Abstract
1α,25-Dihydroxyvitamin D3 (1,25(OH)2D) exerts its biological activities through vitamin D receptor (VDR), which is a member of the superfamily of steroid receptors, that act as ligand-dependent transcription factors. Ligated VDR in complex with retinoid X receptor (RXR) binds to regulatory regions of 1,25(OH)2D-target genes. 1,25(OH)2D is able to induce differentiation of leukemic blasts towards macrophage-like cells. Many different acute myeloid leukemia (AML) cell lines respond to 1,25(OH)2D by increasing CD14 cell surface receptor, some additionally upregulate CD11b and CD11c integrins. In untreated AML cells VDR protein is present in cytosol at a very low level, even though its mRNA is continuously expressed. Ligation of VDR causes protein stabilization and translocation to the cell nuclei, where it regulates transcription of target genes. Several important groups of genes are regulated by 1,25(OH)2D in HL60 cells. These genes include differentiation-related genes involved in macrophage function, as well as a gene regulating degradation of 1,25(OH)2D, namely CYP24A1. We summarize here the data which demonstrate that though some cellular responses to 1,25(OH)2D in AML cells are transcription-dependent, there are many others which depend on intracellular signal transduction, protein trafficking and stabilization. The final effect of 1,25(OH)2D action in leukemic cells requires all these acting together.
1. Introduction
The primary role of 1α,25-dihydroxyvitamin D3 (1,25(OH)2D) is to maintain calcium and phosphate homeostasis in vertebrate organisms via actions in the intestine, bones, kidneys, and parathyroid glands. However, it is well known that physiological roles of 1,25(OH)2D reach much beyond calcium and phosphate homeostasis. For example, 1,25(OH)2D induces differentiation and inhibits proliferation of various normal and cancer cells, including osteoclasts, keratinocytes, and monocytes. In 1981 the group of Suda observed that 1,25(OH)2D was able to induce differentiation in the M1 murine myeloid cell line [1] and that it extended the survival of mice inoculated with leukemia cells [2]. Since then many research projects have been performed in order to prepare ground for clinical use of 1,25(OH)2D or of its low calcemic analogs in leukemia treatment [3–5].
There are two major signal transduction pathways activated by 1,25(OH)2D in target cells. The most important and the best documented is the so-called “genomic pathway,” with its most important player a vitamin D receptor (VDR). The less well described is “nongenomic pathway,” which consists of intracellular signalling molecules, such as mitogen-activated protein kinases (MAPKs), phosphatidylinositol 3-kinase (PI3K), and others, activated by mechanisms that are not fully understood now [6]. It is believed that both pathways need to be activated for full biological activity of 1,25(OH)2D and that the most probable mediator of these actions is a putative membrane VDR (mVDR) [6].
VDR belongs to the superfamily of intracellular receptors for steroid and thyroid hormones. 48 members of the superfamily have been identified in humans; they act as ligand-induced transcription factors [7]. Most of the superfamily members, in order to be biologically active, form homo- or heterodimers. For VDR, retinoid X receptor (RXR) is a dimerization partner. VDR upon ligation undergoes conformational changes that allow binding to specific sequences in promoter regions of target genes, called vitamin D response elements (VDREs). VDREs are composed of two repeated half-sites with the consensus sequence AGGTCAcagAGGTCA (VDRE-DR3). Binding of 1,25(OH)2D to VDR enhances heterodimerization with RXR and allows binding of the coactivator complex, known as vitamin D receptor-interacting protein complex (DRIP) [8] and of other proteins, histone acetylase among them. Acetylated histones relax chromatin structure to make DNA accessible and permit initiation of transcription of target genes [9]. VDR may be ligated not only with 1,25(OH)2D but also with other compounds such as lithocholic acid, docosahexaenoic acid, arachidonic acid, or curcumin [10]. Moreover, there are about 300 compounds closely related to 1,25(OH)2D, called 1,25(OH)2D analogs, which can bind VDR and exert changed biological properties. Subtle conformational changes in VDR structure caused by analogs can produce antagonistic, agonistic, or even superagonistic effects. There are even some analogs that exert semiselective activities, with lowered calcemic and increased antiproliferative and cell differentiating effects [11].
The VDR protein is expressed at low concentrations in target tissues and cultured cells with the level of receptor expression ranging from a few copies of the VDR/cell to 25 000 copies/cell [7]. Among blood cells VDR is expressed in Tcells, Bcells, monocytes, neutrophils, platelets, macrophages, and dendritic cells. Also many different myeloid leukemia cell lines, blocked at various stages of maturation, expressed mRNA for VDR; however, the expression levels were variable [12]. Addition of 1,25(OH)2D to certain acute myeloid leukemia (AML) cells induces dramatic changes in their phenotype and function; however, the extent of these changes is various in various cell lines.
The activation of MAPK/Erk1,2 signal transduction pathway in AML cells in response to 1,25(OH)2D was for the first time reported in 1997 [13], and it was later shown to be important for the process of AML cell differentiation [14]. The exact mechanisms of how MAPK/Erk1,2 participate in the differentiation process are not known; however, they are being connected with a proliferative phase of AML cells differentiation [15]. MAPK/JNK pathway, whose activation was reported later [16], appeared to be involved in a subtle way in regulation of 1,25(OH)2D-dependent transcription factors [17]. Another MAP kinase, p38, has antagonistic effects to both MAPK/Erk1,2 and MAPK/JNK [16, 18]. Also activation of PI3K signal transduction pathway in AML cells exposed to 1,25(OH)2D has been reported [19] and was later shown to be responsible for activation of myeloid zinc finger-1 (MZF-1) transcription factor, which in turn participates in regulation of proteins crucial for macrophage function [20].
2. Nuclear Trafficking of VDR
VDR to fulfil its function of nuclear receptor must be transported into the nucleus [21]. In eukaryotic cells nucleoplasm and genetic material are separated from the cytosol by a double membrane which contains highly selective, bidirectional transporter channel called nuclear pore complex (NPC) [22]. NPC is composed of nearly 30 proteins termed nuclear pore complex components or “nucleoporins” (NUPs) [22] which occur in multiple copies [23]. Nuclear import of proteins through NPC is mediated by transporter proteins, such as importinα and importinβ, which bind cargoes through nuclear localisation signal (NLS) and interact with NUPs. Complex cargo-importinα binds importinβ which interacts with Ran-GDP protein [21]. Ran-GDP exists mostly in cytoplasm, whereas Ran-GTP in nuclei and this GTP gradient ensures the right direction of nuclear transport [21]. High concentration of Ran-GDP promotes the formation of import complex, while high concentration of Ran-GTP dissociates them and promotes formation of export complexes [24]. To complete the cycle importinα and importinβ must be transported back to the cytoplasm. To ensure Ran-GTP gradient, in the cytosol Ran-guanosine triphosphatase activating protein (RanGAP) stimulates the intrinsic GTP-hydrolyzing activity of Ran to form Ran-GDP. Hydrolysis of Ran-GTP to Ran-GDP causes release of importinβ for the next cycle.
