Abstract
8-Nitro-2′-deoxyguanosine (8-nitrodG) is a relatively unstable, mutagenic lesion of DNA that is increasingly believed to be associated with tissue inflammation. Due to the lability of the glycosidic bond, 8-nitrodG cannot be incorporated into oligodeoxynucleotides (ODNs) by chemical DNA synthesis and thus very little is known about its physicochemical properties and base-pairing preferences. Here we describe the synthesis of 8-nitro-2′-O-methylguanosine, a ribonucleoside analogue of this lesion, which is sufficiently stable to be incorporated into ODNs. Physicochemical studies demonstrated that 8-nitro-2′-O-methylguanosine adopts a syn conformation about the glycosidic bond; thermal melting studies and molecular modelling suggest a relatively stable syn-8-nitroG·anti-G base pair. Interestingly, when this lesion analogue was placed in a primer-template system, extension of the primer by either avian myeloblastosis virus reverse transcriptase (AMV-RT) or human DNA polymerase β (pol β), was significantly impaired, but where incorporation opposite 8-nitroguanine did occur, pol β showed a 2:1 preference to insert dA over dC, while AMV-RT incorporated predominantly dC. The fact that no 8-nitroG·G base pairing is seen in the primer extension products suggests that the polymerases may discriminate against this pairing system on the basis of its poor geometric match to a Watson–Crick pair.
INTRODUCTION
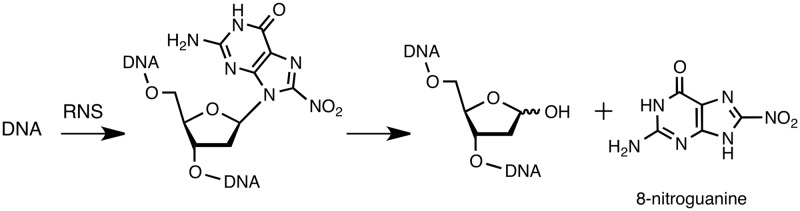
The cause and effect relationship between inflammation and cancer is now well established, with evidence suggesting that more than 20% of human cancers are attributable to various types of chronic inflammation (1–3). Specific examples include: liver cancer, which is often the consequence of inflammation caused by chronic viral hepatitis; lung cancer, which results from inflammation caused by years of inhaled tobacco smoke, and the association between inflammatory stomach infections (such as Helicobacter pylori) and gastric cancer (4,5). The proposed molecular basis for this link is through over-expression of nitric oxide synthase in inflamed tissues, which leads to increased levels of reactive oxygen and nitrogen species (RNS) that damage cellular macromolecules (6,7). The guanine base in DNA, with its low oxidation potential, is particularly sensitive and a large number of guanine lesions, particularly 8-oxodeoxyguanosine (8-oxodG), have been characterized and studied in some detail (8,9). One such modification readily identified is 8-nitro-2′-deoxyguanosine (8-nitrodG); this lesion labilizes the glycosidic bond to release 8-nitroguanine, which can be detected in urine, and leaves an abasic site in the DNA (Scheme 1) (10,11).
Scheme 1.
Nitration of the guanine base in DNA by reactive nitrogen species and release of 8-nitroguanine to leave an abasic site in DNA.
