IL-10 (Reference 4). To investigate whether the CD46-CYT1-mediated stimulation involves subsequent Notch1 signalling, we disrupted Notch1 signalling using either an inhibitory mAb or sN-111-13 to compete for Notch ligands against cell surface-expressed Notch1. Both treatments abrogated CD46-mediated IFN-γ production and reduced IL-10 secretion by >50% in Jurkat BC1 cells (Supplementary Fig. 2c and d), indicating that CD46 and Notch signalling pathways indeed intersect in TH1 cytokine production and IL-10 switching.
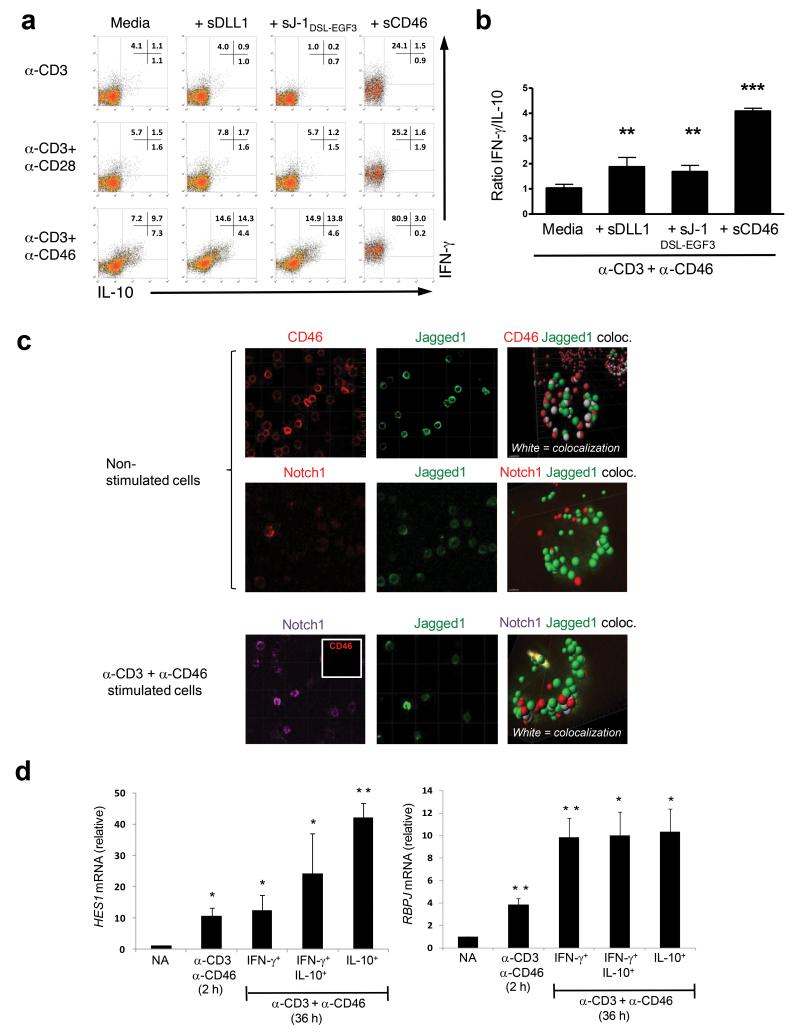
Consistent with this hypothesis, the addition of soluble CD46, DLL1, or Jagged1 also showed a marked reduction in CD3+CD46+IL-2-mediated switching of IFN-γ to IL-10 in purified CD4+ T cells (Fig. 4a and b). This is likely due to interference with temporally regulated changes in CD46 and Notch system member interactions and thus signalling events during T cell activation. Super resolution imaging experiments demonstrated that > 95% of Jagged1 on resting CD4+ T cells co-localizes with CD46, whereas only negligible Jagged1 and Notch interactions can be observed (Fig. 4c, upper two panels). Upon CD3+CD46 activation, which induces CD46 down-regulation (Fig. 4c, lower panel, insert) and ‘release’ of Jagged1, a substantial proportion (> 50%) of Notch1 molecules were found bound to Jagged1 (Fig. 4c, lower panel). These data suggest that our affinity measurements obtained with limited recombinant fragments of CD46, Notch1 and Jagged1 (Fig. 2) extend to intact proteins expressed on the cell surface. Further support for a regulated successive CD46 and Notch system crosstalk during TH1 induction (see model suggestion in Supplementary Fig. 3) comes from the observation that CD46 activation alone, without direct antibody-mediated Notch engagement, induces strong transcription of the Notch target gene hairy and enhancer of split 1 (HES1) and transcription of the Notch signalling mediator recombination signal binding protein for immunoglobulin kappa J region (RBPJ)20 (Fig. 4d). Conversely, inadequate CD46 down-regulation hinders TH1 induction. A member of the E-cadherin network, α-E-catenin, binds to the cytoplasmic portion of CD46 in human intestinal epithelial cells23. Alpha-E-catenin also interacts with CD46 in primary human CD4+ T cells (Supplementary Fig. 4a) and protein knockdown of α-E-catenin (Supplementary Fig. 4b) impaired CD46 down-regulation without an effect on Notch, Jagged1 or DLL1 protein expression (Supplementary Fig. 4c) or on that of additional molecules vital for T cell activation such as CD3, CD25, CD28 or CD69 (Supplementary Fig. 4d). Inhibition of CD46 down-regulation was accompanied by a 50% reduction in IFN-γ and IL-10 production (cell proliferation and viability were unaffected, data not shown) (Supplementary Fig. 4e). Although these data suggest that α-E-catenin participates in CD46-mediated signalling events in CD4+ T cells, we cannot exclude that the changes in cytokine production are secondary to non-apparent additional effects of the α-E-catenin knock down.
Figure. 4. Undisturbed CD46 and Notch system crosstalk is required for normal IFN-γ to IL-10 switching in human TH1 cells.
(a) Addition of sDLL1, sJ-1DSL-EGF3 or sCD46 during CD3, CD3+CD28 or CD3+CD46 activation of CD4+ T cells increases the numbers of IFN-γ-secreting TH1 cells and (b) changes the IFN-γ:IL-10 ratio of cytokines secreted into the media. Results shown are mean ± SD (n = 6). (c) CD46 sequesters Jagged1 on resting T cells and prevents the Jagged1 and Notch1 interaction. Non-activated (upper two rows of panels) or CD3+CD46-activated (lower row of panels) T cells were stained with antibodies to CD46, Notch1 or Jagged1 and then subjected to superresolution confocal microscopy and 3D analysis to assess for molecular co-localization. White areas in the 3D analyses indicate co-localization of assessed proteins. CD3+CD46-activated T cells lose CD46 surface expression and become negative for CD46 staining (see insert in left panel, lower row). Shown are representative results of two independently performed experiments. (d) CD46 co-stimulation of purified CD4+ T cells increases HES1 and RBPJ mRNA transcription. T cells were activated as indicated and the three emerging IFN-γ- and IL-10-secreting populations from the 36hr sample cell sorted. mRNA was purified and then subjected to quantitative PCR analysis for HES1 and RBPJ transcripts. Expression of mRNA is presented relative to 18s mRNA expression in each sample. Data represent mean ± SD (n=3). *, p < 0.05 **, p < 0.005, ***, p < 0 .001 when compared to media (b) or non-activated cells (d).