To overcome the NPC, large molecules, such as nuclear receptors, harbour NLSs recognized by the transporter proteins which interact with NUPs in NPC. In VDR four NLSs were indentified (Figure 1(a)). First (NLS1) is localized between two zinc fingers within DNA-binding domain (DBD) [29]. The second NLS is in the second zinc finger of the DBD but data show that NLS2 does not function as an obligatory NLS. The next two are localized in the hinge region of VDR. NLS3 (102–110) is important for ligand-induced nuclear localisation of VDR but has no effect on unligated VDR nuclear import. NLS4 (154–173) is a short segment without a confirmed function. The RXR which is a partner protein for VDR has two NLSs, the first localized between two zinc fingers in DBD and the second in the second zinc finger (Figure 1(b)) [26]. Some data demonstrated that VDR shuttles between nucleus and cytoplasm in the absence of ligand, but unligated VDR weakly interacts with importinα [27]. However, nuclear trafficking of unligated VDR involves importin 4 through the interaction with the aminoterminus of VDR [21]. Binding the ligand promotes heterodimerization with RXR and enhances nuclear localization. Whereas RXR is predominantly localized in the nuclei even in the absence of ligand, there is also a cytoplasmic fraction of RXR that is translocated to the nucleus in the presence of its ligand. The nuclear import of RXR is mediated through importinβ, while that of VDR is mediated through importinα (Figure 2) [27]. In their publication Prüfer and Barsony showed that VDR is imported using two different pathways: ligand-dependent and ligand-independent. In ligand independent pathway RXR is the dominant partner for nuclear translocation, whereas ligated VDR dominates the mobility of RXR [28]. Binding 1,25(OH)2D stabilizes heterodimer VDR : RXR and its interaction with importinα but inhibits interaction of RXR with importinβ [27].
Figure 1.
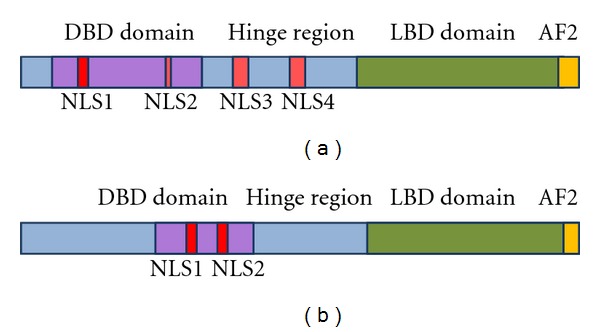
Localization of nuclear import domains (NLS) in VDR and RXR. NLS segments in VDR (a) and in RXR (b). Based on [26].
Figure 2.
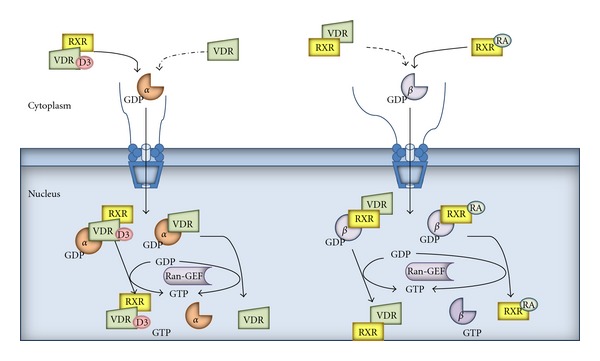
Nuclear import of VDR, RXR, and their heterodimer. GTP: guanosine 5′-triphosphate, GDP: guanosine diphosphate, α: importinα, β: importinβ, and RA: retinoic acid. Based on [27, 28].
In order to let proteins out of the nucleus, exportin named the chromosomal region maintenance 1 protein (CRM1) recognizes proteins containing leucine-rich nuclear export signals (NESs). Export complex formation is favored by high concentrations of RanGTP in the nucleus, which facilitate specific interactions with nucleoporins at the nuclear basket for appropriate translocation through the NPC [25]. VDR utilizes two pathways of nuclear export: ligand dependent and ligand independent. Unligated VDR uses a CRM1 export mechanism using NES localized in position 320–325 in LBD [28]. Mechanism for liganded VDR is CRM1 indepentent and requires DBD which functions as NES [30].
3. Differentiation of Human AML Cell Lines in Response to 1,25(OH)2D
One of the major processes in the array of anticancer actions of 1,25(OH)2D is differentiation of AML cells. Differentiation in AML cells consists of a G0/G1 cell cycle block [32], which is connected to an increase of proteins p21 and p27 [33], an increase in expression of antiapoptotic proteins [34], and acquisition of functional and phenotypic features characteristic for normal macrophages. Functional features are connected with an ability to phagocytose [35], with increased activity of monocyte specific esterase (MSE) [36], and with an ability to generate reactive oxygen species (ROS) and reduce nitroblue tetrazolium (NBT) [37]. Differentiation is also accompanied by upregulation of certain cell surface molecules, which are necessary for macrophage function, such as CD14, which is a coreceptor for lipopolysaccharide (LPS), as well as CD11b, which is a subunit of α M β 2 integrin or CD11c, an integrin αX, both involved in the cell adhesion [31, 38]. The process of 1,25(OH)2D-induced AML cell differentiation is not fast, it requires 3-4 days to reach plateau in expression of cell surface antigens, but differences in cell phenotype and function are spectacular (Figure 3). Since differentiation of blood cells may have beneficial effects, therapeutic applications for 1,25(OH)2D have been postulated. However, a major limitation for therapeutic use of 1,25(OH)2D is its potent calcemic and phosphatemic activity. The doses of 1,25(OH)2D, which are necessary to inhibit cell proliferation and induce differentiation, produce in vivo hypercalcemia and hyperphosphatemia that may be life threatening. Therefore there is a need for new 1,25(OH)2D analogs that retain high differentiating and antiproliferative activities with minimal or tolerable calcemic and phosphatemic effects [39]. Many such analogs were tested in our laboratory and their activities in inducing differentiation of various AML cell lines, as well as differentiation of leukemic blasts from the peripheral blood of AML patients, were studied.
Figure 3.
