Abstract
Pheochromocytomas are chromaffin cell-derived neuroendocrine tumors. There is presently no cure for metastatic pheochromocytoma and no reliable way to distinguish malignant from benign tumors before the development of metastases. In order to successfully manage pheochromocytoma, it is necessary to better understand the biological determinants of tumor behavior. For this purpose, we have recently established a mouse model of metastatic pheochromocytoma using tail vein injection of mouse pheochromocytoma (MPC) cells. We optimized this model modifying the number of cells injected, length of trypsin pre-treatment, and incubation temperature and duration for the MPC cells before injection, and by serial passage and re-selection of tumors exhibiting the metastatic phenotype. We evaluated the effect of these modifications on tumor growth using serial in vivo Magnetic Resonance Imaging studies. These results show that number of cells injected, the pre-injection incubation temperature, and duration of trypsin treatment are important factors to produce faster growing, more aggressive tumors that yielded secondary metastatic lesions. Serial harvest, culture and re-selection of metastatic liver lesions produced even more aggressive pheochromocytoma cells that retained their biochemical phenotype. Microarray gene expression comparison and quantitative real-time PCR of these more aggressive cells to the MPC-parental cell line identified genes that may be important for the metastatic process.
Keywords: Animal model, Cell line, Pheochromocytoma, Magnetic resonance imaging, Microarray, Quantitative real-time PCR
Introduction
Pheochromocytomas are chromaffin cell-derived neuroendocrine tumors arising from the adrenal medulla. It is estimated that up to 14% of pheochromocytomas and a higher percentage of extra-adrenal paragangliomas are malignant [1]. The prognosis of benign and malignant pheochromocytoma differs vastly. Benign tumors are generally treated with surgery, and have a 10 year survival rate of up to 94%. Conversely, no curative treatment exists for metastatic pheochromocytoma, and the 10 year survival rate is less than 20% [2, 3]. Unfortunately, no reliable histological or biochemical markers presently exist that are able to differentiate benign from malignant pheochromocytomas. Thus, the malignant potential of these tumors can only be determined when they spread to sites distant from the primary tumor where chromaffin cells are not present [2, 3].
The availability of preclinical models is crucial to improve our understanding of the biology and pathophysiology of pheochromocytoma, and perhaps more important, to identify and test novel therapies. In spite of few initially promising reports, no human pheochromocytoma cell lines have been definitively established. Preclinical research on this tumor centers on a limited number of rodent cell lines and in vivo models. A mouse model of metastatic pheochromocytoma has recently established using intravenously injected mouse pheochromocytoma (MPC) cells that is characterized by the development of solid organ metastatic lesions, predominantly in the liver by 4 weeks after injection [4, 5]. The MPC cell line derived from an neurofibromatosis1 (Nf1) knockout mouse and resembles human chromaffin cells as they express the enzyme phenylethanolamine- N-methyltransferase (PNMT) that converts norepinephrine to epinephrine as opposed to the rat adrenal pheochromocytoma cell line, PC12 [6].
In the present study, we modified the current metastatic mouse model in an effort to establish a more aggressive metastatic tumor phenotype. Selecting established MPC liver tumors, we generated a highly aggressive cell line designated MTT (mouse tumor tissue-derived). Intravenous injection of MTT cells into athymic nude mice rapidly gave rise to widely disseminated metastases. Using serial magnetic resonance imaging (MRI), we were able to visualize early tumor development in multiple organs and monitor tumor growth over several weeks. Comparative microarray transcriptional profiling of RNA from MTT and MPC cells was performed to identify genomic signatures linked to different metastatic behavior between the two cell lines.
Materials and methods
Cell culture
Mouse pheochromocytoma cells (MPC 4/30PRR) [6] were maintained in RPMI 1640 containing 10% heat-inactivated horse serum (Hyclone Logan, UT), 5% fetal bovine serum (GIBCO, Grand Island, NY), and penicillin (10,000 units/ ml)/streptomycin (10,000 Ug/ml) (GIBCO, Grand Island, NY). MPC cells were grown in tissue culture dishes without collagen. Prior to injection into mice, MPC were incubated in 0.05% trypsin (GIBCO) and gently rocked at 37°C until they detached from the flask. Cells were then transferred to a 15 ml conical tube containing fresh medium, washed and re-suspended in 100 µl of PBS before injection. Rat pheochromocytoma cells, PC12, were cultured as previously described [6, 7].
The conditions under which MPC were prepared for injection into mice were studied for the effect on metastases development. Cell dissociation was carried out according to the following alternative protocols: (1) Cells were trypsinized for 10 min, centrifuged at 4°C and held on ice for up to 60 min prior to tail vein injection (the original protocol used to develop the MPC mouse model); (2) cells were trypsinized for 1 min, pelleted at 4°C and held on ice for up to 60 min; (3) cells were trypsinized for 10 min, centrifuged at 4°C and held at room temperature (24°C) for up to 60 min; or (4) cells were trypsinized for 1 min, centrifuged at 4°C and held in room temperature for up to 60 min. The effect of the different dissociation conditions on cell proliferation was examined by the XTT assay (Cell Proliferation Kit II, Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, IN). Briefly, after dissociation, MPC cells (passage 25 or 26) were seeded in 96-well plates (15,000 cells/well) and incubated at 37°C for approximately 40 h. The XTT labeling mixture was added and the plates were incubated for an additional 24 h, after which spectrophotometric absorbance was measured with a microplate reader (Bio-Rad Laboratories, Philadelphia, PA) according to the manufacturer’s instructions. All experiments were performed in quadruplicate and repeated at least twice.
Animal models
All animal studies were conducted in accordance with the principles and procedures outlined in the National Institute of Health Guide for the Care and Use of Animals, and approved by the Institutional Animal Care and Use Committee. Female athymic nude mice (NCr-nu) were obtained from Taconic Inc. (Germantown, MD) and were housed in a pathogen-free facility. The mice were acclimated for at least 3 days in the animal facility in which the appropriate temperature, humidity and light cycle (6 a.m.–6 p.m.) were controlled, with ad libitum access to food and water.
Each experimental group consisted of 6 to 10 week old mice (n = 5). Tumor cell dissociation prior to injection was carried out according to the protocol outlined above (Table 1). Non-anesthetized mice were intravenously injected in the tail vein using a 1 ml syringe and 30 1/2 gauge needles with 10 × 106 MPC cells in 100 µl of PBS (GIBCO). Animals with the greatest number of metastases in the liver, lungs, ovaries, and adrenal glands seen on microCT [8] at day 20 were selected for harvesting of tumors to generate metastatic cell lines. After the animals were euthanized using CO2 inhalation and cervical dislocation, hepatic metastases were dissected from the liver and dissociated with collagenase (Roche Diagnostics, IN) into a single cell suspension. Dissociated cells were plated onto 75 cm2 culture flasks (1 × 107 cells/flask) in the MPC medium. Medium was replaced every 24–48 h, with the MTT cells usually adhering to the flask within 24 h after plating. For tumorigenicity studies, groups (n = 5) underwent tail vein injection with 5 × 1052 × 106or 10 × 106 MTT cells or an equal number of the parental MPC cells (n = 5) at 5 × 105 or 10 × 106 cells/injection. Mice were monitored daily and maintained until predetermined time points and examined for the number and extent of metastases.