We were not been able to inhibit the observed DLL1 down-regulation during T cell activation without ADAM inhibitor-treatment on CD4+ T cells. However, we noticed that CD46-mediated DLL1 down-regulation on Jurkat-BC1 cells is less efficient compared to that on primary CD4+ T cells (Supplementary Fig. 4f, upper panel), possibly explaining why Jurkat-BC1 cells produce relatively small amounts of IL-10. In agreement with this, transfection of Jurkat-BC1 cells with shRNA targeting DLL1 mRNA reduced DLL1 expression in activated Jurkat BC1 cells (Supplementary Fig. 4f, upper panel) while proportionally increasing IL-10 production (Supplementary Fig. 4f, lower panel).
In sum, these data suggest that CD46 presence on T cell surfaces restricts Notch1 and Jagged1 interactions and that CD46 engagement during T cell activation leads to α-E-catenin-dependent down-regulation of CD46 and α-E-catenin-independent down-regulation of DLL1. Disturbance in this spacially and temporally regulated crosstalk between complement and Notch proteins leads to a deregulated TH1 responses in vitro.
CD46 deficiency causes defective TH1 function in vivo
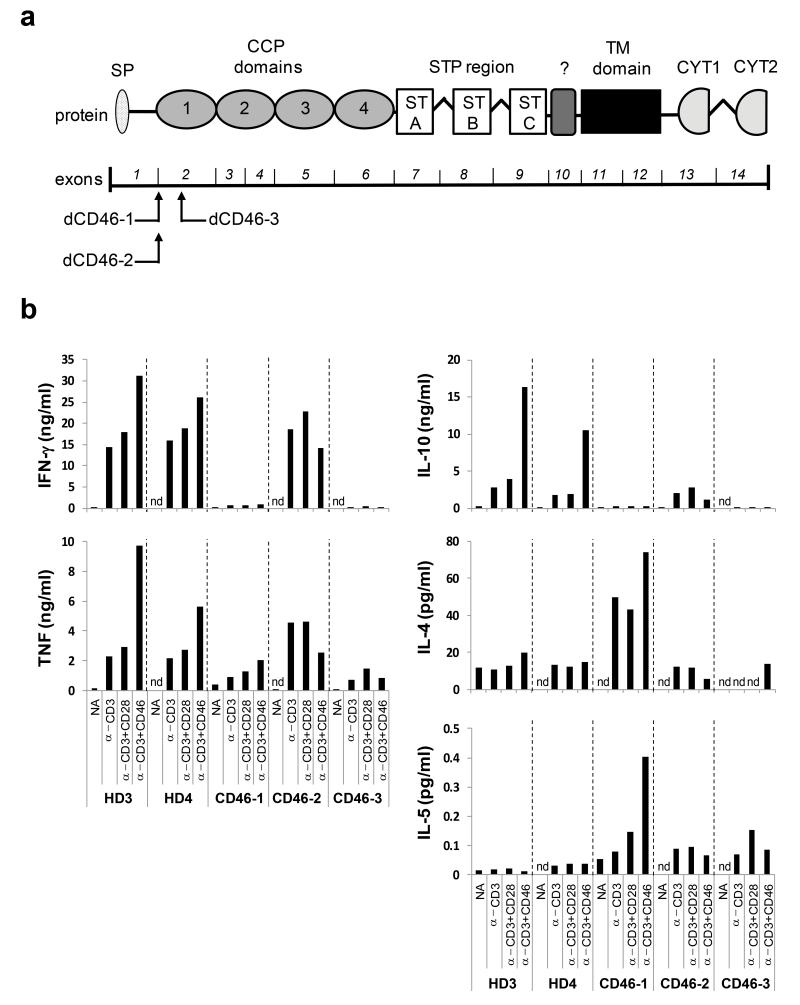
Rodents (mice, rats and guinea pigs) lack CD46 expression on somatic tissues and this restricted expression pattern impedes a direct evaluation of the in vivo importance of the CD46 and Jagged1 interaction using a small animal model15. Although the murine Crry protein compensates for CD46’s complement regulatory function, it does not regulate TH1 responses16. For this reason, we obtained CD4+ T cells from patients with mutations in either CD46 or JAG1 and assessed their capacity to mount TH1 responses in vitro and in vivo. Mutations in the CD46 gene affecting protein expression levels or complement regulatory function cause atypical haemolytic uremic syndrome (HUS)24. There are currently less than ten patients worldwide identified with homozygous mutations in CD46 but, interestingly, over 50% of these patients present with common variable immunodeficiency (CVID) and recurrent chest infections, indicative that CD46 mutations can indeed induce immune defects10,11. Three patients with homozygous CD46 mutations participated in this study (Fig. 5a): (1) Patient CD46-1, who has a splice site alteration between exons 1 and 2 leading to only 10% of normal levels of CD46 expression on peripheral blood mononuclear cells (PBMCs)10, (2) Patient CD46-2, who has a similar splice site alteration causing aberrant mRNA transcripts and loss of CD46 expression on > 90% of PBMC11 and normal expression levels on the remaining 10% and (3) Patient CD46-3, with two mutations in exon 2 and absence of detectable CD46 cell surface expression (Supplementary Table 1 and Supplementary Fig. 5a). Although hospital records for CD46-1 in this regard are lacking, CD46-2 and CD46-3 suffered from confirmed recurrent infections, have been diagnosed with CVID and are under intravenous immunoglobulin infusion (IVIG) regimen. All three patients have normal B cell and CD4+ and CD8+ T cell numbers10,11 (data not shown) and activation of their purified CD4+ T cells showed that they proliferate at normal rates (data not shown) and mount strong TH2 responses (Fig. 5b); this suggests that an intrinsic thymus-derived defect in T cell lineage induction is unlikely. However, neither CD3, CD3+CD28 nor CD3+CD46 activation induced IFN-γ secretion in Patients CD46-1 and CD46-3; consequently, the cells also failed to switch into an IFN-γ and IL-10 co-expressing phenotype and these patients thus are lacking a major T cell population key in the prevention and clearance of infections caused by intracellular pathogens25 (Fig. 5b). In contrast, T cells from Patient CD46-2 produced normal amounts of IFN-γ and IL-10 upon CD3 and CD3+CD28 activation (though they lacked the usual CD46-mediated increase in IFN-γ production and switch to IL-10). The reason for this is currently unclear. It should be noted here that B cells from Patient CD46-1 are fully functional26 but that this has not been confirmed for Patients CD46-2 and CD46-3.
Figure. 5. T cells from CD46-deficient patients present with defective in vitro TH1 induction.