Our understanding of how chemical modification of DNA generates specific mutations has advanced rapidly, largely due to the synthesis and study of oligonucleotides containing site-specific DNA lesions. However, the sensitivity of the 8-nitrodG glycosidic bond means that the synthesis of oligodeoxynucleotides (ODNs) containing 8-nitroguanine by a conventional chemical approach has not been achieved. However, there is growing evidence to suggest that this lesion is highly mutagenic and thus warrants a detailed study of its base pairing and other physicochemical properties. Thus, in primer extension reactions with a template containing 8-nitrodG produced photochemically, Suzuki et al. showed that while DNA synthesis catalyzed by DNA polymerases α and β was partially blocked at the 8-nitrodG lesion, dA was incorporated opposite the lesion (equivalent to G·C to T·A transversion mutation) to a significant extent and a possible 8-nitroG·A base pairing scheme was suggested to explain this observation (12). However, these experiments were conducted with a template-primer system that readily releases 8-nitroguanine to leave abasic sites, which are known cause the same transversion mutation (13,14), although control experiments suggested that depurination was minimal during the experiment. Kaneko et al. also observed the same G·C to T·A mutation, together with increased levels of abasic sites, when a hamster cell line was treated with 8-nitroguanosine (15). Their results were consistent with mutations arising from the incorporation of 8-nitroguanine into DNA (through the salvage pathway), although in this case, the mutational effect was largely attributed to the formation of abasic sites. Despite the documented mutagenicity of this lesion (12,15), there is currently no information on how the physicochemical properties, such as pKa, nucleoside conformation and base pairing preferences of dG are affected by introduction of the 8-nitro group. To obtain a better understanding of the mutagenic effect of this lesion, we now describe the synthesis of ODNs containing 8-nitro-2′-O-methylguanosine, in which the glycosidic bond is stabilized by a ribose sugar (16). The physicochemical properties of this lesion are reported and the data suggest that the most stable base pairing arrangement is an 8-nitroguanine—guanine pair. The mutational behaviour of this lesion has also been investigated in template-directed DNA synthesis using a template containing a stable 8-nitroguanine base, and DNA polymerase β prefers to insert dA opposite the lesion.
MATERIALS AND METHODS
2′-O-Methylguanosine was purchased from ChemGenes Corporation (Wilmington, USA), all reagents and standard nucleoside phosphoramidites for oligonucleotide synthesis were purchased from Link Technologies (Bellshill, Scotland). Unless otherwise stated, all other reagents and solvents for chemical synthesis were purchased from Sigma, TCI or Fisher Scientific and used as supplied.
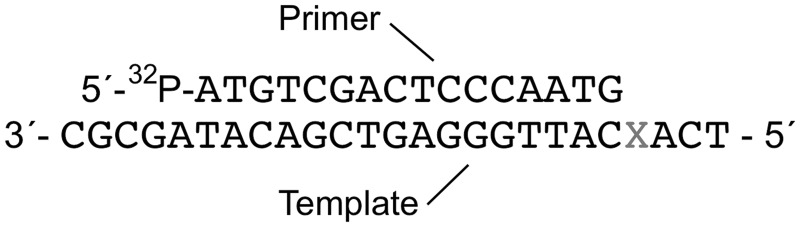
Purified human DNA polymerase β was the kind gift of Dr Joann B. Sweasy, Yale University School of Medicine. Avian Myeloblastosis Virus reverse transcriptase (AMV-RT) and T4 polynucleotide kinase were purchased from New England Biolabs (UK). [γ-32P]ATP (6000 Ci mmol−1) was supplied by Perkin Elmer. Deoxyribonucleotide triphosphates were supplied by Pharmacia. G-25 Sephadex mini Quick Spin Oligo columns were purchased from Roche Applied Science. ZipTipC18 pipette tips were purchased from Millipore. Four 20-mer ODNs serving as DNA sequencing standards (sequence 5′–ATGTCGACTCCCAATGNTGA–3′, containing either A, T, C or G at position N) were synthesized and purified by Eurofins MWG Operon (UK).
Solid-phase oligonucleotide synthesis was carried out on a BioAutomation Corporation MerMade 4® synthesizer equipped with MerMade 12© v2.3.2 software. All syntheses were undertaken using 1.0 μmol, 500 Å controlled-pore glass (CPG) columns purchased from Link Technologies. HPLC was performed on an automated Gilson HPLC system equipped with an autoinjector, a photodiode array detector and dual hydraulic pump. Chromatographic data were controlled and processed using UniPoint© Version 3.0. All oligonucleotides were purified by reverse-phase using a Gemini™ C18 column (110 Å, 250 mm × 4.6 mm) purchased from Phenomenex using a triethylammonium bicarbonate (TEAB) buffer system. A 1 M stock solution of TEAB was prepared by bubbling CO2 through a 1 M aqueous solution of Et3N for 6–8 h until the pH measured 7.5–7.6. Stock solutions of TEAB were refrigerated until needed. All UV measurements were taken on a PerkinElmer Lambda 25 spectrophotometer equipped with a Peltier temperature controller and water circulator using standard quartz cuvettes with a 1 cm path length. TempLab 2.0 software was used to control heating and record absorbencies for thermal melting studies.