Table 1.
Comparison of an animal models under different treatment conditions
| Cell type and number injected |
Cell temperature before injection (°C) |
Trypsin treatment |
Observations |
||
|---|---|---|---|---|---|
| First appearance of lesions (weeks after injection) |
Tumor locations and survival (five mice per group) | ||||
| 1 | MPC 10 × 106 | 4 | Long (10 min) | 4–5 | 4–20 in liver (5/5) median survival 68, range 52–84 days |
| 2 | MPC 10 × 106 | 25 | Short (1–2 min) | 3 | >100 in liver (5/5), 1/5 adrenal and 1/5 ovary lesion *liver tumors used to create MTT variant cell line median survival 25, range 22–28 days |
| 3 | MTT 10 × 106 | 25 | Short (1–2 min) | 2 | Tumor embolism (2/5) mice die within days; multiple (>100) liver lesions and pancreas destroyed (3/5),1/5 ovary lesions, 1/5 kidney lesion, median survival 16, range 13–19 days |
| 4 | MTT 2 × 106 | 25 | Short (1–2 min) | 2 | Liver lesions (5/5) on 2 weeks Lung (2/5), ovaries (2/5), and adrenal (3/5) lesions on 3 weeks median survival 28, range 26–30 days |
| 5 | MTT 5 × 105 | 25 | Short (1–2 min) | 2.5 | >40 liver lesions on 3 weeks (5/5), at 4 weeks 3/5 mice with adrenal lesions, 2/5 with lung lesions, 2/5 with ovary lesions, 3/5 mice with bone lesions, median survival 29, range 27–31 days |
| 6 | MPC 5 × 105 | 25 | Short (1–2 min) | 4.5 | 20 in liver on 4.5 weeks (5/5), at 5.5 weeks 1/5 to ovary and lungs, median survival 43, range 41–45 days |
Starting with the same concentration of MPC injected intravenously and conditions (1) We found storing cells at room temperature prior to injection and reducing the trypsin time yielded more liver tumors which also developed sooner (2) Liver tumors were cultured which yielded MTT cells. The MTT were injected at the same conditions but was lethal in most mice (3) By reducing the number of MTT cells injected, we found optimal conditions for aggressive tumor presentation: tumors appearing after 2 weeks and lesions in multiple organs (4) We also did a comparison of a reduced number of MTT (5) and MPC (6) cells and found tumors developed earlier than when more MPC cells are injected
Histological, biochemical and molecular characterization
Selected tumors and organs were dissected free, formalin-fixed, and paraffin-embedded and sectioned (10 µm). Hematoxylin and eosin (H&E) staining was performed. Remaining tumors were frozen on dry ice and stored at −80°C for subsequent catecholamine level measurements.
Plated MPC and MTT cells were washed three times with PBS, and fixed in 4% glutaraldehyde for 1 h. Cells were then scraped off the flask and centrifuged at 2,500 rpm for 10 min. Afterwards, cells were double-fixed in PBS-buffered glutaraldehyde (2.5%) and osmium tetroxide (0.5%), dehydrated and embedded into Spurr’s epoxy resin. Ultrathin sections (90 nm) were made, double-stained with uranyl acetate and lead citrate, and examined with a Philips CM10 transmission electron microscope (Electronic Instruments, Mahwah, NJ) [9, 10].
Intracellular catecholamine levels were measured in both MPC and MTT cells using HPLC after batch alumina extraction as previously described [11], and expressed as pg/1,000 cells. Total RNA from MPC, MTT, and PC12 cells and in vivo tumors was extracted using TRIzol (Invitrogen, Carlsbad, CA) followed by RNeasy RNA purification (Qiagen, Valencia, CA) and reverse transcribed to cDNA using random hexamers. Quantitative real-time PCR (qRT-PCR) with primers and TaqMan probes provided as pre-made sets through Applied Biosystems (Foster City, CA) for tyrosine hydroxylase (TH), PNMT and normalized to 18S ribosomal RNA using 7500 Sequence Detector (Applied Biosystems, Foster City, CA), as previously described [11].
Microarray gene expression profiles
Oligo microarray chips were generated from either a 31,769 70-mer probe set obtained from the Qiagen Mouse Genome Oligo Set Version 3.0 (Valencia, CA) or a 35,000 mouse oligo set from Illumina HEEBO Set (San Diego, CA). Microarray generation and processing was as described [12, 13]. Briefly, 1 µg of total RNA was reverse-transcribed with an oligo dT-T7 system and amplified using the Ambion MessageAmp™ II Kit (Ambion, Austin, TX). The generated aminoallyl-UTP-labeled RNA was then coupled with either Cy3 or Cy5 mono NHS ester CyDyes obtained from GE Healthcare (Piscataway, NJ). After purification, the labeled antisense RNA was hybridized overnight to the oligo chips in 5× SSC, 25% formamide and 0.2% SDS buffer at 45°C in a Maui Mixer FL hybridization chambers (BioMicro Systems, Salt Lake City, UT). Hybridized slides were washed at room temperature in a series of SSC/SDS buffers and dried by centrifugation. Hybridizations were done in triplicate, including one dye swap.
The newly generated MTT and MPC microarray data were mined against a previously generated human pheochromocytoma microarray database [12]. Genes were selected based on a two fold differential expression (up or down) between the MPC and MTT, and then compared to genes whose expression was statistically different (P < 0.05) in benign versus malignant human pheochromocytomas. The expression profiles were generated on over 69,000 gene transcripts (Qiagen and Illumina sets). Only genes that are up-or-down by two fold in both platforms were accounted for.
Quantitative real-time PCR validation was performed on seven genes that were associated with the same Ingenuity pathway (Ingenuity Systems, Inc., Redwood City, CA) generated from the 47 genes list that were differentially expressed between MTT versus MPC and also significantly differentially expressed between benign versus malignant human pheochromocytomas. All qRT-PCR were performed using an Applied Biosystems 7500 Real Time PCR System (Applied Biosystems, Foster City, CA). For the genes, matrix metallopeptidase 14 (MMP14), v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS1), Fyn-related kinase (FRK), GATA binding protein 2 (GATA2), keratin 8 (KRT8), matrix metallopeptidase 2 (MMP2), and neurotensin (NTS1) using QuantiTect Primer Assay (Qiagen, Valencia, CA) with QuantiFast SYBER Green PCR (Qiagen) using the manufacture’s parameters. Mouse genes on three MPC passages and four MTT passages were normalized to GAPDH. Human genes on seventeen tumors (4 non-adrenal metastatic and 13 adrenal non-metastatic) were normalized to beta-actin. Data were analyzed using a Two-tailed unpaired T-test with P value < 0.05 considered significant.