(a) Localization of the CD46 gene mutations in the three CD46-deficient patients (CD46-1 to CD46-3) assessed for TH1 induction. The lower part shows the exon structure of the CD46 gene and above the corresponding protein domains. (b) Comparison of cytokine expression by CD4+ T cells from healthy donors and CD46-deficient patients. T cells from two healthy donors (HD3 and HD4) or patients were purified from freshly-drawn blood samples and either left non-activated (NA), or activated with immobilized antibodies to CD3 and CD46 in the presence of 25 U/ml rhIL-2. Indicated cytokine secretion into the cell culture media was assessed 36h post activation using the TH1 and TH2 CBA Cytokine Secretion Assay. Data shown are the mean value of each condition performed in duplicate. Note, that though values for only two HD are shown, these are representative for 12 age- and gender-matched donors assessed over the course of the study. CCP, complement control protein; CYT1 or CYT2, cytoplasmic tail 1 or −2; nd, not detectable; SP, signal peptide; STP, serine threonine proline-rich; TM, transmembrane;?, region of unknown function
As predicted, lack of CD46 resulted in deregulated Notch1 and Jagged1 expression regulation as non-activated T cells from Patients CD46-2 and CD46-3 expressed higher levels of Jagged1 but then failed to upregulate the protein upon activation. Similarly, T cells from Patient CD46-3 were defective in activation-induced Notch1 upregulation while T cells from Patient CD46-2 overexpressed Notch1 upon activation (data on Notch1 and Jagged1 expression are not available for CD46-1) (Table 1 and Supplementary Fig. 5a). We also compared the expression of additional key cell surface markers required for normal TH1 responses between healthy donors and the patients with CD46 mutations on resting and activated T cells. No significant differences were found in the expression and regulation of CD3, CD11a (α chain of LFA-1), CD28, CD69, CD122, and CCR7 (data not shown). Although all three patients showed a trend towards reduced CD25 upregulation and CD62L down-regulation upon CD3 and CD3+CD28 activation as well as a small memory T cell pool (CD4+CD45RA−CD45RO+ cells), these results were within the range of normal donor variation (data not shown). We discovered though, that T cells from all patients showed a marked deregulation of CD127 and CD132, which together form the IL-7 receptor27: T cells from patients lacked the CD127 down-regulation usually induced by activation while CD132 was overexpressed on resting or activated T cells (Table 1 and Supplementary Fig. 5a).
Table 1. Surface marker deregulated on resting or activated CD4+ T cells from CD46-deficient patients.
| Jagged1 | Notch1 | CD46 | CD127 | CD132 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NA | CD3 CD28 |
CD3 CD46 |
NA | CD3 CD28 |
CD3 CD46 |
NA | CD3 CD28 |
CD3 CD46 |
NA | CD3 CD28 |
CD3 CD46 |
NA | CD3 CD28 |
CD3 CD46 |
|
| HD3 | + | ↑ | ↑ | + | ↑ | ↑ | + | ↔ | ↓ | + | ↓ | ↓ | + | ↑ | ↑ |
| HD4 | + | ↑ | ↑ | + | ↑ | ↑ | + | ↔ | ↓ | + | ↓ | ↓ | + | ↑ | ↑ |
| CD46-1 | nda | nda | nda | nda | nda | nda | 10% of normal levels | ↔ | ↓ | + ( ↑) |
↓ | ↔ c | + (↑↑) |
↑ | ↔ a |
| CD46-2 | + (↑) | ↔ | ↔ a | + | ↑ | ↑ | On 10% of cells | ↔ | ↓ | + | ↔ | ↑ | +
(↑↑) |
↔ | ↔ a |
| CD46-3 | +(↑) | ↔ | ↔ a | + | ↔ | ↔ b | − | − | − | + | ↓ | ↔ c | + | ↑↑ c | ↑↑ c |
+, present on resting T cells; ↑, increased expression upon activation; ↓, decreased expression upon activation; ↔, no change in expression; −, not present on resting (or activated) T cells; (↑)(≤ 250%) and (↑↑)(> 250%), increased baseline expression compared to HD3 and 4; ↑(≤ 250%) and ↑↑(> 250%), increased upregulation on activated cells compared to HD3 and 4; ↔, lack of change in expression compared to nonactivated sample;
expression is comparable to activated T cells from HDs
expression remains decreased compared to HD3 and 4
expression remains increased compared to HD3 and 4;(Supplementary Fig. 5)
CD46-1, -2, -3, CD46-deficient patients 1,2 and 3; HD, healthy donor; NA, non-activated; nda, no data available
The observed defect in in vitro TH1 induction by T cells lacking normal CD46 expression extends also to an in vivo model of GvHD28. CD3+CD28-activated T cells from three healthy donors or Patient CD46-1 were injected into NOD-SCID-B2M−/− mice and human T cell engraftment was monitored through the presence of human CD45RA+ cells in blood and hIFN-γ in serum. Cell engraftment between T cells from CD46-1 and healthy donors was comparable. In contrast, and in line with the in vitro generated data, hIFN-γ could only be detected in sera from mice injected with T cells from healthy donors (Supplementary Fig. 5b and c). Further, while T cells from the latter induced GvHD as indicated by weight loss (Supplementary Fig. 5d) and immunohistochemistry analysis of intestinal tissue (data not shown), mice injected with T cells from Patient CD46-1 developed no disease. Blood samples from Patients CD46-2 and CD46-3 could not be obtained for this experiment.
Thus, CD46-mediated signalling events are not only required for the switch of TH1 cells into an IL-10-co-expressing phenotype4 but also for the initial induction of a TH1 phenotype in CD4+ T cells in vivo. Further, CD46 participates in the expression regulation of CD127 and CD132 on TH1 cells but is not required for normal TH2 effector function or cell proliferation.