All NMR spectra were recorded on a Bruker 400 MHz spectrometer; 1H (400 MHz), 13C (100 MHz) and 31P (162 MHz). All samples were run in deuterated chloroform (CDCl3) or deuterated dimethyl sulfoxide (D6-DMSO) as specified under Preparation of Nucleosides section. All chemical shifts are reported in p.p.m. and coupling constants (J) in Hz. 13C and 31P spectra were 1H decoupled. 1H and 13C chemical shifts are relative to an internal standard of tetramethylsilane, 31P spectra are relative to an external standard of 85% H3PO4. Mass spectra of nucleosides and chemically synthesized ODNs were recorded on a MicroMass LCT ™ mass spectrometer using electrospray ionization (ES) and direct infusion syringe pump sampling. All small molecules were dissolved in methanol. Oligonucleotides were dissolved in water and analysed using negative mode ES ionization (ES-). Theoretical masses for oligonucleotides were calculated using a spreadsheet created in Microsoft® Excel. Mass spectra of primer extension assays were collected using a Bruker UltrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, UK). The instrument was operated in a reflectron, positive ion mode. A panel of six oligonucleotides, in the Mr range 4400–7100, was used to calibrate the mass spectrometer. Baseline subtraction and peak detection were performed using Bruker Daltonics flexAnalysis software.
Preparation of nucleosides
2′,3′,5′-Tri-O-acetylguanosine (2a) and 8-bromo-2′,3′,5′-tri-O-acetylguanosine (3a) were prepared as previously described (17,18).
3′,5′-Di-O-acetyl-2′-O-methylguanosine (2b)
To a stirred suspension of 2′-O-methylguanosine (4.00 g, 13.50 mmol) in MeCN (50 ml) was added Et3N (4.95 ml, 35.50 mmol) and DMAP (0.16 g, 1.35 mmol). The resulting suspension was cooled in an ice bath prior to addition of Ac2O (3.19 ml, 33.75 mmol). Once the addition was complete the mixture was allowed to reach room temperature and was stirred for a further 2 h. The reaction was quenched by the addition of MeOH (5 ml) and concentrated in vacuo to a white semi-solid which was triturated with boiling iPrOH to afford the title product as a white solid (4.80 g, 93%). 1H NMR (400 MHz, D6-DMSO): δ (ppm) 10.70–10.80 (1H, bs, H1); 8.00 (1H, s, H8); 6.55–6.70 (2H, bs, C2 NH2); 5.80–5.85 (1H, d, J = 7.6 Hz, H1′); 5.35–5.40 (1H, d, J = 5.0 Hz, H2′); 4.60–4.65 (1H, dd, J = 7.6 and 5.0 Hz, H3′); 4.20–4.35 (3H, m, H4′, H5′ and H5′′); 3.25–3.27 (3H, s, 2′-OMe); 2.15–2.16 (3H, s, 3′-OAc); 2.05–2.06 (3H, s, 5′-OAc). 13C NMR (100 MHz, D6-DMSO): δ (ppm) 170.56, 169.99, 157.05, 154.28, 151.76, 135.50, 117.04, 84.48, 80.14, 80.11, 71.05, 63.74, 58.61, 21.01, 20.96. HRMS (ES+) (m/z): Found, 404.1181 [M+Na]+; C15H19N5O7Na requires 404.1182.