Magnetic resonance imaging
Respiratory-triggered MRI was performed in mice anesthetized with inhaled isoflurane/O2 at a dose of 1.5–5% v/v adjusted to produce a respiratory rate of approximately 40 breaths/minute (bpm). Mice were placed in the prone position and kept warm during the scanning as well as during post-scan recovery from anesthesia. Signals produced during respiration and transduced by the respiratory pillow were converted and processed through a computer software package (SA Instruments, Inc., Stony Brook, NY). Respiratory-triggered T2-weighted MRI spin echo images were acquired on a clinical Philips Intera 3.0 T system, using a dedicated 40 mm inner diameter solenoid coil (Philips, Best, Netherlands). A total of 40 slices were acquired with TE/TR 65/4,500 ms, slice thickness 0.5 mm, 0.16 × 0.16 mm2 in-plane resolutions and a scan time of 3–7 min for two signal averages. Tumors were imaged in both MPC (n = 5) and MTT (n = 5) injected (0.5 × 106 cells) mice using this T2-weighted protocol [8] once or twice a week for 3 weeks to monitor liver tumor growth.
Magnetic resonance imaging data analysis
Magnetic resonance imaging data were acquired and reconstructed using the scanner software (Intera, Philips Medical System, Best, Netherlands). Quantitative measurements of volume changes were obtained using the ImageJ and MIPAV [14] software. Because the size of tumors varied, we divided liver lesions into small (volume < 0.8 mm3) and large (volume ≥0.8 mm3) tumors based on their rate of differential growth rate [8]. Results on the volumes of liver lesions are presented as mean ± standard error of the mean (SEM). The statistical significance of the effect of the different dissociation conditions tested on MPC cell proliferation potential was assessed by un-paired T-test, with P value < 0.01 set as significant.
Results
Generation of a highly metastatic mouse model of pheochromocytoma
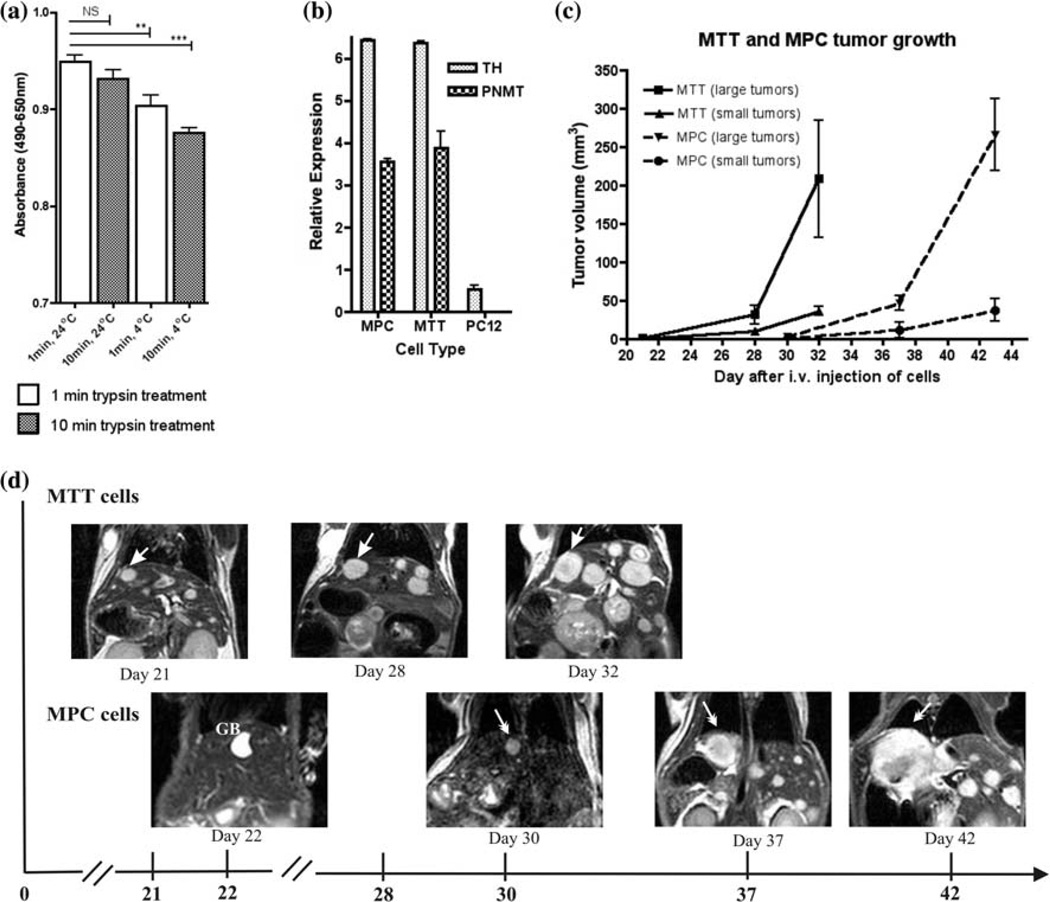
In order to optimize our previously established model of malignant pheochromocytoma to produce a more aggressive metastatic phenotype, we studied a number of modifications to our experimental conditions (Table 1). First, we studied the effect of the length of trypsin exposure (1–2 vs. 10 min.) used during cell dissociation, as well as of the temperature at which cells were incubated after dissociation (4 vs. 24°C) prior to animal injection on cell proliferation in vitro (Fig. 1a). Proliferation rates were significantly higher in the MPC cells exposed to trypsin for 1 min and incubated at room temperature (P < 0.0001). Consistently, injection of MPC cells exposed to trypsin for the shorter period and incubated at room temperature after dissociation resulted in greater number of hepatic metastases (>100 vs. 4–20 lesions), earlier appearance of metastatic lesions (3 vs. 4–5 weeks), and overall decreased median survival (median 68, range 52–84 days vs. median 25, range 22–28 days) (Table 1).
Fig. 1.