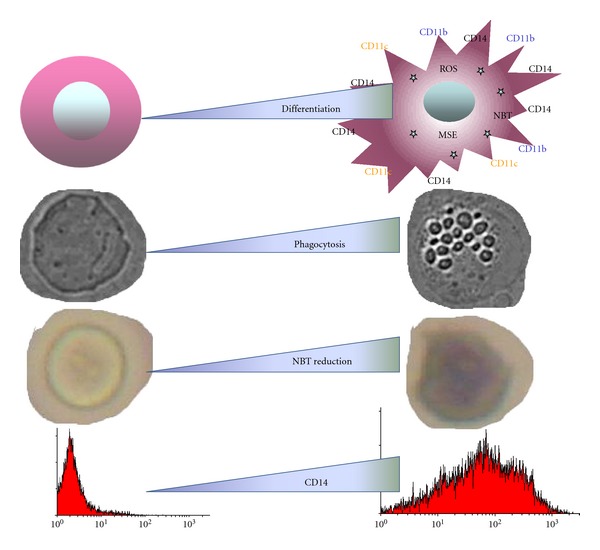
1,25(OH)2D-induced changes in AML cells. MSE: monocyte specyfic esterase; ROS: reactive oxygen species; NBT: nitroblue tetrazolium, CD14: co-receptor for LPS, CD11b: subunit of α M β2 integrin, CD11c: integrin αX. Based on [31].
4. Side-Chain Modified Analogs of Vitamin D2 and Vitamin D3
In our previous papers we described prodifferentiating activities of various side-chain modified analogs of vitamin D3, PRI-2191, PRI-2201, PRI-2202, and PRI-2205 and of vitamin D2, PRI-1906, PRI-1907 PRI-1908, and PRI-1909 [40, 41, 45–48]. We studied their pro-differentiating activities towards various human AML cell lines with genetic lesions characteristic for AML. Our studies indicated that some of the tested analogs were more potent than 1,25(OH)2D in induction of cell differentiation. The most efficient were the two analogs of vitamin D3, named PRI-2191 and PRI-2201, and the two analogs of vitamin D2, PRI-1906 and PRI-1907 [40–42]. Interestingly, one of the most active analogs tested by our group was PRI-2191, which in fact is a 1,25(OH)2D metabolite, already in use for treatment of psoriasis [41, 49] and a potential drug for vitiligo [50]. The four analogs mentioned above were tested in vivo for their calcemic activity. Results of these studies showed that PRI-2191, PRI-2201, and PRI-1906 were less calcemic than 1,25(OH)2D, while PRI-1907 was comparable to 1,25(OH)2D [48, 51, 52].
The cell lines used in our studies were derived from various AML subtypes. HL60 cells originate from M2 subtype of AML and are the most sensitive to 1,25(OH)2D-induced differentiation out of all cell lines used in our tests. NB-4 cells carry the t(15;17) PML-RARA fusion gene, which is characteristic for AML M3, U937 cells carry translocation t(10;11) often seen in AML M5, MV4-11 cells express fusion gene MLL-AF4, and MOLM13 cells express fusion gene MLL-AF9. Moreover, MOLM13 and MV4-11 have an internal tandem duplication in Flt3 gene (Flt3-ITD), in one or in both alleles, respectively [53]. Our studies revealed that in all cell lines studied, 1,25(OH)2D and its active analogs were able to upregulate CD14 cell surface antigen, while only in some cell lines CD11b integrin level was elevated (Figure 4). These data demonstrate that the expressions of CD14 and CD11b are controlled by two different signal transduction pathways, and in some AML cells CD11b pathway is blocked and cannot be overcome by 1,25(OH)2D.
Figure 4.
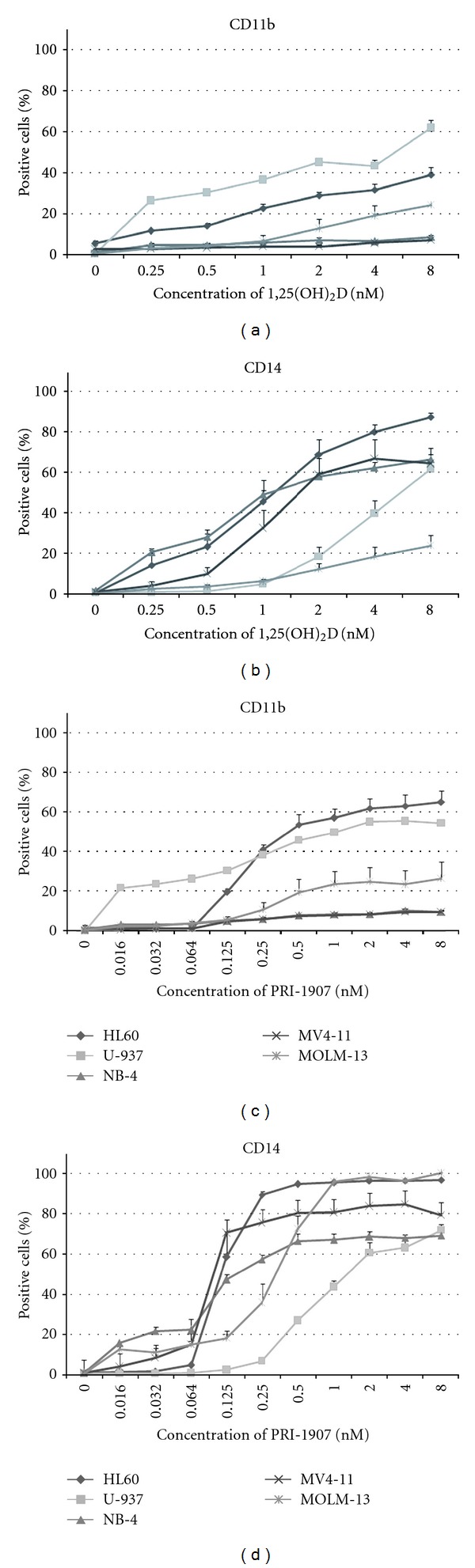
Expression of monocytic differentiation markers in AML cell lines exposed to either 1,25(OH)2D or to PRI-1907. HL60, U-937, NB-4, MV4-11, and MOLM-13 cells were exposed either to 1,25(OH)2D (a, b) or to PRI-1907 (c, d) for 96 h, and then the expression of CD11b and CD14 was tested in flow cytometry. The graphs show mean percentages (±SEM) of cells expressing cell differentiation markers. Based on [40].