8-Bromo-3′,5′-diacetyl-2′-O-methylguanosine (3b)
3′,5′-Di-O-acetyl-2′-O-methylguanosine (2b) (4.75 g, 12.48 mmol) was suspended in distilled water (30 ml). Saturated bromine water (ca. 20 ml) was added with vigorous stirring at room temperature at such a rate that the yellow colour dissipated between additions. Once the yellow colour persisted the mixture was allowed to stir for a further 30 min before filtering. The filter cake was washed with the liquors followed by ice cold iPrOH before drying under vacuum for 12 h to yield the title product as an off-white amorphous solid (5.20 g, 90%). Rf = 0.31 (15% MeOH in DCM). 1H NMR (400 MHz, D6-DMSO): δ (ppm) 10.90–10.95 (1H, s, H1); 6.50–6.65 (2H, bs, C2 NH2); 5.75–5.80 (1H, d, J = 5.5 Hz, H1′); 5.50–5.55 (1H, m, H2′); 5.00–5.05 (1H, dd, J = 5.5 and 5.7 Hz, H3′); 4.35–4.40 (1H, dd, J = 3.4 and 10.9 Hz, H5′) 4.20–4.34 (2H, m, H4′ and H5′′); 3.25 (3H, s, 2′-OMe); 2.15–2.16 (3H, s, 3′-OAc); 2.00–2.01 (3H, s, 5′-OAc). 13C NMR (100 MHz, D6-DMSO): δ (ppm) 170.54, 169.99, 155.81, 154.07, 152.45, 120.53, 117.62, 88.19, 79.95, 78.50, 71.18, 63.49, 58.66, 21.01, 20.94. HRMS (ES+) (m/z): Found, 482.0282 and 484.0267 [M+Na]+; C15H18N5O7BrNa requires 482.0287 and 484.0267.
N2-Dimethoxytrityl-8-bromo-3′,5′-di-O-acetyl-2′-O-methylguanosine (4b) and N2-dimethoxytrityl-8-bromo-2′,3′,5′-tri-O-acetylguanosine (4a)
8-Bromo-3′,5′-di-O-acetyl-2′-O-methylguanosine (3b) (5.18 g, 11.30 mmol) was co-evaporated to dryness with anhydrous pyridine before re-dissolving in anhydrous pyridine (35 ml). The resultant solution was stirred at room temperature under an atmosphere of N2 and DMTCl (4.20 g, 12.40 mmol) was added in ca. 0.5 g portions over 5 min. The mixture was stirred for a further 8 h at room temperature before removing most of the pyridine under reduced pressure. The isolated sticky solid was partitioned between DCM (50 ml) and water (50 ml), the DCM layer was further washed with water (2 × 50 ml) and saturated aqueous NaHCO3 (30 ml) before drying over Na2SO4, filtering and evaporating to a yellow solid. Flash column chromatography on silica gel (DCM, MeOH and Et3N 19:1:0.1) led to isolation of product 4b as a pale yellow solid (6 g, 70%). Rf = 0.21 (4:6 EtOAc and DCM). 1H NMR (400 MHz, CDCl3): δ (ppm) 11.10–11.45 (1H, bs, H1); 7.65–7.80 (1H, bs, C2 NH); 7.05–7.45 (9H, m, DMT Ar-H); 6.75–6.90 (4H, m, DMT Ar-H); 5.55–5.60 (1H, d, J = 6.4 Hz, H1′); 5.05–5.10 (1H, m, H2′); 4.15–4.20 (1H, m, H3′); 4.05–4.10 (1H, m, H4′); 4.00–4.10 (1H, m, H5′); 3.78–3.80 (6H, s, 2 x Ar-OMe); 3.70–3.78 (1H, m, H5′′); 2.80–2.82 (3H, s, 2′-OMe); 2.15–2.16 (3H, s, 3′-OAc); 2.05–2.06 (3H, s, 5′-OAc). 13C NMR (100 MHz, CDCl3): δ (ppm) 170.75, 170.27, 158.88, 158.83, 152.32, 152.11, 130.34, 128.81, 128.38, 127.49, 122.57, 119.05, 113.77, 113.67, 88.87, 80.03, 71.46, 70.50, 63.23, 58.86, 55.54, 21.47, 21.21. MS (ES+) (m/z): Found, 784/786 [M+Na]+ and 863/865 [M+Et3NH]+.