In vitro and in vivo characterization of MPC and MTT cells. a Cell proliferation assay showing the effect of different condition for MPC cells. Trypsin did not show a significant effect if the cells were treated at room temperature, but was significant at 4°C. An un-paired t-test analysis of these data with P value less than 0.01 as significant (*P < 0.01; **P < 0.001; ***P < 0.0001) was performed. b Expression of TH and PNMT mRNA in MPC, MTT, and PC12 cells determined by qRT-PCR relative to levels of 18S RNA. In vivo characterization of metastatic pheochromocytoma with MRI; c Small and large tumor growth in mice injected with MPC (n = 5) or MTT (n = 5). Mice injected with MTT developed tumors approximately 10 days earlier than mice injected with MPC. d Serial MRI images in same mouse scanned on days 21, 28, and 32 for MTT and on days 22, 30, 37 and 42 for MPC cells. Arrows indicate the same liver tumor
Next, we disaggregated and cultured tumor cells from liver metastases resected from mice, inoculated with the MPC cells pretreated under the optimal conditions indicated above in order to generate the highly metastatic MTT cells. We then generated the MTT mouse model by injecting 10 × 106 MTT cells (dissociated under the same low trypsin exposure/room temperature incubation conditions) back into mice. In the MTT model, two of five mice died within days, possibly from tumor embolism. The other three mice displayed large numbers of liver lesions at necropsy, with almost all of the liver parenchyma was replaced by tumors at 16 ± 3 days after the i.v. injection of MTT cells. The lungs and other organs were filled with fluid. In addition, two out five mice developed hind limb paralysis 2 weeks after injection suggesting the development of spine lesions.
Under these conditions the extremely aggressive nature of this model would not lend itself for studying the disease pathophysiology, as well as for testing novel therapeutics due to the rapid progression and short life span of the mice. Therefore, we reduced the number of cells injected to 2 × 106 and found that the MTT model metastatic tumors formed sooner than in the optimized MPC model (2 vs. 3 weeks), with enhanced metastases to other organs including lungs, adrenals and ovaries.
We also compared injection of lower numbers MPC and MTT cells (0.5 × 106) and confirmed the latter cells are indeed more aggressive, producing metastases sooner in mice at these lower inoculums. Figure 1c represents a graph of MRI measurements of liver tumors in five serially imaged animals over 3 weeks. Figure 1d demonstrates comparison MRI images of the same mice from either the MPC or MTT injected groups.
In vitro characterization of MPC versus MTT cells
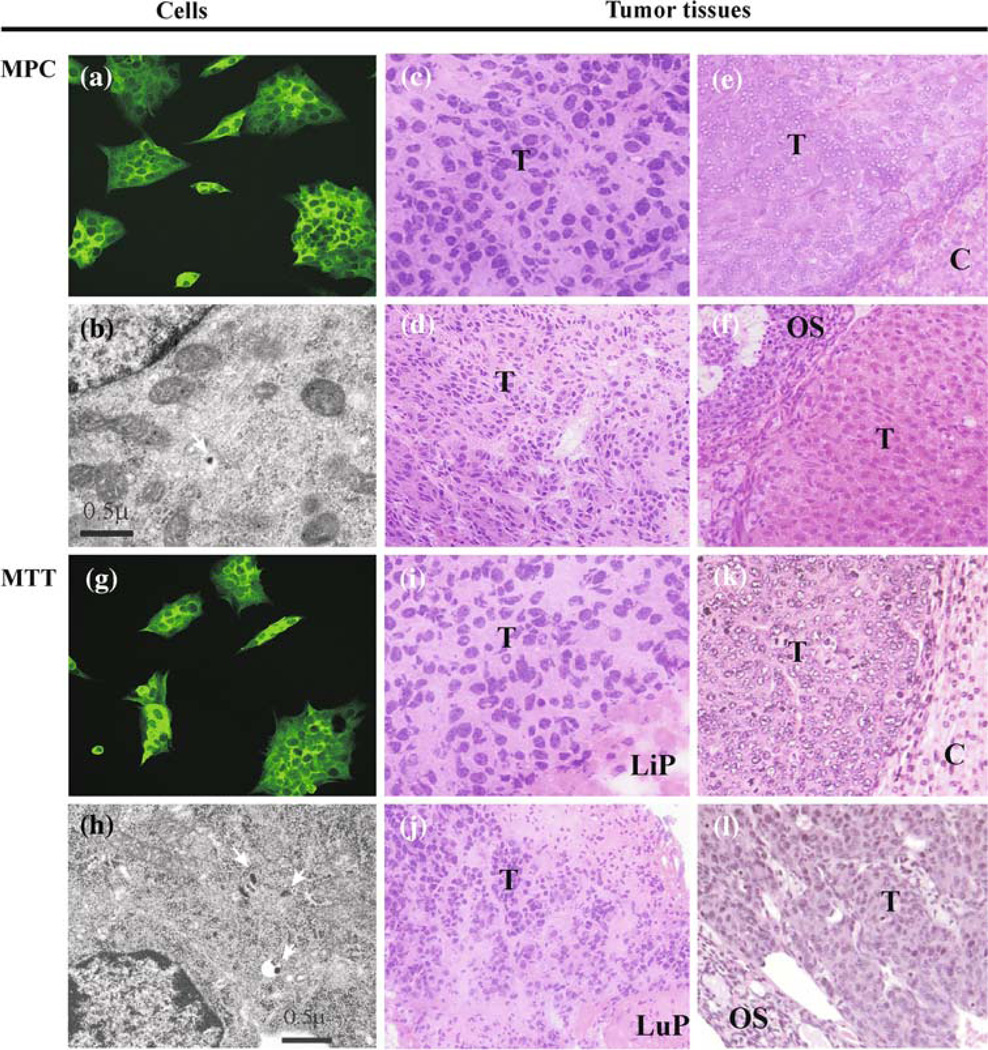
We confirmed that the MTT cells derived from MPC maintain a pheochromocytoma phenotype by measuring intracellular catecholamines, the expression of TH by immunocytochemistry, TH and PNMT gene expression by qRT-PCR (Fig. 1b) and morphologic presence of dense-core secretory granules by electron microscopy (Fig. 2). The presence of the secretory granules on ultrastructure is a diagnostic feature of the pheochromocytomas.
Fig. 2.
Characterization of MPC and MTT tumor histopathology. Immunocytochemistry reveals expression of TH in MPC (a) and MTT (g). Ultrastructure by electron microscopy reveals secretory dense cores granules (arrows) typically seen in pheochromocytomas in both cell lines (b, h); scale bar = 0.5 µm. Multiple organ lesions from MPC and MTT models: liver (ci); adrenal gland (ek); lung (dj); ovary (fl) reveal similar histopathology (H&E) suggestive of neuroendocrine tumor origin, magnification 40×. Elevated tissue catecholamine levels were demonstrated in these lesions confirming pheochromocytoma. Abbreviations T tumor, Li liver parenchyma, A adrenal cortex, Lu lung parenchyma, O ovarian Stroma
Expression profiling of over 69,000 gene transcripts identified a total of 565 (109 up regulated and 456 down regulated) transcripts differentially expressed by at least two folds between MTT and MPC, these 565 transcripts encodes for 338 genes. Of these 338 genes, 47 genes (9 up and 38 down) were also statistically differentially expressed between benign and malignant human pheochromocytomas. Several genes from the original mouse MTT versus MPC list were not accounted for because of homology gap between human and mouse genes, or because of lack of functional annotation (Table 2).