5. VDR in Human AML Cell Lines
Experiments made by our group showed that AML cell lines have very low constitutive level of VDR protein, which increases significantly after exposure of the cells to 1,25(OH)2D. Therefore if the proposed nuclear import and export of unligated VDR exists in AML cells, it remains at a very low level, which is difficult to detect. 1,25(OH)2D-induced changes in nuclear trafficking of VDR could be observed using confocal microscopy, but western blotting of cell lysates fractionated into cytosol and nuclei appeared to be a much more sensitive method of VDR detection [54]. The kinetics of VDR accumulation in AML cells is surprisingly fast after exposure of the cells to 1,25(OH)2D. In HL60 cells, VDR starts to accumulate in the cell nuclei after few minutes from exposure to 1,25(OH)2D [42] and after half of an hour the difference is significant (Figure 5(a)). However, most of our studies were performed using HL60 cell line; also in other cell lines (Figure 5(b)) and in AML blasts (Figure 5(c)) accumulation of VDR in cell nuclei was observed [42, 54]. Obviously, the increase has not been caused through transcription of new mRNA for VDR since mRNA levels remained almost unchanged by 1,25(OH)2D, as confirmed by real-time PCR (not shown). Therefore the mechanism of accumulation of VDR must be regulated at post transcriptional level, and according to our data it is caused by ligation-induced protection of VDR protein from degradation [42]. It seems that VDR is continuously produced in the cytosol, and as long as it is unligated, it undergoes degradation. Ligation of VDR with 1,25(OH)2D induces rapid translocation of the receptor to the cell nuclei, where degradation process is slower. For effective nuclear trafficking, active p42,44/MAPK pathway and active PI3-K pathway are needed; however, the mechanism of this effect is not known [54]. It was surprising that in most of freshly isolated AML cells from patients constitutive level of VDR in cytosol was higher than in established cell lines [41]. Unfortunately we were unable to test this phenomenon in a more detailed manner, because neither of the patient's samples was possible to be cultured in vitro for longer than two-three weeks. It is known; however, that other nuclear receptors become stabilized in the cytosol by heat shock proteins (Hsp) [55, 56], so the involvement of this class of proteins was addressed. We demonstrated recently that, in HL60 cells, VDR interacts with Hsp90 and that activation of Hsp90 is necessary for the differentiation process [57], but our new experiments documented that activation of Hsp90 is not necessary for nuclear translocation of VDR. Geldanamycin, which inhibits activity of Hsp90, was not able to block 1,25(OH)2D-induced nuclear accumulation of VDR (Figure 6(a)). VDR protein appears in the nuclei of 1,25(OH)2D-treated cells very fast but disappears slowly. Our experiments in which HL60 cells were exposed to 1,25(OH)2D and an inhibitor of CRM1, namely, leptomycin B, confirmed that nuclear export of ligated VDR is CRM1 independent. As presented in Figure 6(b), presence of leptomycin B did not cause further accumulation of VDR in cells exposed to 1,25(OH)2D. As it was presented before, VDR protein levels remain elevated even after 4 days from exposure of HL60 cells to 1,25(OH)2D [42]. During this time VDR activates transcription of its target genes. One of them is CYP24A1, which encodes an enzyme, 24-hydroxylase of 1,25(OH)2D, responsible for degradation of 1,25(OH)2D to calcitrioic acid. As presented in our previous publication, 1,25(OH)2D increased in HL60 cells levels of CYP24A1 mRNA significantly; however, kinetics of induction was very slow [40]. CYP24A1 protein is localized exclusively in an inner membrane of mitochondria, where its levels also increase slowly after exposure to 1,25(OH)2D (not shown here).
Figure 5.
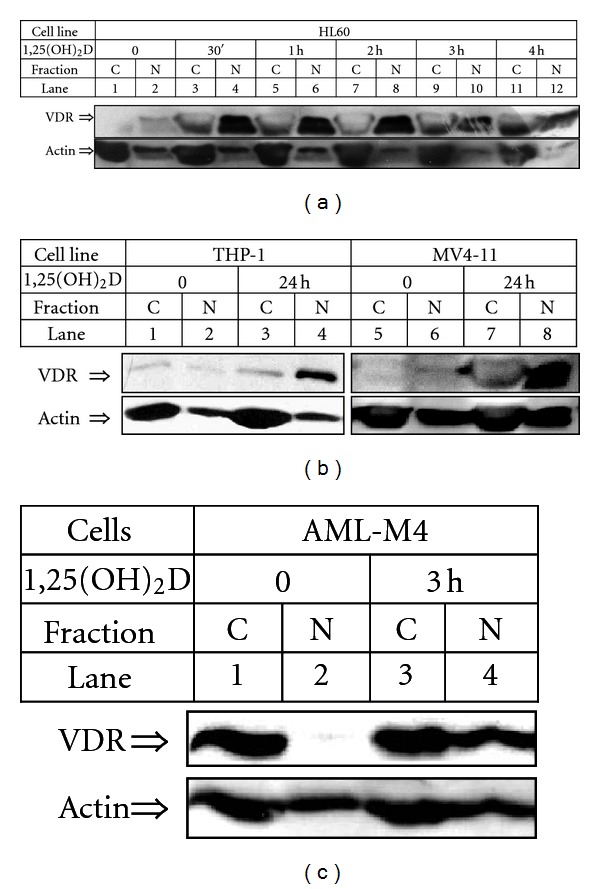
Expression of VDR protein in AML cells exposed to 1,25(OH)2D. AML cells after incubation for indicated times with 10 nM 1,25(OH)2D were lysed and fractionated into the cytoplasmic (C) and nuclear (N) fractions. The lysates from equal numbers of cells were separated in SDS-PAGE and blotted to the membrane. The membrane was probed with anti-VDR. Actin was probed as a control of equal loading and transfer of proteins. (a) HL60 cells, (b) THP-1, MV4-11 cells, and (c) AML-M4 blasts from patient's peripheral blood. Based on [41, 42] and on unpublished data.
Figure 6.
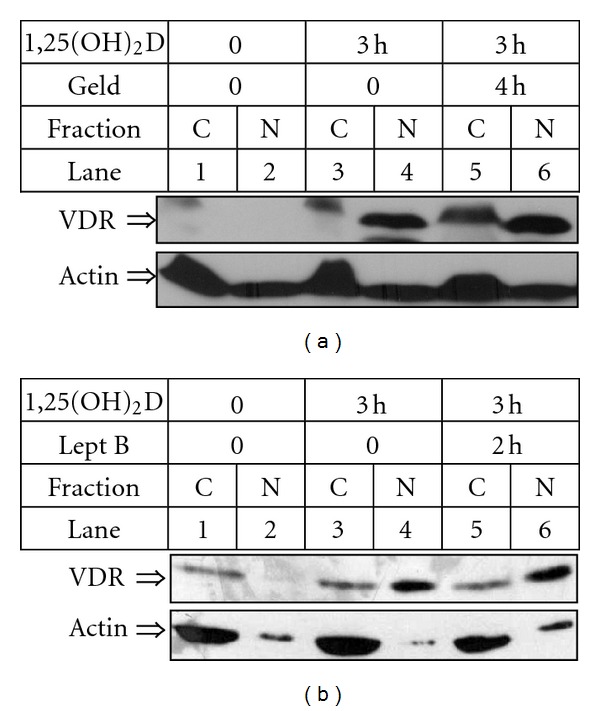
Nuclear trafficking of VDR in presence of geldanamycin (a) or leptomycin B (b). HL60 cells were exposed to 10 nM 1,25(OH)2D for 3 h. One sample was pretreated with 1 μM geldanamycin (geld) for 1 h before exposure to 1,25(OH)2D (a). Another sample was treated with 5 mg/ml of leptomycin B (lept B) for the last two hours of incubation (b). After incubation the cells were lysed and fractionated into the cytoplasmic (C) and nuclear (N) fractions. The lysates from equal numbers of cells were separated in SDS-PAGE and blotted to the membrane. The membrane was probed with anti-VDR. Actin was probed as a control of equal loading and transfer of proteins.