Compound (4a) was prepared in a similar fashion to compound (4b) using (3a) (1 g, 2.05 mmol), DMTCl (0.83 g, 2.49 mmol) and pyridine (10 ml). The (4a) was isolated as a pale yellow foam after flash column chromatography (CHCl3 to 98:2 CHCl3 and MeOH), (1.53 g, 94%). Rf = 0.48 (1:9 MeOH and DCM).1H NMR (400 MHz, CDCl3): δ (ppm) 8.21–8.70 (1H, bs, H1); 7.21–7.35 (9H, m, DMT Ar-H); 6.86–6.90 (1H, bs, C2 NH) 6.79–6.87 (4H, m, DMT Ar-H); 6.03–6.08 (1H, m, H1′); 5.84–5.89 (2H, m, H2′ and H3′); 4.26–4.37 (2H, m, H4′ and H5′); 4.16–4.22 (1H, dd, J = 5.9 and 11.4 Hz, H5′′); 3.78–3.80 (6H, s, 2 x Ar-OMe); 2.11–2.12, 2.05–2.06 and 1.98–1.99 (9H, 3 x s, 2′,3′,5′-OAc). 13C NMR (100 MHz, CDCl3): δ (ppm) 170.91, 169.93, 169.71, 159.37, 152.21, 151.77,135.48, 130.16, 128.99, 128.59, 128.12, 118.70, 114.35, 88.12, 79.35, 72.08, 70.70, 70.69, 63.28, 55.67, 21.08, 20.99, 20.87. HRMS (ES+) (m/z): Found, 812.1512 and 814.1514 [M+Na]+ C37H36N5O10BrNa requires 812.1543 and 814.1514.
N2-Dimethoxytrityl-8-nitro-3′,5′-di-O-acetyl-2′-O-methylguanosine (5b) and N2-dimethoxytrityl-8-nitro-2′,3′,5′-tri-O-acetylguanosine (5a)
Protected bromoguanosine (4b) (2.00 g, 3.58 mmol), was dissolved in a mixture of DMF (5 ml) and 18-crown-6 (9.40 g, 35.80 mmol) heated to 60°C. KNO2 (3.00 g, 35.80 mmol) was added and the resultant mixture was stirred at 100°C for 6 h under an inert atmosphere before cooling to room temperature and pouring directly into water (50 ml). The precipitate that formed was extracted with EtOAc (50 ml) and the aqueous solution was further extracted with EtOAc (2 × 50 ml). The combined organic solutions were washed with saturated aqueous NaHCO3 (50 ml), dried (Na2SO4), filtered and concentrated to a yellow foam. Flash column chromatography on silica gel (1:1 EtOAc and hexane) afforded the title product as a yellow amorphous solid (1.12 g, 43%). Rf = 0.44 (4:6 EtOAc and DCM). 1H NMR (400 MHz, CDCl3): δ (ppm) 11.25–11.55 (1H, bs, H1); 8.05–8.20 (1H, bs, C2 NH); 7.05–7.45 (9H, m, DMT Ar-H); 6.75–6.90 (4H, m, DMT Ar-H); 5.55–5.60 (1H, m, H1′); 5.05–5.10 (1H, m, H2′); 4.20–4.25 (1H, m, H3′); 4.14–4.20 (1H, dd, J = 3.3 and 12.3 Hz, H5′); 4.00–4.08 (1H, dd, J = 6.0 and 9.7 Hz, H4′); 3.90–3.96 (1H, dd, J = 6.0 and 12.3 Hz H5′′); 3.78–3.80 (6H, s, 2 x Ar-OMe); 2.80–2.82 (3H, s, 2′-OMe); 2.15–2.16 (3H, s, 3′-OAc); 2.05–2.07 (3H, s, 5′-OAc). 13C NMR (100MHz, CDCl3): δ (ppm) 170.54, 169.99, 159.14, 158.90, 158.87, 153.39, 152.29, 145.03, 139.38, 136.02, 130.63, 130.42, 129.55, 128.94, 128.19, 127.40, 115.38, 113.65, 113.52, 90.33 79.53, 79.03, 71.29, 70.66, 62.51, 58.60, 55.61, 21.47, 21.21. HRMS (ES+) (m/z): Found, 751.2346 [M+Na]+; C36H36N6O11Na requires 751.2340.