Table 2.
Genes identified to be differentially expressed between more aggressive MTT and parental MPC
| Gene name | Gene symbol | Entrez gene ID for human |
Entrez gene ID for mouse |
Fold MTT/MPC |
Clinical data |
|||
|---|---|---|---|---|---|---|---|---|
| Qiagen | Illumina | P value (malignant vs. benign) |
Fold change (malignant vs. benign) |
|||||
| A UP IN MTT | ||||||||
| 1 | Advillin | AVIL | 10,677 | 11,567 | 2.5,085 | 2.1467 | 0.007016 | 1.1728 |
| 2 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 | BACH1 | 571 | 12,013 | 2.1617 | 2.13845 | 0.004032 | 1.1810 |
| 3 | Branched chain keto acid dehydrogenase E1, alpha polypeptide | BCKDHA | 593 | 12,039 | 2.0296 | 2.0608 | 0.033830 | 1.0943 |
| 4 | Glutamate receptor, ionotropic delta 2 | GRID2 | 2,895 | 14,804 | 2.8176 | 2.8997 | 0.010286 | 1.1408 |
| 5 | Jub, ajuba homolog (Xenopus laevis) | JUB | 84,962 | 16,475 | 3.2328 | 4.0456 | 0.000306 | 1.1975 |
| 6 | MAM domain containing 2 | MAMDC2 | 256,691 | 71,738 | 2.07725 | 2.32625 | 0.003226 | 1.2924 |
| *7 | Matrix metallopeptidase 14 (membrane-inserted) | MMP14 | 4,323 | 17,387 | 2.1091 | 2.22115 | 0.008550 | 1.1975 |
| 8 | Transforming, acidic coiled-coil containing protein 1 | TACC1 | 6,867 | 320,165 | 2.0344 | 3.0971 | 0.047422 | 1.1251 |
| 9 | T-cell lymphoma invasion and metastasis 2 | TIAM2 | 26,230 | 24,001 | 3.66385 | 2.95735 | 0.019213 | 1.1329 |
| B DOWN IN MTT | ||||||||
| 1 | Adenylate cyclase 7 | ADCY7 | 113 | 11,513 | 0.1082 | 0.4664 | 0.014868 | 0.8888 |
| 2 | ASF1 anti-silencing function 1 homolog B (S. cerevisiae) | ASF1B | 55,723 | 66,929 | 0.31185 | 0.4842 | 0.000022 | 0.6462 |
| 3 | Asparagine synthetase transcript variant 3 | ASNS | 440 | 27,053 | 0.49995 | 0.47725 | 0.014519 | 0.9013 |
| 0.028100 | 0.8827 | |||||||
| 4 | Chromosome 4 open reading frame 32 | C4ORF32 | 132,720 | 70,617 | 0.37295 | 0.3323 | 0.038578 | 0.9266 |
| 5 | Coiled-coil domain containing 99 | CCDC99 | 54,908 | 70,385 | 0.22785 | 0.3449 | 0.012122 | 0.9013 |
| 6 | Calsyntenin 1 transcript variant 2 | CLSTN1 | 22,883 | 65,945 | 0.27065 | 0.284 | 0.000602 | 0.8645 |
| 7 | COMM domain containing 10 | COMMD10 | 51,397 | 69,456 | 0.4655 | 0.4514 | 0.000320 | 0.8526 |
| 0.000364 | 0.8526 | |||||||
| 8 | DNA segment on chromosome 4 (unique) 234 expressed sequence transcript variant 2 |
D4S234E | 27,065 | 18,196 | 0.3841 | 0.43325 | 0.002952 | 0.8827 |
| 9 | Dermokine | DMKN | 93,099 | 73,712 | 0.4168 | 0.4386 | 0.025554 | 0.9138 |
| 0.005339 | 0.8766 | |||||||
| 10 | Formin homology 2 domain containing 3 | FHOD3 | 80,206 | 225,288 | 0.2479 | 0.4387 | 0.001161 | 0.9013 |
| 0.017076 | 0.8066 | |||||||
| *11 | v-fos FBJ murine osteosarcoma viral oncogene homolog | FOS | 2,353 | 14,281 | 0.17845 | 0.1554 | 0.033596 | 0.7220 |
| *12 | Fyn-related kinase | FRK | 2,444 | 14,302 | 0.2495 | 0.19155 | 0.030182 | 0.9330 |
| *13 | GATA binding protein 2 | GATA2 | 2,624 | 14,461 | 0.4903 | 0.42275 | 0.036644 | 0.9266 |
| 14 | G protein-coupled receptor 56 transcript variant 3 | GPR56 | 9,289 | 14,766 | 0.333 | 0.22535 | 0.012741 | 0.8766 |
| 15 | G protein-coupled receptor 155 transcript variant 1 | GPR155 | 151,556 | 68,526 | 0.35165 | 0.40795 | 0.003324 | 0.8526 |
| 0.013478 | 0.9075 | |||||||
| 16 | Hedgehog interacting protein | HHIP | 64,399 | 15,245 | 0.064 | 0.12915 | 0.000163 | 0.8293 |
| 0.025741 | 0.8888 | |||||||
| 17 | High-mobility group box 2 | HMGB2 | 3,148 | 97,165 | 0.33305 | 0.2812 | 0.000063 | 0.8011 |
| 18 | ISL1 transcription factor, LIM/homeodomain, (islet-1) | ISL1 | 3,670 | 16,392 | 0.4416 | 0.4616 | 0.000238 | 0.8123 |
| *19 | Keratin 8 | KRT8 | 3,856 | 16,691 | 0.24445 | 0.24095 | 0.018072 | 0.9266 |