6. Regulation of C/EBP Transcription Factors by 1,25(OH)2D in AML Cells
There are several important groups of genes regulated by 1,25(OH)2D in AML cells, including differentiation-related genes that encode proteins important for function of monocytes or macrophages. The examples of such are genes encoding CAAT-enhancer binding proteins (C/EBPs), belonging to the family of basic leucine zipper (bZIP) transcription factors [44, 58, 59]. There are six genes for different C/EBPs (α, β, γ, δ, ε, and ζ) which are expressed in hematopoietic cells, hepatocytes, adipocytes, spleen, kidney, brain, and others. They can form homodimers and heterodimers with other family members and with other transcription factors. The C/EBP proteins contain highly conserved bZIP domain at the C-terminus, an activation domain at the N-terminus and some other regulatory domains (Figure 7) [43]. In hematopoietic cells C/EBPα is necessary mainly for differentiation from lymphoid-myeloid progenitors to granulocytes [60], while C/EBPβ is crucial for specialization of normal and 1,25(OH)2D-induced monocytes and macrophages, as well as for their proper functions [61–64], as it regulates transcription of many monocyte-specific proteins, such as CD14, lactoferrin, or lysozyme [43]. Recently an involvement of C/EBPβ in differentiation-related inhibition of proliferation was reported [65]. Because of alternative translation initiations sites, two different products of C/EBPα (42 kDa, 30 kDa) and three products of C/EBPβ genes are translated (55 kDa, 49 kDa, 20 kDa) [43, 66]. As presented in Figure 7, 30 kDa form of C/EBPα and 20 kDa form of C/EBPβ are devoid of portions of N-termini where transactivation domains are localized. They are, however, still able to dimerize, and therefore they can play an inhibitory function. Experiments done by groups of Studzinski and ours have shown that after exposure of HL60 to 1,25(OH)2D C/EBPα was only transiently upregulated in an early phase of differentiation, whilst upregulation of C/EBPβ was strong, long-lasting and correlated with the differentiation process [67]. Specially two shorter C/EBPβ isoforms were abundant in differentiating cells and their increase correlated with acquisition of monocytic differentiation markers, such as CD11b, CD14 [67], or CD11c (not shown).
Figure 7.
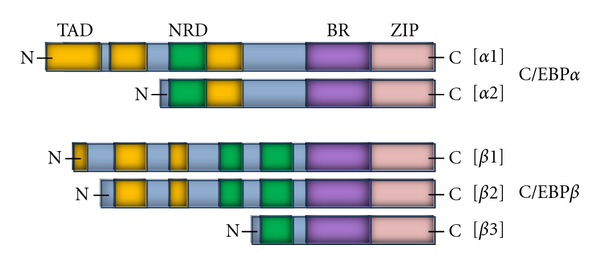
Schematic representation of the C/EBPα and C/EBPβ isoforms. TAD: transcription activation domain, NRD: negative regulatory domain, BR: basic region, and ZIP: leucine zipper. Based on [43, 44].
Transcriptional activity of C/EBP proteins is regulated not only by their length and dimerization but most importantly by their intracellular localization. As transcription factors, C/EBPs must enter the cell nuclei to bind CCAAT box motif in their target gene promoters [44, 66]. Therefore, cellular trafficking of C/EBPs was extensively studied by our group. Again, studied cells were fractionated into cytosol and nuclei. As shown in Figure 8, after exposure of the cells to 1,25(OH)2D, the full length isoform of C/EBPβ-1 (55 kDa) is present in either cytoplasmic or nuclear fraction, whilst the majority of shorter isoforms C/EBPβ-2 (49 kDa) and C/EBPβ-3 (20 kDa) are placed in the nuclei of the cells. Similar results were observed in various AML cell lines, such as THP-1, MV4-11, or MOLM-13 and in some samples of AML blasts isolated from patients. But when localization of C/EBPα was tested in fractionated cells, it appeared to be cytoplasmic, and no translocation was noticed after various times of exposure of the cells to 1,25(OH)2D. As examples, HL60, THP-1 cell lines, and AML blasts from patient's peripheral blood are presented (Figure 9). These findings suggest that, in AML cells, even if not mutated, C/EBPα is transcriptionally inactive what leads to the disturbances in granulopoiesis. Elevated expression of C/EBPβ and its nuclear translocation induced by 1,25(OH)2D can possibly allow the cells to bypass this block and switch differentiation into monocyte/macrophage pathway. The above hypothesis was presented before [68] and is further supported by the findings shown here, that in AML cells C/EBPα is localized in the cytosol, where it cannot exert its transcriptional activity.
Figure 8.
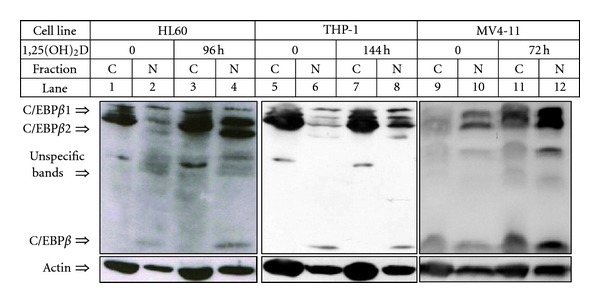
Subcellular localization of C/EBPβ isoforms in AML cells. The cells were exposed to 10 nM 1,25(OH)2D for given times and then were fractionated into cytoplasmic (C) and nuclear (N) fractions. The lysates from equal numbers of cells were separated in SDS-PAGE and blotted to the membrane. The membranes were probed with anti-C/EBPβ antibody. Actin was probed as a control of equal loading and transfer of proteins.
Figure 9.
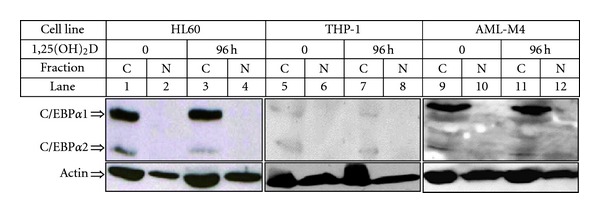
Subcellular localization of C/EBPα isoforms in AML cells. HL60 and AML-M4 blasts from patient's peripheral blood were exposed to 10 nM 1,25(OH)2D for 96 h and then were fractionated into cytoplasmic (C) and nuclear (N) fractions. The lysates from equal numbers of cells were separated in SDS-PAGE and blotted to the membrane. The membrane was probed with anti-C/EBPα antibody. Actin was probed as a control of equal loading and transfer of proteins.