Compound (5a) was prepared from compound (4a) (1 g, 1.32 mmol), KNO2 (1.11 g, 13.2 mmol), 18-Crown-6 (3.49 g, 13.2 mmol) and DMF (20 ml) in an identical manner to compound (5b) affording a yellow amorphous solid (0.36 g, 36%). Rf = 0.49 (4:6 EtOAc and DCM). 1H NMR (400 MHz, CDCl3): δ (ppm) 11.06–11.50 (1H, bs, H1); 7.80–7.99 (1H, bs, C2 NH); 7.20–7.34 (9H, m, DMT Ar-H); 6.82–6.87 (4H, m, DMT Ar-H); 6.01–6.06 (1H, m, H1′); 5.83–5.90 (2H, m, H2′ and H3′); 4.26–4.39 (2H, m, H4′ and H5′); 4.16–4.24 (1H, dd, J = 5.9 and 11.5 Hz, H5′′); 3.78–3.80 (6H, s, 2 x Ar-OMe); 2.11–2.12, 2.05–2.06 and 1.98–1.99 (9H, 3 x s, 2′,3′,5′-OAc). 13C NMR (100MHz, CDCl3): δ (ppm) 170.96, 169.75, 169.46, 158.96, 153.58, 152.41, 143.84, 130.50, 130.45, 129.54, 128.90, 128.37, 127.62, 115.52, 113.80, 88.81, 78.98, 72.37, 71.45, 69.47, 62.44, 55.62, 21.48, 21.02, 20.83. HRMS (ES+) (m/z): Found, 779.2292 [M+Na]+; C37H36N6O12Na requires 779.2289.
N2-Dimethoxytrityl-2′-O-methyl-8-nitroguanosine (7)
Protected 2′-O-methylnitroguanosine (5b) (0.43 g, 0.59 mmol) was dissolved in 7M NH3 (anhydrous) in MeOH (5 ml) and stirred in a sealed vessel for 48 h. The solvent was removed in vacuo and residue purified by flash column chromatography on silica gel (1:1 EtOAc, DCM), yielding the deacylated nucleoside as a yellow amorphous solid (0.36 g, 94%). Rf = 0.19 (40% EtOAc in DCM). 1H NMR (400 MHz, MeOD): δ (ppm) 7.17–7.30 (9H, m, DMT Ar-H); 6.83–6.87 (4H, m, DMT Ar-H); 5.81–5.82 (1H, d, J = 3.0 Hz, H1′); 3.72–3.81 (m, 8H, 2 x Ar-OMe, H2′, H3′); 3.58–3.64 (1H, dt, J = 7.2 and 2.9 Hz, H4′); 3.51–3.56 (1H, dd, J = 12.2 and 2.9 Hz, H5′); 3.36–3.42 (1H, dd, J = 12.2 and 7.2 Hz, H5′′); 3.00–3.01 (3H, s, 2′-OMe). 13C NMR (100 MHz, MeOD): δ (ppm) 160.26, 159.13, 154.57, 152.81, 146.25, 145.38, 138.05, 137.45, 131.39, 131.29, 129.98, 129.37, 129.28, 116.69, 114.57, 91.32, 85.60, 81.46, 72.07, 70.75, 63.66, 58.69, 55.96. HRMS (ES+) (m/z): Found, 667.2154 [M+Na]+; C32H32N6O9Na requires 667.2128.