Alagille Syndrome causes defective TH1 function in vivo
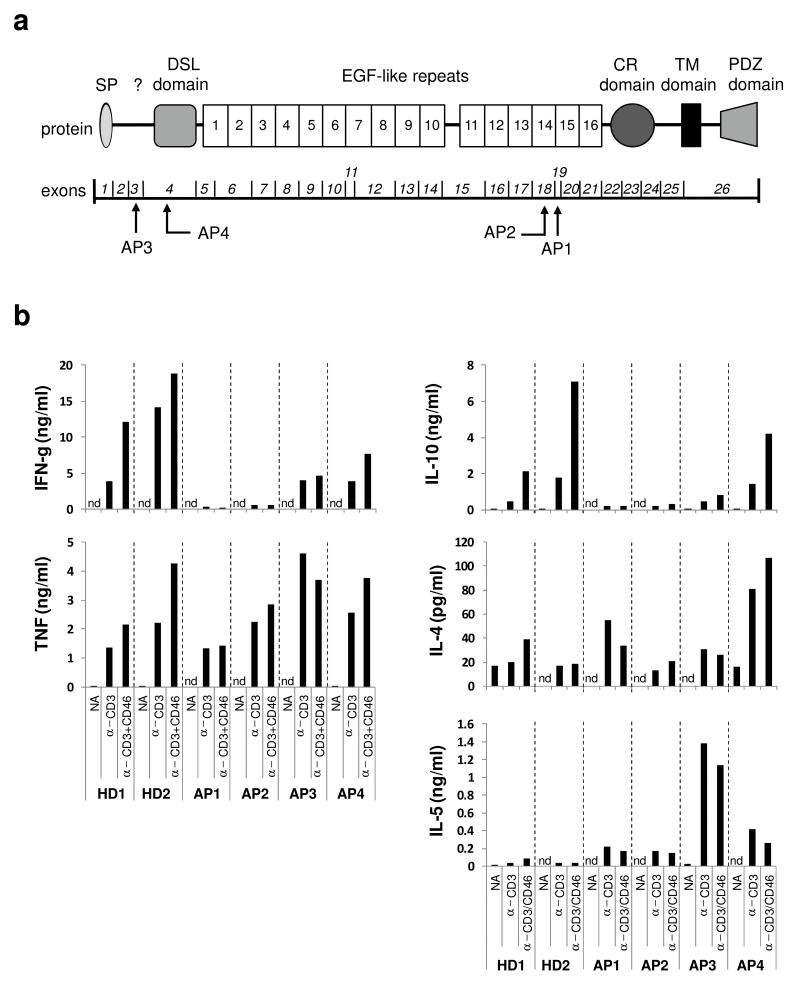
Complete deficiencies of Notch family members have not been described yet; this is probably due to the fact that the Notch system plays a central role in cell-cell communication during tissue morphogenesis and organ development29. Heterozygous JAG1 mutations are inherited in an autosomal dominant fashion and cause Alagille Syndrome (AS), a condition characterized by developmental problems affecting primarily liver, heart, eye and skeleton12,30. Although it is known that ~25% of AS patients also suffer from recurrent ear and respiratory tract infections 31, the immunological defect(s) underlying these infections are entirely unexplored. Given the discovery of an interaction between CD46 and Jagged1 and the reduced or absent TH1 response in CD46-deficient individuals, we hypothesized that recurrent infections in AS patients are also rooted in altered TH1 function caused by a deregulated CD46-Notch system crosstalk. In this study, we enrolled four AS patients (Patients AP1 to AP4) that had mutations in either exon 3, 4, 18 or 19 of the JAG1 gene and suffered from recurrent and persistent otitis media and chest infections (Supplementary Table 2 and Fig. 6a). Because Notch and Notch ligand interactions participate in CD4 and CD8 T cell lineage development in the thymus9 we first assessed the cell population composition of PBMCs from AP1 to AP4 and found no deviations in the percentages of key lymphocyte subpopulations when compared to healthy donors32 (Supplementary Table 2). However, when we activated the purified CD4+ T cells from these patients and compared the TH1 and TH2 cytokine expression upon activation with those from healthy control T cells, we observed a profile reminiscent of that in CD46-deficient patients (Fig. 6b). Cell proliferation and tumor necrosis factor (TNF) secretion was normal in all patients, but T cells from Patients AP1 and AP2 produced no IFN-γ (and did not switch to IL-10 production) upon either CD3, CD3+CD28 (not shown) or CD3+CD46 activation, while AP3 and AP4 presented with a marked decrease in TH1 induction (≤ 50%). T cells from these last two patients also gave a noticeably increased TH2 response (Fig. 6b).
Figure. 6. T cells from Alagille Syndrome patients present with defective in vitro TH1 induction.
(a) Localization of the JAG1 gene mutations in the four Alagille Syndrome patients (AP1 to AP4) assessed for TH1 induction. The lower part depicts the exon structure of the JAG1 gene with exons 1 to 26 and the upper part the corresponding Jagged1 protein domains. Note that all four JAG1 mutations result likely in the retention of the altered protein in the endoplasmic reticulum. (b) Comparison of cytokine expression by CD4+ T cells from healthy donors and APs. T cells from two healthy donors (HD1 and HD2) or APs1-4 were purified from freshly-drawn blood samples and either left non-activated (NA), or activated with immobilized antibodies to CD3 and CD46 in the presence of 25 U/ml rhIL-2. Indicated cytokine secretion into the cell culture media was assessed 36h post activation using the TH1 and TH2 CBA Cytokine Secretion Assay. Data shown are the mean value of each condition performed in duplicate. CR, cysteine rich; nd, not detectable; PDZ, post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (Dlg1), and zonula occludens-1 protein (zo-1); SP, signal peptide; TM, transmembrane;?, region of unknown function
Although Notch1 expression and up-regulation upon activation was unaffected, all patients had unexpectedly increased Jagged1 protein expression on resting T cells. In contrast to the CD46-deficient patients though, Jagged1 was further up-regulated upon activation (Table 2 and Supplementary Fig. 6a). Whereas expression and regulation of CD3, CD11a, CD25, CD28, CD69, CD122, and CCR7 was also normal in this patient group (data not shown), we found significant deviations (comparable to those observed in the CD46-deficient patients) in the regulation of CD127 and CD132 by T cells from the AS patients. AP1 and AP4 entirely lacked CD127 down-regulation upon CD3+CD46 activation and while all patients had increased amounts of CD132 on resting T cells compared to healthy donors, upon activation T cells from AP1, AP3 and AP4 up-regulated this molecule well beyond normal levels. Furthermore, we observed that T cells from AP2 and AP3 were unable to down-regulate CD46 upon CD3+CD46 activation efficiently (Table 2 and Supplementary Fig. 6a).
Table 2. Surface marker deregulated on resting or activated CD4+ T cells from Alagille Syndrome patients.
| Jagged1 | Notch1 | CD46 | CD127 | CD132 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NA | CD3 CD46 |
NA | CD3 CD46 |
NA | CD3 CD46 |
NA | CD3 CD46 |
NA | CD3C D46 |
|
| HD1 | + | ↑ | + | ↑ | + | ↓ | + | ↓ | + | ↑ |
| HD2 | + | ↑ | + | ↑ | + | ↓ | + | ↓ | + | ↑ |
| AP1 | + (↑) | ↑(↑) | + | ↑ | + | ↓ | + | ↑ | + (↑↑) | ↑↑ |
| AP2 | + (↑) | ↑(↑) | + | ↑ | + | ↓ (50%) | + | ↓ | + (↑) | ↔ |
| AP3 | + (↑) | ↑(↑) | + (↓) | ↑ | + | ↔ | + | ↓ | + (↑) | ↔ |
| AP4 | + (↑) | ↔ | + (↓) | ↑ | + | ↓ | +(↑↑) | ↔ | + (↑↑) | ↔ a |
+, present on resting T cells; ↑, increased expression upon activation; ↓, decreased expression upon activation; (↑); increased baseline expression compared to HD1 and 2 (≤ 250%); (↑↑), increased baseline expression compared to HD1 and 2 (< 250%); (↓), decreased baseline expression compared to HD1 and 2; ↔, no change in expression compared to non-activated sample;
expression remains increased compared to HDs (Supplementary Fig. 6)
AP, Alagille Syndrome patient; HD, healthy donor; NA, non-activated
Similar to T cells from Patient CD46-1, T cells from AS Patients AP1 and AP3 were also unable to induce TH1 responses in vivo and cause GvHD when injected into NOD-SCID-IL2RG−/− mice (Supplementary Fig. 6b-d). However, this failure to cause GvHD might be a result of poor engraftment because in contrast to T cells from CD46-1, which engrafted at a reasonable rate (Supplementary Fig. 5b), T cells from AS patients failed to engraft into the animals. Notch1 signalling on human CD4+ T cells has recently been shown to regulate cell adhesion, migration and chemotaxis in these cells via modulation of Rho family GTPases33. Thus, defects in the Notch system signalling may not only impact on TH1 cytokine production in AS patients but also on the homing capacity of their T cells.