| 20 | L1 cell adhesion molecule | L1CAM | 3,897 | 16,728 | 0.2159 | 0.31755 | 0.0008848 | 0.7846 |
| 0.000580 | 0.8888 | |||||||
| 21 | Leucine rich repeat neuronal 1 | LRRN1 | 57,633 | 16,979 | 0.3495 | 0.1858 | 0.009137 | 0.8467 |
| 22 | Lymphocyte antigen 6 complex, locus E | LY6E | 4,061 | 17,069 | 0.0847 | 0.2604 | 0.001543 | 0.8586 |
| *23 | Matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase) |
MMP2 | 4,313 | 17,390 | 0.34175 | 0.38105 | 0.013771 | 0.9395 |
| 24 | MAX dimerization protein 3 | MXD3 | 83,463 | 17,121 | 0.08465 | 0.1876 | 0.045603 | 0.9013 |
| 25 | Myosin VIIA | MYO7A | 4,647 | 17,921 | 0.38115 | 0.27035 | 0.000156 | 0.8766 |
| 0.000123 | 0.9013 | |||||||
| 26 | Nucleotide-binding oligomerization domain containing 1 | NOD1 | 10,392 | 107,607 | 0.3392 | 0.45665 | 0.019006 | 0.8827 |
| *27 | Neurotensin | NTS (includes E.G:4922) |
4,922 | 67,405 | 0.072 | 0.2086 | 0.015360 | 0.9075 |
| 28 | PCTAIRE protein kinase 1 transcript variant 1 | PCTK1 | 5,127 | 18,555 | 0.42975 | 0.4852 | 0.043295 | 0.9266 |
| 29 | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae)transcript variant 1 | RAD51 | 5,888 | 19,361 | 0.41035 | 0.40715 | 0.025319 | 0.9138 |
| 30 | Regulating synaptic membrane exocytosis 3 | RIMS3 | 9,783 | 242,662 | 0.4441 | 0.4079 | 0.000003 | 0.6690 |
| 31 | SERTA domain containing 4 | SERTAD4 | 56,256 | 214,791 | 0.4773 | 0.35545 | 0.017776 | 0.9075 |
| 32 | Solute carrier family 14 (urea transporter), member 1 (Kidd blood group) | SLC14A1 | 6,563 | 108,052 | 0.0984 | 0.1137 | 0.000506 | 0.8706 |
| 33 | Solute carrier family 18 (vesicular monoamine), member 2 | SLC18A2 | 6,571 | 214,084 | 0.2226 | 0.2073 | 0.010239 | 0.8950 |
| 34 | Spindle pole body component 24 homolog (S. cerevisiae) | SPC24 | 147,841 | 67,629 | 0.3539 | 0.4108 | 0.017555 | 0.9266 |
| 35 | Spindle pole body component 25 homolog (S. cerevisiae) | SPC25 | 57,405 | 66,442 | 0.14695 | 0.1806 | 0.000174 | 0.8293 |
| 36 | Tropomodulin 1 | TMOD1 | 7,111 | 21,916 | 0.28235 | 0.30885 | 0.043568 | 0.8011 |
| 37 | Thiopurine S-methyltransferase | TPMT | 7,172 | 22,017 | 0.48925 | 0.47005 | 0.022231 | 0.8706 |
| 38 | Zinc finger protein 35 | ZNF35 | 7,584 | 22,646 | 0.40275 | 0.49375 | 0.000002 | 0.5510 |
A Genes with sequence homology in human and mouse up-regulated by at least two folds (both Qiagen and Illumina system) in MTT cells compared to MPC cells and significantly (P<0.05) up-regulated in malignant human microarray. B Genes with sequence homology in human and mouse down-regulated by at least two fold (both Qiagen and Illumina system) in MTT compared to MPC and significantly (P<0.05) down-regulated in malignant human microarray.
Genes selected by Ingenuity pathway analysis for qRT-PCR validation
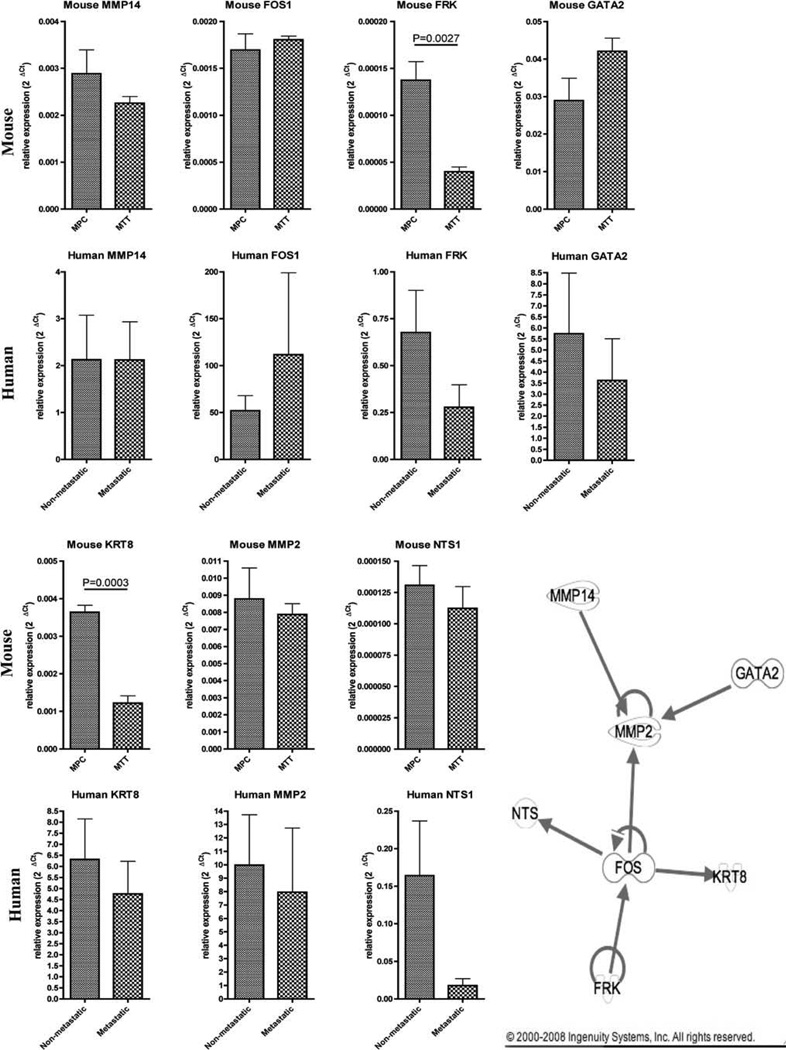
The 47 genes were put in the Ingenuity Pathway Analysis to determine if these genes are part of any biological pathway (Fig. 3). As a result, seven genes were found to be part of a network including matrix metallopeptidase 14 (MMP14), v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS1), Fyn-related kinase (FRK), GATA binding protein 2 (GATA2), keratin 8 (KRT8), matrix metallopeptidase 2 (MMP2), and neurotensin (NTS1). These seven genes were then selected for qRT-PCR validation (Fig. 3). qRT-PCR was performed on the mouse MPC and MTT cells and also on a different set of human metastatic and non-metastatic pheochromocytomas to compare the in vitro/animal data to the human samples. This validation revealed two genes (FRK, P = 0.0027, and KRT8, P = 0.0003) that were significantly down regulated in MTT cells compared to MPC confirming the microarray analysis. In human pheochromocytoma samples the same seven genes did not show statistically significant expression.
Fig. 3.