7. Conclusions
Exposure of AML cells to 1,25(OH)2D or to its analogs triggers a long series of events which eventually lead to acquisition of monocyte/macrophage phenotype and function (some of them presented in Figure 10). The detailed description of 1,25(OH)2D-induced differentiation in cell line models, as well as in AML blasts isolated from patients might be important for future therapeutic applications of 1,25(OH)2D analogs. It is especially significant to learn which differentiation pathways are blocked in certain AML subtypes and how they could be bypassed with help of pharmacological agents.
Figure 10.
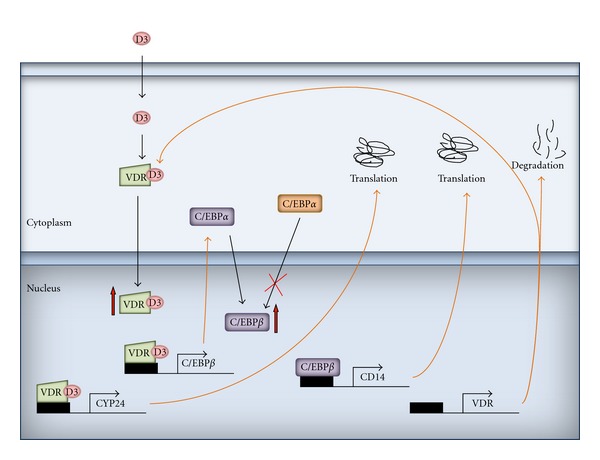
Overview of 1,25(OH)2D-induced intracellular events in AML cells. Description is in the text.
Acknowledgment
Part of the research described in this paper was supported by Wroclaw Research Center EIT+ within the project Biotechnologies and Advanced Medical Technologies—BioMed (POIG 01.01.02-02-003/08-00) financed by the European Regional Development Fund (Operational Program Innovative Economy, 1.1.2).
References
- 1.Abe E, Miamura C, Sakagami H, et al. Differentiation of mouse myeloid leukemia cells induced by 1-alpha,25-dihydroxyvitamin D3 . Proceedings of the National Academy of Sciences of the United States of America. 1981;78:4990–4994. doi: 10.1073/pnas.78.8.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honma Y, Hozumi M, Abe E, et al. 1-Alpha,25-dihydroxyvitamin D3 and 1-alpha-hydroxyvitamin D3 prolong survival time of mice inoculated with myeloid leukemia cells. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:201–204. doi: 10.1073/pnas.80.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto R, Akagi T, Koeffler P. Vitamin D compounds and myelodysplastic syndrome. Leukemia and Lymphoma. 2008;49(1):12–13. doi: 10.1080/10428190701757827. [DOI] [PubMed] [Google Scholar]
- 4.Nowak D, Stewart D, Koeffler HP. Differentiation therapy of leukemia: 3 Decades of development. Blood. 2009;113(16):3655–3665. doi: 10.1182/blood-2009-01-198911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luong QT, Koeffler HP. Vitamin D compounds in leukemia. Journal of Steroid Biochemistry and Molecular Biology. 2005;97(1-2):195–202. doi: 10.1016/j.jsbmb.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Mizwicki M, Norman A. Vitamin D sterol/VDR conformational dynamics and nongenomic actions. In: Feldman D, Pike J, Adams J, editors. Vitamin D. 3rd edition. Academic Press; 2011. [Google Scholar]
- 7.Pike W, Meyer M, Lee S. The vitamin D receptor: biochemical, molecular, biological, and genomic era investigations. In: Feldman D, Pike J, Adams J, editors. Vitamin D. 3rd edition. Academic Press; 2011. pp. 97–135. [Google Scholar]
- 8.Rachez C, Lemon BD, Suldan Z, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398(6730):824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 9.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiological Reviews. 2001;81(3):1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 10.Haussler MR, Haussler CA, Bartik L, et al. Vitamin D receptor: molecular signaling and actions of nutritional ligands in disease prevention. Nutrition Reviews. 2008;66(supplement 2):S98–S112. doi: 10.1111/j.1753-4887.2008.00093.x. [DOI] [PubMed] [Google Scholar]
- 11.Rochel N, Moras D. Structural basis for ligand activity in VDR. In: Feldman D, Pike J, Adams J, editors. Vitamin D. 3rd edition. Academic Press; 2011. pp. 171–191. [Google Scholar]
- 12.Kizaki M, Norman AW, Bishop JE, Lin CW, Karmakar A, Koeffler HP. 1,25-dihydroxyvitamin D3 receptor RNA: expression in hematopoietic cells. Blood. 1991;77(6):1238–1247. [PubMed] [Google Scholar]
- 13.Marcinkowska E, Wiedlocha A, Radzikowski C. 1,25-Dihydroxyvitamin D3 induced activation and subsequent nuclear translocation of MAPK is upstream regulated by PKC in HL-60 cells. Biochemical and Biophysical Research Communications. 1997;241:419–426. doi: 10.1006/bbrc.1997.7832. [DOI] [PubMed] [Google Scholar]
- 14.Marcinkowska E. Evidence that activation of MEK1,2/erk1,2 signal transduction pathway is necessary for calcitriol-induced differentiation of HL-60 cells. Anticancer Research. 2001;21(1):499–504. [PubMed] [Google Scholar]
- 15.Wang X, Studzinski G. Activation of extracellular signal-regulated kinases (ERKs) defines the first phase of 1,25-dihydroxyvitamin D3-induced differentiation of HL60 cells. Journal of Cellular Biochemistry. 2001;80:471–482. doi: 10.1002/1097-4644(20010315)80:4<471::aid-jcb1001>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Rao J, Studzinski GP. Inhibition of p38 MAP kinase activity up-regulates multiple MAP kinase pathways and potentiates 1,25-dihydroxyvitamin D3-induced differentiation of human leukemia HL60 cells. Experimental Cell Research. 2000;258(2):425–437. doi: 10.1006/excr.2000.4939. [DOI] [PubMed] [Google Scholar]
- 17.Chen-Deutsch X, Garay E, Zhang J, Harrison JS, Studzinski GP. c-Jun N-terminal kinase 2 (JNK2) antagonizes the signaling of differentiation by JNK1 in human myeloid leukemia cells resistant to vitamin D. Leukemia Research. 2009;33(10):1372–1378. doi: 10.1016/j.leukres.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velders MP, Fatima Macedo M, Provenzano M, et al. Inhibition of p38MAP kinase potentiates the JNK/SAPK pathway and AP-1 activity in monocytic but not in macrophage or granulocytic differentiation of HL60 cells. Journal of Cellular Biochemistry. 2001;82(1):68–77. doi: 10.1002/jcb.1141. [DOI] [PubMed] [Google Scholar]
- 19.Hmama Z, Nandan D, Sly L, Knutson KL, Herrera-Velit P, Reiner NE. 1α,25-dihydroxyvitamin D3-induced myeloid cell differentiation is regulated by a vitamin D receptor-phosphatidylinositol 3-kinase signaling complex. Journal of Experimental Medicine. 1999;190(11):1583–1594. doi: 10.1084/jem.190.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moeenrezakhanlou A, Shephard L, Lam L, Reiner NE. Myeloid cell differentiation in response to calcitriol for expression CD11b and CD14 is regulated by myeloid zinc finger-1 protein downstream of phosphatidylinositol 3-kinase. Journal of Leukocyte Biology. 2008;84(2):519–528. doi: 10.1189/jlb.1207833. [DOI] [PubMed] [Google Scholar]