In summary, AS patients with recurrent infections present with a comparable T effector cell phenotype as observed in CD46-deficient patients, characterized by defective TH1 induction, and deregulation of CD127 and CD132 expression but unaffected TH2 function.
DISCUSSION
Here, we identify Jagged1 as a novel physiological ligand for CD46 and demonstrate that a coordinated CD46-Jagged1 crosstalk is required for TH1 responses. Notch system activation is controlled by spatial and temporal restriction of receptor and ligand availability during cell-cell interactions9,29 and our study suggests that CD46 participates in this process. We propose a model in which CD46 sequesters Jagged1 on resting T cells, thereby limiting Jagged1 and Notch1 interactions and favouring Notch1 and DLL1 cis interactions, which inhibit T cell activation34. Thus, similar to DLL1, in the absence of antigen or danger signals, CD46 expression on T cells may function as ‘brake’. Upon TCR engagement, the CD46 ligand C3b4 is generated locally. Binding of C3b to CD46 initiates CD46-mediated signalling events, including T cell migration and cluster formation35, CD46 and DLL1 down-regulation and the maintenance of Notch1 and Jagged1 surface availability. This change in surface expression of CD46 and Notch proteins releases the ‘brake’ and allows for orchestrated Notch1 and DLL1 interactions in trans (generating IFN-γ9) as well as for Notch1 and Jagged1 binding in cis or trans (necessary for IL-10 induction 9). The role of IL-2 and potential functions of generated soluble CD46 and Notch family members remains to be integrated into this model. In support of this model is our observation that both CD46-deficient individuals and patients with JAG1 mutations suffering from recurrent infections cannot generate normal TH1 responses. Both patient groups also share additional key features in their T cell phenotype as additional evidence that an overlapping CD46 and Notch pathway is affected. Firstly, in line with previous observations that CD46-mediated signals are specifically needed for TH1 induction, TH2 responses and TNF can be induced19. Moreover, both patient groups show a trend towards exaggerated TH2 responses, possibly explaining why AS patients also suffer from increased prevalence of TH2-driven conditions including otitis media, asthma and eczema12,25. Further, and consistent with our model, CD4+ T cells from C3-deficient patients (these T cell can not produce the CD46 ligand C3b locally), are also unable to assume a TH1 phenotype, present with deregulated IL-2R expression but produce high amounts of TH2 cytokines (manuscript submitted).
The mutations in AP2 and AP3 are predicted to lead to nonsense-mediated decay of mutant JAG1 mRNA resulting in expression of wild type Jagged1 on the cell surface. Further, expression studies of additional AS-associated missense mutations in cell lines lead to retention of the mutated protein in the endoplasmic reticulum (unpublished data), suggesting that haploinsufficiency is the pathogenic mechanism operating in most cases. The AS patients here unexpectedly show rather increased Jagged1 expression on resting T cells and we have currently no explanation for this observation. Nonetheless, each AS and CD46-deficient patient presents with clear expression deviations either of Jagged1, Notch1 or CD46 or a combination thereof. However, the most striking phenotype in regards to cell surface receptors involved in TH1 biology is the marked deregulation of CD127 and CD132 (which together form the IL-7 receptor [IL-7R]) on T cells from both patient groups. Importantly, IL-7 is not only required for T cell homeostasis and enhancement of TH1 and TH17 responses36 but CD127 has also been identified as a strong non-major histocompatibility complex-linked risk locus for the T cell-driven disease condition multiple sclerosis (MS)37,38. Similarly, deregulation of CD46 isoform expression has previously been connected with progression of MS 39. It would thus be interesting to assess if T cells from patients with CD46 mutations have an altered response to IL-7. However, CD132 is also an essential component of the receptors for IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21 (IL-2 receptor family)27. Most members of this family are involved in normal T and B cell as well as natural killer (NK) cell function and CD132 deficiency is linked to X-linked severe combined immunodeficiency (XSCID)27. Deregulation of CD132 will therefore also affect the responsiveness of T cells towards other members of the IL-2 cytokine family beside IL-7. IL-2-mediated signalling through the high affinity IL-2 receptor (CD25, CD122 and CD132; IL-2R) is needed for cell activation induction of TH1 responses 27 and chemical inhibition of Notch1 signalling impedes normal IL-2R expression and TH1 induction40. IL-2 is also linked to CD46: CD46 negatively regulates IL-2 expression but also integrates IL-2R signals for IL-10 co-expression in TH1 cells4,5. It is therefore tempting to speculate that the T cells from CD46-deficient and AS patients may be unable to induce TH1 responses, at least in part because of aberrant IL-2R signalling.
The immunomodulatory function of CD46 is likely one reason why several human pathogens use CD46 as a receptor. CD46-interacting viruses target CCP1-2 of CD462,6,7, which contains the Jagged1 binding site. Structures of CCP1-2 in complex with viral proteins binding CD46 demonstrated substantial reorientation of these two CCPs with respect to each other, suggesting that their arrangement is highly ligand-specific. The interaction surfaces identified here for Jagged1 binding within CD46 suggest that a Jagged1-specific conformation of CCP1-2 is required for binding and implies that Jagged1-bound CD46 will not be able to bind the viral ligands simultaneously without displacement of Jagged1 from CD46. Hence, the observation that binding of adenovirus type 35 (rAd35) to CD46 on human CD4+ T cells induces down-regulation of CD46 but reduced IL-2 and IFN-γ production could be due to interference with the coordinated CD46-Notch system signalling events during T cells activation41,42.
Our observations may provide a platform to advance understanding of the complex signalling networks underlying TH1 biology as well as differences in the human and murine systems. Future analyses of the impact of virus binding to CD46 and its effect on the Jagged1 interaction may also provide novel insights into pathogen-mediated immune deviation. Further, identifying CD46 surface expression as a ‘stop signal’ offers an intriguing hypothesis for the counterintuitive down-regulation of CD46 on most activated cell types; it provides a ‘go signal’ when immune activation is apparent5,23. And lastly, as the Notch system also plays fundamental roles in tissue morphogenesis and renewal, we anticipate that the CD46-Jagged1 interaction may be important in these biological processes as well.
ONLINE METHODS
Healthy donors and patients
Purified T cells were obtained from buffy coats (National Blood Service) or blood samples from healthy volunteers. Informed consent was obtained from all subjects included in the study and blood was collected and processed with the approval and in accordance of the King’s College Ethics Committee guidelines (Reference No: 06/Q0705/20). Adult patients with CD46-deficiencies were recruited in France under appropriate institutional guidelines; two cases had previously been described10,11. Six Caucasian children aged between 2 and 12 years were recruited of whom 4 were diagnosed with Alagille Syndrome and two were healthy (Review Board of National Research Ethics Services (NRES) Committee London Reference No: 09/H0711/38). Patients with Alagille Syndrome all exhibited repeated infections and/or allergies and food intolerances. None of the patients were taking immunosuppressants or were post liver transplant. Blood samples were processed with in a maximum of 3 hours from the time of collection.