Quantitative real-time PCR of mouse and human genes. qRT-PCR of MMP14, FOS1, FRK, GATA2, KRT8, MMP2, and NTS1 in mouse MPC versus MTT cells and human non-metastatic versus metastatic tumors (mean ± SEM). Ingenuity pathway analysis network showing interaction of seven genes selected for validation
In vivo characterization of MPC versus MTT-induced metastatic tumors
All MPC and MTT metastatic tumors were confirmed to be pheochromocytomas by histology and intratumoral catecholamine measurements. Elevated norepinephrine levels were found in MPC hepatic metastases and ranged between 78,082 and 1,225,792 pg/mg tissue and in the metastatic MTT subclone were 42,307–150,767 pg/mg tissue compared to normal liver which was 25–125 pg/mg tissue. Epinephrine levels were below detectable levels in the liver lesions. The growth of tumors resulting from injections of equivalent numbers of MPC and MTT cells prepared under the same conditions was monitored by serial MRI. From previous studies, we expected tumors as small as 1 mm in diameter to become visible in the liver 4 weeks after injection [4]. In the parental MPC model, animals lived ten or more weeks after injection; however, their survival strongly correlated with the number of liver lesions. Liver tumors were monitored by MRI from day 21 to day 42 after tail vein injection of 5 × 105 MTT or MPC cells (Fig. 1c, d). In these groups, we found that injected MTT cells resulted in liver lesions visible by MRI on day 21 compared to day 30 in mice injected with MPC cells. We divided MTT and MPC liver lesions into small (0.3–0.8 mm3) and large (≥0.8 mm3) tumors because of their considerable difference in tumor growth rate. Aggressiveness of MTT cell line was confirmed by rapid liver tumor growth. By day 32, all mice had extensive tumor burden and had to be euthanized. In comparison, three of five mice of the MPC-inoculated animals survived until day 44.
Discussion
The diagnosis of malignant pheochromocytoma is difficult to make in its early stages before any spread and continues to rely on histological confirmation of tumor metastases to sites where chromaffin cells are not normally present. No reliable human pheochromocytoma cell lines are available. The murine MPC were first described in 1998 and are derived from a transplantable pheochromocytoma from heterozygous Nf1 gene knockout mice [6, 15]. The tumors produced by this cell line histologically and functionally resemble human pheochromocytoma [4, 5]. Similarities exist to conclude that MPC-developed tumors resemble human malignant tumors. First, in our models the sites involved in tumor development are in the agreement with the definition of malignant pheochromocytomas spread to sites where chromaffin cells are not normally present. Furthermore, we assume that delivery of the human pheochromocytoma and MPC cells are both mainly occurring through the bloodstream. However, what is different in our model is the absence of a primary tumor. Our group, therefore, adapted the MPC line to generate a mouse model of aggressive metastatic pheochromocytoma to study possible different gene expression profiles.
In this study, we optimized our previously established mouse model by adjusting three parameters of MPC cell preparation for injection: cell number, duration of exposure to trypsin, and post-dissociation incubation temperature. We concluded that tail vein injection of 5 × 105 MPC cells pre-treated with trypsin for 1–2 min incubated at room temperature for 60 min after dissociation increased the number of solid organ metastases in nude mice. These findings are likely related to improved cell viability.
By culturing disaggregated tumor cells isolated from hepatic metastases generated in the optimized MPC model, we were able to develop a more aggressive subline, MTT cells. These cells were injected via tail vein to produce metastatic disease that retained the histological and biochemical feature of pheochromocytomas, while at the same time representing a closer convergence with the aggressiveness seen in the human disease. The MTT cells retained their biochemical phenotype and expression of the catecholamine biosynthetic enzymes TH and PNMT compared to MPC. Elevated norepinephrine levels were also detected in MPC and MTT liver tumors compared to normal liver tissue. Monitoring of tumor growth by MRI in the MTT model clearly demonstrated its practicality, in particular for pre-clinical testing of novel treatment, because of the greater number of lesions achieved and shorter development time. We have observed that animals with fewer tumors in liver would live longer; however, liver tumors can get as big as 1.5 cm in diameter. Remaining amount of liver tissue was a predictive factor for survival, rather than single bigger lesions or numerous smaller lesions. For the purpose of possible drug experimental testing, both need to be considered.
Gene expression array analysis revealed genes that may be important in the development of an aggressive and more malignant phenotype of pheochromocytoma. Multiple genes were down regulated in the highly metastatic MTT cells compared to MPC cells and correlated with more aggressive behavior. This also correlated with clinical findings that loss of gene transcription correlated with a metastatic phenotype [16].
Forty-seven genes, nine upregulated and thirty-eight downregulated, were selected from our gene expression array we analyzed by using Ingenuity pathway analysis to identify a network interaction of genes for validation by qRT-PCR in both mouse cell and human tumor samples. This network of seven genes also represents a biological pathway in which these genes may be differentially expressed in malignant pheochromocytoma. Of the genes identified, FRK and KRT8 were significantly down-regulated (P < 0.01) in MTT compared to MPC. Coincidently, human tumors did not reveal any statistically significant differences for these genes.
Importantly, some limitations of this validation that may account for discrepancy in microarray and qRT-PCR findings are that (1) we selected genes in mouse cell lines which had varying metastatic potential and referenced to human transcripts selected comparing malignant versus benign tumors. Thus, while the genes may give insight for metastatic potential of mouse and human cells, it is not ideal because both mouse cell lines may produce metastasis. (2) We selected genes for validation based on pathway analysis versus most significance or fold-change. Increasing the cut-off of our initial microarray analysis (e.g. P < 0.001 vs. P < 0.05) would certainly select for genes with most robust differential expression; however, we aimed to identify pathways which may be important in development of metastatic potential. (3) The microarray data was conducted with oligos addressing mostly the 3’ of each gene, while all qRT-PCR used primers within internal exons (human NTS only) or between exons (all other genes). Thus, splice variants may be responsible for these discrepancies. (4) Finally, we have shown previously that in vivo behavior may differ from in vitro cell pathways as alluded to in our previous study comparing MPC with organ metastatic lesions [5].
In summary, this study presents a novel enhanced metastasis model in which cells taken from MPC metastasis were re-injected to form more aggressive, less differentiated metastasis. This animal model of metastatic pheochromocytoma improves upon previous results used to generate primarily liver lesions. Furthermore, we developed variant MTT cell line that demonstrates a more aggressive phenotype in vivo, as well as displaying a specific gene signature by microarray analysis. Rapid tumor growth and the development of numerous multi-organ metastases were verified by serial anatomical imaging. This study establishes how the mouse model of metastatic pheochromocytoma can be enhanced, monitored, and utilized in the identification of genes involved in aggressive tumorgencity or metastasis. This model will also be useful for future testing of new therapies for metastatic pheochromocytoma.