- 21.Miyauchi Y, Michigami T, Sakaguchi N, et al. Importin 4 is responsible for ligand-independent nuclear translocation of vitamin D receptor. The Journal of Biological Chemistry. 2005;280(49):40901–40908. doi: 10.1074/jbc.M509347200. [DOI] [PubMed] [Google Scholar]
- 22.Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. Journal of Cell Biology. 1992;119(4):705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada J, Phillips JL, Patel S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Molecular and Cellular Proteomics. 2010;9(10):2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. Journal of Cell Science. 2000;113(10):1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- 25.Faustino RS, Nelson TJ, Terzic A, Perez-Terzic C. Nuclear transport: target for therapy. Clinical Pharmacology and Therapeutics. 2007;81(6):880–886. doi: 10.1038/sj.clpt.6100141. [DOI] [PubMed] [Google Scholar]
- 26.Barsony J, Prufer K. Vitamin D receptor and retinoid X receptor interactions in motion. Vitamins and Hormones. 2002;65:345–IN3. doi: 10.1016/s0083-6729(02)65071-x. [DOI] [PubMed] [Google Scholar]
- 27.Yasmin R, Williams RM, Xu M, Noy N. Nuclear import of the retinoid X receptor, the vitamin D receptor, and their mutual heterodimer. The Journal of Biological Chemistry. 2005;280(48):40152–40160. doi: 10.1074/jbc.M507708200. [DOI] [PubMed] [Google Scholar]
- 28.Prüfer K, Barsony J. Retinoid X Receptor dominates the nuclear import and export of the unliganded vitamin D receptor. Molecular Endocrinology. 2002;16(8):1738–1751. doi: 10.1210/me.2001-0345. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh J, Shimizu Y, Minoshima S, et al. Novel nuclear localization signal between the two DNA-binding zinc fingers in the human vitamin D receptor. Journal of Cellular Biochemistry. 1998;70:94–109. [PubMed] [Google Scholar]
- 30.Black BE, Holaska JM, Rastinejad F, Paschal BM. DNA binding domains in diverse nuclear receptors function as nuclear export signals. Current Biology. 2001;11(22):1749–1758. doi: 10.1016/s0960-9822(01)00537-1. [DOI] [PubMed] [Google Scholar]
- 31.Gocek E, Studzinski GP. Vitamin D and differentiation in cancer Signaling differentiation E. Gocek and G. P. Studzinski. Critical Reviews in Clinical Laboratory Sciences. 2009;46(4):190–209. doi: 10.1080/10408360902982128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Studzinski G, Chen F, Coffman F, Harrison L. p53/56(lyn) antisense shifts the 1,25-dihydroxyvitamin D3-induced G1/S block in HL60 cells to S phase. Journal of Cellular Physiology. 2000;183:238–246. doi: 10.1002/(SICI)1097-4652(200005)183:2<238::AID-JCP10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 33.Coffman FD, Studzinski GP. Differentiation-related mechanisms which suppress DNA replication. Experimental Cell Research. 1999;248(1):58–73. doi: 10.1006/excr.1999.4457. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang J, Studzinski GP. AKT pathway is activated by 1,25-dihydroxyvitamin D3 and participates in its anti-apoptotic effect and cell cycle control in differentiating HL60 cells. Cell Cycle. 2006;5(4):447–451. doi: 10.4161/cc.5.4.2467. [DOI] [PubMed] [Google Scholar]
- 35.Marcinkowska E, Kutner A, Radzikowski C. Cell differentiating and anti-proliferative activity of side-chain modified analogues of 1,25-dihydroxyvitamin D3 . Journal of Steroid Biochemistry and Molecular Biology. 1998;67(1):71–78. doi: 10.1016/s0960-0760(98)00065-x. [DOI] [PubMed] [Google Scholar]
- 36.Uphoff CC, Drexler HG. Biology of monocyte-specific esterase. Leukemia and Lymphoma. 2000;39(3-4):257–270. doi: 10.3109/10428190009065825. [DOI] [PubMed] [Google Scholar]
- 37.Steiner M, Priel I, Giat J, Levy J, Sharoni Y, Danilenko M. Carnosic acid inhibits proliferation and augments differentiation of human leukemic cells induced by 1,25-dihydroxyvitamin D3 and retinoic acid. Nutrition and Cancer. 2001;41(1-2):135–144. doi: 10.1080/01635581.2001.9680624. [DOI] [PubMed] [Google Scholar]
- 38.Studzinski GP, Rathod B, Wang QM, Rao J, Zhang F. Uncoupling of cell cycle arrest from the expression of monocytic differentiation markers in HL60 cell variants. Experimental Cell Research. 1997;232(2):376–387. doi: 10.1006/excr.1997.3484. [DOI] [PubMed] [Google Scholar]
- 39.Brown AJ, Slatopolsky E. Vitamin D analogs: therapeutic applications and mechanisms for selectivity. Molecular Aspects of Medicine. 2008;29(6):433–452. doi: 10.1016/j.mam.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Baurska H, Klopot A, Kielbinski M, et al. Structure-function analysis of vitamin D2 analogs as potential inducers of leukemia differentiation and inhibitors of prostate cancer proliferation. Journal of Steroid Biochemistry and Molecular Biology. 2011;126(1-2):46–54. doi: 10.1016/j.jsbmb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Gocek E, Kiełbiński M, Baurska H, Haus O, Kutner A, Marcinkowska E. Different susceptibilities to 1,25-dihydroxyvitamin D3-induced differentiation of AML cells carrying various mutations. Leukemia Research. 2010;34(5):649–657. doi: 10.1016/j.leukres.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Gocek E, Kiełbiński M, Wyłób P, Kutner A, Marcinkowska E. Side-chain modified vitamin D analogs induce rapid accumulation of VDR in the cell nuclei proportionately to their differentiation-inducing potential. Steroids. 2008;73(14):1359–1366. doi: 10.1016/j.steroids.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochemical Journal. 2002;365(3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54(1):6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 45.Opolski A, Wietrzyk J, Chrobak A, et al. Antiproliferative activity in vitro of side-chain analogues of calcitriol against various human normal and cancer cell lines. Anticancer Research. 1999;19(6):5217–5222. [PubMed] [Google Scholar]