5′-O,N2-bis-dimethoxytrityl-2′-O-methyl-8-nitroguanosine (8)
N2-Dimethoxytrityl-2′-O-methyl-8-nitroguanosine (7) (0.29 g, 0.45 mmol) was co-evaporated with anhydrous pyridine before re-dissolving in pyridine (3 ml) and adding DMTCl (0.20 g, 0.60 mmol). The solution was stirred for 16 h under an inert atmosphere at room temperature. The pyridine was removed in vacuo and the residue subject to flash column chromatography on silica gel (1:4 EtOAc, hexane) affording the title compound as a yellow solid (0.34 g, 80%). Rf = 0.67 (40% EtOAc in DCM). 1H NMR (400 MHz, CDCl3): δ (ppm) 11.30–11.55 (1H, bs, H1); 7.90–8.00 (1H, bs, C2 NH); 6.95–7.30 (18H, m, DMT Ar-H); 6.64–6.89 (8H, m, DMT Ar-H); 5.50–5.55 (1H, m, H1′); 3.74–3.84 (2H, m, H2′ and H3′); 3.41–3.70 (13H, m, H4′ and 4 x Ar-OMe); 3.05–3.20 (2H, m, H5′ and H5′′); 2.91–2.95 (3H, s, 2′-OMe); 2.41–2.55 (1H, bs, 3′-OH). 13C NMR (100 MHz, CDCl3): δ (ppm) 159.24, 153.38, 152.21, 145.24, 136.57, 130.98, 130.60, 129.57, 128.91, 128.51, 127.39, 127.11, 126.96, 115.42, 113.74, 111.75, 90.14, 86.54, 83.43, 80.61, 71.34, 69.72, 60.88, 58.21, 55.67. HRMS (ES+) (m/z): Found, 969.3481 [M+Na]+; C53H50N6O11Na requires 969.3435.
Phosphoramidite of 5′-O,N2-bis-dimethoxytrityl-2′-O-methyl-8-nitroguanosine (9)
5′-O,N2-bis-dimethoxytrityl-2′-O-methyl-8-nitroguanosine (8) (0.51 g, 0.53 mmol) was dissolved in anhydrous DCM (5 ml) and cooled to 0°C under an inert atmosphere. Diisopropyl ethylamine (1.15 ml, 6.61 mmol) was added followed by drop-wise addition of 2-cyanoethyl diisopropylchlorophosphoramidite (0.15 ml, 0.66 mmol). The solution was allowed to warm to room temperature and stir for 3 h before adding water (10 ml) and DCM (20 ml). The phases were separated and the organic solution dried (Na2SO4), filtered and concentrated to a yellow solid prior to flash column chromatography (1:4 EtOAc, hexane). Phosphoramidite 9 was isolated as a yellow foam containing both diastereoisomers (0.41 g, 67%). Rf = 0.40 (20% EtOAc in DCM). 31P NMR (160 MHz, CDCl3): δ (ppm) 150.81, 149.25. HRMS (ES-) (m/z): Found, 1145.4517 [M-H]-; C62H66N8O12P requires 1145.4538.