Mice and GvHD model
CD46-transgenic mice were generated by backcrossing an established CD46-transgenic line onto outbred MF1 mice14. Animals were handled and samples obtained and processed under UK Home Office license 70/6906. NOD-SCID-B2M−/− animals were used for the injection of T cells from the CD46-deficient Patient CD46-1 and obtained from Taconic Farms (Germantown, NY, USA). NOD-SCID-IL2−/− mice were used for the injection of T cells from patients with Alagille Syndrome (AP-1 and AP-3) and were purchased form Charles River and maintained under pathogen-specific sterile conditions. GvHD was induced as previously described28. Briefly, PBMCs from healthy donors, CD46-deficient or Alagille Syndrome patients were activated for 72h with immobilized mAbs to CD3 and CD28 prior to adoptive transfer via tail vein injection (1 × 107 T cells; 80-85% CD4+ T cells and 15-20% CD8+ T cells in all cases). Human cell engraftment was monitored by enumeration of huCD45+ cell (ratio of human vs. mouse CD45+ cells) and measurement of hIFN-γ in mouse blood at different time points. Body weight was monitored and mice culled when they reached the humane end point of 15% decrease in body weight. Disease was further confirmed by immunohistological analysis of intestinal tissue.
T cell isolation and activation
T cells were isolated and activated as previously described4. The HEK293T and Jurkat cell lines was purchased from the American Tissue Culture Centre (ATCC, Manassas, VA) and cultured according to the manufacturer’s protocol. Jurkat cells (including those stably transfected with either CD46-BC1 or CD46-BC2)4 were activated as described for purified CD4+ T cells but for 5d with IL-2 supplementation every 2d.
Recombinant proteins
Serum purified C3b was bought from Complement Technologies Inc. (Tyler, TX) and rCR1 was a produced as previously published43. Human rDelta-like1 (DLL1), human rJagged1 (rhJ-1), and murine rJagged1 (rmJ-1) were obtained from R&D Systems (Minneapolis, MN). The generation of c-terminally biotinylated protein fragments corresponding to EGF-like domains 11-13 of Notch1 (N-111-13) or the DSL and first three EGF-like domains of Jagged1 (J-1DSL-EGF3) were produced as described previously13. CD46 constructs (Adprotech Ltd., Saffron Walden, UK) were subcloned into the pET14b vector and transformed into B834 cells. Labeled protein was produced as described previously44 and refolding of protein was performed using the established protocol45. rCrry (CCP1-4) was generated as previously described46.
Antibodies and inhibitors
Cell-stimulating mAbs were bought from BD Biosciences, San Diego, CA (anti-human CD28, CD28.2; anti-mouse CD3, 145-2C11; anti-mouse CD28, 37.51), purified from a specific hybridoma line (anti-CD3; OKT-3) or generated in house (anti-CD46; TRA-2-1047). Notch1 expression/binding was assessed using mAbs 527425 (R&D systems, Minneapolis, MN) or A6 (Thermo Fisher Scientific, Fremont, CA). hJagged1 was detected with mAb 188331 (R&D systems) and DLL1 with mAb 251127 (R&D Systems). Biotinylated J-1DSL-EGF3 and N-111-13 were detected utilizing APC-labelled streptavidin (BD Biosciences, San Jose, CA). CD46 expression was assessed using anti-CD46 (clone E4.3, BD Biosciences). Human C3b and CR1 were detected with mAb ab17453 (Abcam, Cambridge, UK) and mAb E11 (BD Biosciences), respectively. The mAb to α-E-catenin (ab19446) was purchased from Abcam. Antibodies to CD122 (FAB224A) and CCR7 (FAB197F) were obtained from R&D Systems and antibodies to CD4 (555349), CD8 (555635), CD11a (555379), CD25 (555431), CD46RA (555488), CD45RO (559865), CD62L (559772), CD69 (555530), CD127 (557938), CD132 (555900) and to mouse CD45RB (16A) were all obtained from BD Biosciences. The broad-spectrum matrix metalloproteinase (MMP) inhibitor TAPI-2 was bought from Merck Chemicals Ltd, (Nottingham, UK), Marimastat from Tocris Bioscience (Bristol, UK) and the Presenilin/γ-secretase inhibitor L-685,458 was obtained fromSigma-Aldrich (Saint Louis, MO, USA).
ELISA
96-well microplates were coated with 5μg/ml of protein over night at 4 °C (e.g. sCD46, hC3b, J-1DSL-EGF3 or rCR1), blocked with 1% BSA and then incubated for 1.5 h at 37°C with different protein samples diluted to 0.5 μg/ml (and roughly equimolar amounts) in 4% BSA/0.005% Tween20/0.25% NP-40/20mM HEPES/10mM CaCl, pH7.4. Because rmC3b and rmC4b are commercially unavailable, 5% mouse serum was used as source for mouse C3b and C4b. Bound proteins were detected using the appropriate primary mAbs followed by horseradish-peroxidase (HRP)-linked secondary Abs or HRP-linked streptavidin and subsequent visualization with O-phenylenediamine Dihydrochloride (OPD) substrate (Sigma-Aldrich).
Surface Plasmon Resonance
All data were collected using a Biacore T100 (GE Healthcare, Buckinghamshire, UK) with Jagged1 or CD46 immobilized through primary amine-coupling to the Biacore™ CM5 Chip surface. CD46 or J-1DSL-EGF3 constructs were passed over the chip in 10mM HEPES (pH 7.4), 150mM NaCl, 3mM EDTA, 0.005% surfactant P20. Multiple titrations were performed over a concentration range of 0.1μM to 40μM, with flow rates from 25 μl/min to 40 μl/min at 25°C. Data were processed using the manufacturer’s BIAevaluation software and fit either using a Langmuir 1:1 equilibrium model or, where possible, a kinetic analysis, employing simultaneous fits of the kon and koff rates.
NMR Spectroscopy
A sample containing 65μM of a CD4612 construct uniformly enriched in 15N in 25mM sodium acetate (pH 5.5), 5% D2O was used to collect a sensitivity enhanced 1H,15N-heteronuclear single quantum coherence (HSQC)48 on a 500MHz Bruker Avance (Bruker UK Limited, Coventry, UK) equipped with a cryoprobe. Another 1H,15N-HSQC was collected with the addition of 38μM unlabelled Jagged1 BirA (previously described13). In addition the 1H,15N-HSQC was repeated with a sample of unlabelled Jagged1 without the BirA tag (data not shown). For the 1H,15N-HSQC acquisition times for t1 were 42 ms with 256 complex data points. Data were processed and analysed using NMRPipe and Sparky. Chemical shift perturbation values were calculated using the following equation:
Assignments were performed on [U-15N, 13C, 1H] CD4612 using standard triple resonance correlation experiments. HNCA/HN(CO)CA, HNCO/HN(CA)CO and HNCACB/HNCA(CO)CB experiments were performed as described previously49.