- 46.Chodyński M, Wietrzyk J, Marcinkowska E, Opolski A, Szelejewski W, Kutner A. Synthesis and antiproliferative activity of side-chain unsaturated and homologated analogs of 1,25-dihydroxyvitamin D2 (24E)-(1S)-24-Dehydro-24a-homo-1,25-dihydroxyergocalciferol and congeners. Steroids. 2002;67(9):789–798. doi: 10.1016/s0039-128x(02)00038-7. [DOI] [PubMed] [Google Scholar]
- 47.Marcinkowska E, Kutner A. Side-chain modified vitamin D analogs require activation of both PI 3-K and erk1,2 signal transduction pathways to induce differentiation of human promyelocytic leukemia cells. Acta Biochimica Polonica. 2002;49(2):393–406. [PubMed] [Google Scholar]
- 48.Wietrzyk J, Nevozhay D, Milczarek M, Filip B, Kutner A. Toxicity and antitumor activity of the vitamin D analogs PRI-1906 and PRI-1907 in combined treatment with cyclophosphamide in a mouse mammary cancer model. Cancer Chemotherapy and Pharmacology. 2008;62(5):787–797. doi: 10.1007/s00280-007-0666-6. [DOI] [PubMed] [Google Scholar]
- 49.Marquez Balbas G, Sanchez Regana M, Umbert Millet P. Tacalcitol ointment for the treatment of nail psoriasis. Journal of Dermatological Treatment. 2009;1:1–3. doi: 10.1080/09546630902787585. [DOI] [PubMed] [Google Scholar]
- 50.Rodríguez-Martín M, García Bustínduy M, Sáez Rodríguez M, Noda Cabrera A. Randomized, double-blind clinical trial to evaluate the efficacy of topical tacalcitol and sunlight exposure in the treatment of adult nonsegmental vitiligo. British Journal of Dermatology. 2009;160(2):409–414. doi: 10.1111/j.1365-2133.2008.08906.x. [DOI] [PubMed] [Google Scholar]
- 51.Ji Y, Kutner A, Verstuyf A, Verlinden L, Studzinski GP. Derivatives of vitamins D2 and D3 activate three MAPK pathways and upregulate pRb expression in differentiating HL60 cells. Cell Cycle. 2002;1(6):410–415. doi: 10.4161/cc.1.6.269. [DOI] [PubMed] [Google Scholar]
- 52.Wietrzyk J, Chodyński M, Fitak H, Wojdat E, Kutner A, Opolski A. Antitumor properties of diastereomeric and geometric analogs of vitamin D3 . Anti-Cancer Drugs. 2007;18(4):447–457. doi: 10.1097/CAD.0b013e3280143166. [DOI] [PubMed] [Google Scholar]
- 53.Quentmeier H, Reinhardt J, Zaborski M, Drexler HG. FLT3 mutations in acute myeloid leukemia cell lines. Leukemia. 2003;17(1):120–124. doi: 10.1038/sj.leu.2402740. [DOI] [PubMed] [Google Scholar]
- 54.Gocek E, Kiełbiński M, Marcinkowska E. Activation of intracellular signaling pathways is necessary for an increase in VDR expression and its nuclear translocation. FEBS Letters. 2007;581(9):1751–1757. doi: 10.1016/j.febslet.2007.03.055. [DOI] [PubMed] [Google Scholar]
- 55.Picard D. Chaperoning steroid hormone action. Trends in Endocrinology and Metabolism. 2006;17(6):229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Grad I, Picard D. The glucocorticoid responses are shaped by molecular chaperones. Molecular and Cellular Endocrinology. 2007;275(1-2):2–12. doi: 10.1016/j.mce.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 57.Marcinkowska E, Gocek E. Heat shock protein 90 interacts with vitamin D receptor in human leukemia cells. Journal of Steroid Biochemistry and Molecular Biology. 2010;121(1-2):114–116. doi: 10.1016/j.jsbmb.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90(2):489–519. [PubMed] [Google Scholar]
- 59.Studzinski GP, Garay E, Patel R, Zhang J, Wang X. Vitamin D receptor signaling of monocytic differentiation in human leukemia cells: role of MAPK pathways in transcription factor activation. Current Topics in Medicinal Chemistry. 2006;6(12):1267–1271. doi: 10.2174/156802606777864935. [DOI] [PubMed] [Google Scholar]
- 60.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26(47):6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 61.Nagamura-Inoue T, Tamura T, Ozato K. Transcription factors that regulate growth and differentiation of myeloid cells. International Reviews of Immunology. 2001;20(1):83–105. doi: 10.3109/08830180109056724. [DOI] [PubMed] [Google Scholar]
- 62.Ji Y, Studzinski GP. Retinoblastoma protein and CCAAT/enhancer-binding protein β are required for 1,25-dihydroxyvitamin D3-induced monocytic differentiation of HL60 cells. Cancer Research. 2004;64(1):370–377. doi: 10.1158/0008-5472.can-03-3029. [DOI] [PubMed] [Google Scholar]
- 63.Hackanson B, Bennett KL, Brena RM, et al. Epigenetic modification of CCAAT/enhancer binding protein α expression in acute myeloid leukemia. Cancer Research. 2008;68(9):3142–3151. doi: 10.1158/0008-5472.CAN-08-0483. [DOI] [PubMed] [Google Scholar]
- 64.Akagi T, Thoennissen NH, George A, et al. In vivo deficiency of both C/EBPβ and C/EBP results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015419. Article ID e15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gutsch R, Kandemir JD, Pietsch D, et al. CCAAT/enhancer-binding protein β inhibits proliferation in monocytic cells by affecting the retinoblastoma protein/E2F/cyclin E pathway but is not directly required for macrophage morphology. The Journal of Biological Chemistry. 2011;286(26):22716–22729. doi: 10.1074/jbc.M110.152538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nerlov C. Transcriptional and translational control of C/EBPs: the case for “deep” genetics to understand physiological function. BioEssays. 2010;32(8):680–686. doi: 10.1002/bies.201000004. [DOI] [PubMed] [Google Scholar]
- 67.Marcinkowska E, Garay E, Gocek E, Chrobak A, Wang X, Studzinski GP. Regulation of C/EBPβ isoforms by MAPK pathways in HL60 cells induced to differentiate by 1,25-dihydroxyvitamin D3 . Experimental Cell Research. 2006;312(11):2054–2065. doi: 10.1016/j.yexcr.2006.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marcinkowska E, Wang X, Studzinski G. C/EBPb: a candidate for a major player in vitamin D-induced monocytic differentiation of human leukemia cells. In: Stolzt V, editor. Vitamin D: New Research. New York, NY, USA: Nova Science; 2006. pp. 25–40. [Google Scholar]