Acknowledgments
We would like to acknowledge the valuable assistance of the colleagues in our group, Dr. Graeme Eisenhofer, Dr. Nancy Nader, and Sinnie Ng. Special thank belong to Dr. David Thomasson for introducing us to MRI. This research was supported (in part) by the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Human Genome Research Institute, National Cancer Institute, National Institute of Mental Health, and NS37685 (to A.S.T.) and APVV-0148-06. The authors have no conflict of interest to disclose.
Abbreviations
- FOS1
v-fos FBJ murine osteosarcoma viral oncogene homolog
- FRK
Fyn-related kinase
- GATA2
GATA binding protein 2
- KRT8
Keratin 8
- MMP2
Matrix metallopeptidase 2
- MMP14
Matrix metallopeptidase 14
- MPC
Mouse pheochromocytoma
- MTT
Mouse tumor tissue
- Nf1
Neurofibromatosis1
- NTS1
Neurotensin
- PNMT
Phenylethanolamine N-methyltransferase
- qRT-PCR
Quantitative real-time PCR
- TH
Tyrosine hydroxylase
Contributor Information
Lucia Martiniova, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA; Institute of Experimental Endocrinology, Slovak Academy of Sciences, 83306 Bratislava, Slovakia.
Edwin W. Lai, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA
Abdel G. Elkahloun, National Human Genome Research Institute, Bethesda, MD 20892, USA
Mones Abu-Asab, Laboratory of Pathology, National Cancer Institute, Bethesda, MD 20892, USA.
Andrea Wickremasinghe, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA.
Daniel C. Solis, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA
Shiromi M. Perera, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA
Thanh-Truc Huynh, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA.
Irina A. Lubensky, Cancer Diagnosis Program, Division of Cancer Treatment and Diagnosis National Cancer Institute, National Institutes of Health, Rockville, MD 20892, USA
Arthur S. Tischler, Department of Pathology, Tufts Medical Center, Tufts University School of Medicine, Boston, MA 02111, USA
Richard Kvetnansky, Institute of Experimental Endocrinology, Slovak Academy of Sciences, 83306 Bratislava, Slovakia.
Salvatore Alesci, Clinical Neuroendocrinology Branch, National Institute of Mental Health, Bethesda, MD 20892, USA.
John C. Morris, Metabolism Branch, Center for Cancer Research, National Cancer Institute, Bethesda, MD 20892, USA
Karel Pacak, Section on Medical Neuroendocrinology, Reproductive and Adult Endocrinology Program, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD, NIH), Building 10 Room 1E-3140, 10 Center Drive MSC-1109, Bethesda, MD 20892-1109, USA, karel@mail.nih.gov.
References
- 1.Lack EE. Atlas of human pathology. Washington: Armed Forces Institute of Pathology; 1997. Tumors of the adrenal gland and extra-adrenal paraganglia. [Google Scholar]
- 2.Eisenhofer G, Bornstein SR, Brouwers FM, et al. Malignant pheochromocytoma: current status and initiatives for future progress. Endocr Relat Cancer. 2004;11:423–436. doi: 10.1677/erc.1.00829. [DOI] [PubMed] [Google Scholar]
- 3.John H, Ziegler WH, Hauri D, et al. Pheochromocytomas: can malignant potential be predicted? Urology. 1999;53:679–683. doi: 10.1016/s0090-4295(98)00612-8. [DOI] [PubMed] [Google Scholar]
- 4.Martiniova L, Ohta S, Quion P. Anatomical and functional imaging of tumors in animal models: focus on pheochromocytoma. Ann N Y Acad Sci. 2006;1073:392–404. doi: 10.1196/annals.1353.043. [DOI] [PubMed] [Google Scholar]
- 5.Ohta S, Lai EW, Morris JC, et al. Metastasis-associated gene expression profile of liver and subcutaneous lesions derived from mouse pheochromocytoma cells. Mol Carcinog. 2008;47:245–251. doi: 10.1002/mc.20388. [DOI] [PubMed] [Google Scholar]
- 6.Powers JF, Evinger MJ, Tsokas P, et al. Pheochromocytoma cell lines from heterozygous neurofibromatosis knockout mice. Cell Tissue Res. 2000;302:309–320. doi: 10.1007/s004410000290. [DOI] [PubMed] [Google Scholar]
- 7.Tischler AS, Greene LA, Kwan PW, et al. Ultrastructural effects of nerve growth factor on PC 12 pheochromocytoma cells in spinner culture. Cell Tissue Res. 1983;228:641–648. doi: 10.1007/BF00211480. [DOI] [PubMed] [Google Scholar]
- 8.Martiniova L, Kotys S, Thomasson D, et al. Non-invasive monitoring of a murine model of metastatic pheochromocytoma: a comparison of contrast enhanced microCT and non-enhanced MRI. J Magn Reson Imaging. 2008 doi: 10.1002/jmri.21654. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alesci S, Abu-Asab M, Perera SM, et al. Mitochondrial localization of human recombinant adenovirus: from evolution to gene therapy. Neuroimmunomodulation. 2007;14:221–223. doi: 10.1159/000113065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alesci S, Perera SM, Lai EW, et al. Adenoviral gene transfer in bovine adrenomedullary and murine pheochromocytoma cells: potential clinical and therapeutic relevance. Endocrinology. 2007;148:3900–3907. doi: 10.1210/en.2007-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenhofer G, Walther MM, Huynh TT, et al. Pheochromocytomas in von Hippel-Lindau syndrome and multiple endocrine neoplasia type 2 display distinct biochemical and clinical phenotypes. J Clin Endocr Metab. 2001;86:1999–2008. doi: 10.1210/jcem.86.5.7496. [DOI] [PubMed] [Google Scholar]
- 12.Brouwers FM, Elkahloun AG, Munson PJ, et al. Gene expression profiling of benign and malignant pheochromocytoma. Ann N Y Acad Sci. 2006;1073:541–556. doi: 10.1196/annals.1353.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JL, Snir M, Noushmehr H, et al. Transcriptional profiling of endogenous germ layer precursor cells identifies dusp4 as an essential gene in zebrafish endoderm specification. Proc Natl Acad Sci USA. 2008;105:12337–12342. doi: 10.1073/pnas.0805589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazin PL, Cuzzocreo JL, Yassa MA, et al. Volumetric neuroimage analysis extensions for the MIPAV software package. J Neurosci Methods. 2007;165:111–121. doi: 10.1016/j.jneumeth.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tischler AS, Powers JF, Alroy J. Animal models of pheochromocytoma. Histol Histopathol. 2004;19:883–895. doi: 10.14670/HH-19.883. [DOI] [PubMed] [Google Scholar]
- 16.Ohta S, Lai EW, Pang AL, et al. Downregulation of metastasis suppressor genes in malignant pheochromocytoma. Int J Cancer. 2005;114:139–143. doi: 10.1002/ijc.20670. [DOI] [PubMed] [Google Scholar]