High resolution microscopy and co-localization analysis
T cells were treated as indicated and then stained with antibodies to CD46, Jagged1 or Notch1 for 25 minutes at 4°C. Cells were then mounted with Fluoromount-G (SouthernBiotech). Images were obtained by confocal fluorescence microscopy with a laser scanning microscope (Fluorview 1000, Olymbus, Hamburg, Germany) with a 60x NA1.35 oil objective. For 3-D image analysis Z-stacks were taken with an interval of 0.1 μm using the confocal microscope with 20 to 30 slices per stack in order to show cells in their full extension. Stacks were then used for image analysis by IMARIS software (version 7.4.2, Bitplane AG, Zürich, Switzerland), operating with IMARIS Surpass (volume and isosurface rendering analysis), to visualize and locate points of interest (Jagged1, CD46 and Notch1 expression). For co-localization studies, data sets were analyzed using the IMARIS software. Data of co-localization events were determined with the statistical modules in the co-localization software of the IMARIS package.
Cytokine measurements
Cytokines from cell cultures or mouse serum were measured using the human TH1/TH2 Cytometric Bead Arrays (BD Biosciences) or the human IFN-γ, and IL-10 Cytokine Secretion Assay Kits (Miltenyi Biotec, Bergisch-Gladbach, Germany) in combination as per manufacturer’s protocol.
Quantitative real-time RT-PCR
Primers used to quantify mRNA transcription in CD4+ T cells: NOTCH1 Forward (F) 5′ - CGC ACA AGG TGT CTT CCA G - 3′ and Reverse (R) 5′ - AGG ATC AGT GGC GTC GTG - 3′, NOTCH2 (F) 5′ - TTG AGA GTT ATA CTT GCT TGT GTG C - 3′ and (R) 5′ - GAT ACA CTC GTC AAT GTC AAT GG - 3′, JAG1 (F) 5′ - AGC CTT GTC GGC AAA TAG C - 3′ and (R) 5′ - AGC CTT GTC GGC AAA TAG C - 3′, JAG2 (F) 5′ - CGA CCA GTA CGG CAA CAA - 3′ and (R) 5′ - GGA GCA AAT TAC ACC CTT GTT TA - 3′, DLL1 (F) 5′ - GTG GGG AGA AAG TGT GCA A - 3′ and (R) 5′ - TCA CAA AAT CCA TGC TGC TC - 3′, HES1 (F) 5′ - GAA GCA CCT CCG GAA CCT - 3′ and (R) 5′ - GTC ACC TCG TTC ATG CAC TC - 3′, RBPJ (F) 5′ - GAA GTA CCA TGG CGT GGA TT- 3′ and (R) 5′ - TTT CGC ATA GCT TCC CTA GTA AGT- 3′.
RNA silencing
Cy3-labeled siRNA targeting human α-E-catenin (s3718) and negative control siRNA were purchased from Ambion, Austin, TX and siRNA experiments performed as previously described4. Transfection efficiency and cell viability was consistently above 80% and 75%, respectively, and protein knockdown peaked at 24-36h post-transfection. For shRNA-mediated silencing of DLL1 in Jurkat T cells, an appropriate Lentivirus was generated by cotransfecting HEK293T cells with packaging plasmid psPAX2 (Addgene, Cambridge, MA), envelope plasmid pMD2.G (Addgene), and pLKO.1 vector containing a shRNA targeting DLL1 (Abgene, Epsom, UK) using FuGENE® 6 Transfection Reagent (Roche Diagnostics Ltd, Burgess Hill, UK). After 48h, media was harvested, filtered, and added to Jurkat cell cultures. Virus-infected cells were selected by puromycin. DLL1 protein knockdown was consistently above 50%.
Statistical analysis
Statistical analyses were performed using the Student’s one-tailed t-test and Bonferroni correction for multiple comparisons (Excel software [Microsoft, Redmond, WA]).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the CD46-deficient and Alagille Syndrome patients for their support and thank Adrian Hayday for data discussion. This work is supported by an MRC Research Grant (G1002165 to CK), EU FP7 IMI BTCURE (CK as lead researcher), the MRC Centre for Transplantation, Guy’s Hospital, King’s College and the Department of Health, NIHR comprehensive Biomedical Research Centre award to Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust, a Wellcome Trust Project Grant (097928/A/08/Z to SML and PAH) and the German Research Foundation (GRK1727 TP8 and SFB/TR22 A21 to JK). SNW is funded by the European Research Council (“SomaBio”) and holds a honorary position at Witswatersrand University, South Africa.
Footnotes
Address reagent requests to: PAH (penny.handford@bioch.ox.ac.uk) and SML (susan.lea@path.ox.ac.uk)
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interest
AUTHOR CONTRIBUTIONS
G.LF. designed and performed experiments and wrote the manuscript; D.S. performed SPR and NMR Spectroscopy experiments; P.W. and C.C. generated recombinant Notch/Jagged1 proteins; C.M.K and J.K. performed the super resolution microscope studies and edited the manuscript. S.S., A.B., C.D., L.C. and V.F-B. provided blood samples from the patients and discussed the data; A.L. performed the GvHD experiments and discussed data; L.B. and M.J.D. designed the RT-PCR experiments and discussed the data; T.M. and R.A.S. generated sCD46 and sCR1 and discussed data; S.N.W. provided hCD46-transgenic mice and edited the paper; J.M.M. performed SPR experiments and edited the paper; P.A.H. provided recombinant Notch/Jagged proteins, designed experiments and edited the paper; S.M.L. designed the SPR and NMR experiments, provided recombinant CD46 proteins and edited the manuscript; C.K., perceived and designed the study, performed experiments and edited the manuscript.
REFERENCES
- 1.Liszewski MK, Post TW, Atkinson JP. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu Rev Immunol. 1991;9:431–455. doi: 10.1146/annurev.iy.09.040191.002243. [DOI] [PubMed] [Google Scholar]
- 2.Cattaneo R. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens’ magnet. J Virol. 2004;78:4385–4388. doi: 10.1128/JVI.78.9.4385-4388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astier A, Trescol-Biemont MC, Azocar O, Lamouille B, Rabourdin-Combe C. Cutting edge: CD46, a new costimulatory molecule for T cells, that induces p120CBL and LAT phosphorylation. J Immunol. 2000;164:6091–6095. doi: 10.4049/jimmunol.164.12.6091. [DOI] [PubMed] [Google Scholar]
- 4.Cardone J, et al. Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nat Immunol. 2010;11:862–871. doi: 10.1038/ni.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cope A, Le Friec G, Cardone J, Kemper C. The Th1 life cycle: molecular control of IFN-gamma to IL-10 switching. Trends Immunol. 2011;32:278–286. doi: 10.1016/j.it.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Casasnovas JM, Larvie M, Stehle T. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 1999;18:2911–2922. doi: 10.1093/emboj/18.11.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnberg N. Adenovirus receptors: implications for tropism, treatment and targeting. Rev Med Virol. 2009;19:165–178. doi: 10.1002/rmv.612. [DOI] [PubMed] [Google Scholar]
- 8.Riley RC, Tannenbaum PL, Abbott DH, Atkinson JP. Cutting edge: inhibiting measles virus infection but promoting reproduction: an explanation for splicing and tissue-specific expression of CD46. J Immunol. 2002;169:5405–5409. doi: 10.4049/jimmunol.169.10.5405. [DOI] [PubMed] [Google Scholar]
- 9.Amsen D, Antov A, Flavell RA. The different faces of Notch in T-helper-cell differentiation. Nat Rev Immunol. 2009;9:116–124. doi: 10.1038/nri2488. [DOI] [PubMed] [Google Scholar]
- 10.Fremeaux-Bacchi V, et al. Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2006;17:2017–2025. doi: 10.1681/ASN.2005101051. [DOI] [PubMed] [Google Scholar]
- 11.Couzi L, et al. Inherited deficiency of membrane cofactor protein expression and varying manifestations of recurrent atypical hemolytic uremic syndrome in a sibling pair. Am J Kidney Dis. 2008;52:e5–9. doi: 10.1053/j.ajkd.2008.02.359. [DOI] [PubMed] [Google Scholar]
- 12.Vajro P, Ferrante L, Paolella G. Alagille syndrome: An overview. Clin Res Hepatol Gastroenterol. 2012;36:275–277. doi: 10.1016/j.clinre.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Cordle J, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greig JA, et al. Influence of coagulation factor x on in vitro and in vivo gene delivery by adenovirus (Ad) 5, Ad35, and chimeric Ad5/Ad35 vectors. Mol Ther. 2009;17:1683–1691. doi: 10.1038/mt.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujimura A, et al. Molecular cloning of a murine homologue of membrane cofactor protein (CD46): preferential expression in testicular germ cells. Biochem J. 1998;330(Pt 1):163–168. doi: 10.1042/bj3300163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Centeno E, de Ojeda G, Rojo JM, Portoles P. Crry/p65, a membrane complement regulatory protein, has costimulatory properties on mouse T cells. J Immunol. 2000;164:4533–4542. doi: 10.4049/jimmunol.164.9.4533. [DOI] [PubMed] [Google Scholar]
- 17.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 18.Kemper C, Atkinson JP. T-cell regulation: with complements from innate immunity. Nat Rev Immunol. 2007;7:9–18. doi: 10.1038/nri1994. [DOI] [PubMed] [Google Scholar]
- 19.Kemper C, et al. Activation of human CD4+ cells with CD3 and CD46 induces a T-regulatory cell 1 phenotype. Nature. 2003;421:388–392. doi: 10.1038/nature01315. [DOI] [PubMed] [Google Scholar]
- 20.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutz S, et al. Notch regulates IL-10 production by T helper 1 cells. Proc Natl Acad Sci U S A. 2008;105:3497–3502. doi: 10.1073/pnas.0712102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni Choileain S, et al. The dynamic processing of CD46 intracellular domains provides a molecular rheostat for T cell activation. PLoS One. 2011;6:e16287. doi: 10.1371/journal.pone.0016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardone J, Al-Shouli S, Kemper C. A novel role for CD46 in wound repair in Front Immun. 2011;2:28. doi: 10.3389/fimmu.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang CJ, et al. Membrane cofactor protein mutations in atypical hemolytic uremic syndrome (aHUS), fatal Stx-HUS, C3 glomerulonephritis, and the HELLP syndrome. Blood. 2008;111:624–632. doi: 10.1182/blood-2007-04-084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romagnani S. Th1/Th2 cells. Inflamm Bowel Dis. 1999;5:285–294. doi: 10.1097/00054725-199911000-00009. [DOI] [PubMed] [Google Scholar]
- 26.Fuchs A, Atkinson JP, Fremeaux-Bacchi V, Kemper C. CD46-induced human Treg enhance B-cell responses. Eur J Immunol. 2009;39:3097–3109. doi: 10.1002/eji.200939392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: new insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr Opin Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nervi B, et al. Factors affecting human T cell engraftment, trafficking, and associated xenogeneic graft-vs-host disease in NOD/SCID beta2mnull mice. Exp Hematol. 2007;35:1823–1838. doi: 10.1016/j.exphem.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- 30.Oda T, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 31.Quiros-Tejeira RE, et al. Variable morbidity in alagille syndrome: a review of 43 cases. J Pediatr Gastroenterol Nutr. 1999;29:431–437. doi: 10.1097/00005176-199910000-00011. [DOI] [PubMed] [Google Scholar]
- 32.Robinson M, et al. An analysis of the normal ranges of lymphocyte subpopulations in children aged 5-13 years. Eur J Pediatr. 1996;155:535–539. doi: 10.1007/BF01957900. [DOI] [PubMed] [Google Scholar]
- 33.Bhavsar PJ, Infante E, Khwaja A, Ridley AJ. Analysis of Rho GTPase expression in T-ALL identifies RhoU as a target for Notch involved in T-ALL cell migration. Oncogene. 2012 doi: 10.1038/onc.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.del Alamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21:R40–47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 35.Alford SK, Longmore GD, Stenson WF, Kemper C. CD46-induced immunomodulatory CD4+ T cells express the adhesion molecule and chemokine receptor pattern of intestinal T cells. J Immunol. 2008;181:2544–2555. doi: 10.4049/jimmunol.181.4.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bikker A, Hack CE, Lafeber FP, van Roon JA. Interleukin-7: a key mediator in T cell-driven autoimmunity, inflammation, and tissue destruction. Curr Pharm Des. 2012;18:2347–2356. doi: 10.2174/138161212800165979. [DOI] [PubMed] [Google Scholar]
- 37.Gregory SG, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 38.Hafler DA, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 39.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J Clin Invest. 2006;116:3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adler SH, et al. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol. 2003;171:2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- 41.Adams WC, et al. Attenuation of CD4+ T-cell function by human adenovirus type 35 is mediated by the knob protein. J Gen Virol. 2012;93:1339–1344. doi: 10.1099/vir.0.039222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams WC, et al. Adenovirus type-35 vectors block human CD4+ T-cell activation via CD46 ligation. Proc Natl Acad Sci U S A. 2011;108:7499–7504. doi: 10.1073/pnas.1017146108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibb AL, Freeman AM, Smith RA, Edmonds S, Sim E. The interaction of soluble human complement receptor type 1 (sCR1, BRL55730) with human complement component C4. Biochim Biophys Acta. 1993;1180:313–320. doi: 10.1016/0925-4439(93)90056-7. [DOI] [PubMed] [Google Scholar]
- 44.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 45.White J, et al. Biological activity, membrane-targeting modification, and crystallization of soluble human decay accelerating factor expressed in E. coli. Protein Sci. 2004;13:2406–2415. doi: 10.1110/ps.03455604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roversi P, et al. Structures of the rat complement regulator CrrY. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:739–743. doi: 10.1107/S1744309111016551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G, Liszewski MK, Chan AC, Atkinson JP. Membrane cofactor protein (MCP; CD46): isoform-specific tyrosine phosphorylation. J Immunol. 2000;164:1839–1846. doi: 10.4049/jimmunol.164.4.1839. [DOI] [PubMed] [Google Scholar]
- 48.Schleucher J, et al. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J Biomol NMR. 1994;4:301–306. doi: 10.1007/BF00175254. [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A Suite of Triple-Resonance Nmr Experiments for the Backbone Assignment of N-15, C-13, H-2 Labeled Proteins with High-Sensitivity. Journal of the American Chemical Society. 1994;116:11655–11666. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.