8-Nitroguanosine (6a)
Compound (5a) (40 mg, 0.05 mmol) was dissolved in CHCl3 before adding a solution of TsOH (10 mg, 0.06 mmol) in MeOH (0.5 ml). After 10 min the resulting orange solution was evaporated to dryness and triturated with cold Et2O before dissolving the residue in 7 M NH3 in MeOH, in a sealed vessel and stirring at room temperature for 48 h. The solvent and NH3 were removed in vacuo leaving an orange solid which was washed with Et2O and identified as the deprotected ribonucleoside (12 mg, 73%). 1H NMR (400 MHz, D6-DMSO): δ (ppm) 9.73–9.98 (1H, bs, H1); 7.12–7.30 (2H, bs, C2 NH2); 6.24–6.26 (1H, d, J = 5.2 Hz, H1′); 5.35–5.40 (1H, bs, OH); 5.01–5.07 (1H, bs, OH); 4.86–4.90 (1H, m, H2′); 4.17–4.21 (1H, m, H3′); 3.81–3.86 (1H, dd, J = 4.8 and 10.2 Hz, H4′); 3.64–3.69 (1H, dd, J = 11.9 and 3.0 Hz, H5′); 3.47–3.53 (1H, dd, J = 11.9 and 5.5 Hz, H5′′). 13C NMR (100 MHz, D6-DMSO): δ (ppm) 158.11, 157.09, 153.60, 143.10, 115.54, 88.71, 85.24, 80.50, 70.64, 60.31. HRMS (ES-) (m/z): Found, 327.0698 [M-H]-; C11H13N6O7.requires 327.0689. λmax (10 mM Tris-HCl pH 7.4) 231 nm, ε = 9798 M-1; 398 nm, ε = 8109 M-1
8-Nitro-2′-O-methylguanosine (6b)
Compound (5b) (0.12 g, 0.16 mmol) was dissolved in CHCl3 (2 ml) before adding a solution of TsOH (32 mg, 0.17 mmol) in 5:1 CHCl3 and MeOH (1 ml). After 15 min the solvent was removed in vacuo leaving an orange gum, which was triturated with Et2O to afford the crude base deprotected nucleoside as an orange solid. This was dissolved in 7 M NH3 in MeOH (2 ml) and stirred in a sealed vessel at room temperature for 48 h. The precipitated solid was isolated by drawing off the solution with a syringe and was triturated with Et2O three times to afford compound 6b as an orange solid (44 mg, 81%). 1H NMR (400 MHz, D6-DMSO): δ (ppm) 9.90–10.1 (1H, bs, H1); 7.15–7.39 (2H, bs, C2 NH2); 6.32–6.34 (1H, d, J = 4.8 Hz, H1′); 5.02–5.20 (1H, bs, 3′-OH); 4.86–4.97 (1H, bs, 5′-OH); 4.63–4.67 (1H, dd, J = 5.8 and 4.8 Hz, H2′); 4.35–4.40 (1H, dd, J = 5.9 and 5.8 Hz, H3′); 3.79–3.84 (1H, ddd, J = 6.0, 5.9 and 3.9 Hz, H4′); 3.64–3.69 (1H, dd, J = 11.8 and 3.9 Hz, H5′); 3.47–3.53 (1H, dd, J = 11.8 and 6.0 Hz, H5′′); 3.58–3.60 (3H, s, 2′-OMe). 13C NMR (100 MHz, D6-DMSO): δ (ppm) 157.81, 156.33, 152.60, 143.10, 115.29,88.74, 85.64, 80.50, 69.14, 61.64, 57.86. HRMS (ES-) (m/z): Found, 341.0843 [M-H]-; C11H13N6O7.requires 341.0843. λmax (10 mM Tris-HCl pH 7.4) 231 nm, ε = 9874 M−1; 396 nm, ε = 8147 M−1
2′-O-Methyl-8-bromoguanosine
Compound (3b) (0.1 g, 0.21 mmol) was dissolved in 7 M NH3 (anhydrous) in MeOH (3 ml) and stirred in a sealed vessel for 48 h. The resulting precipitate was isolated by drawing off the solution with a syringe and was triturated with Et2O three times to afford compound 9 as a white solid (76 mg, 90%). 1H NMR (400 MHz, D6-DMSO): δ (ppm) 9.70–9.85 (1H, bs, H1); 6.59–6.71 (2H, bs, C2 NH2); 5.76–5.79 (1H, d, J = 6.4 Hz, H1′); 5.07–5.24 (1H, bs, 3′-OH); 4.90–5.05 (1H, bs, 5′-OH); 4.73–4.78 (1H, dd, J = 6.4 and 5.7 Hz, H2′); 4.35–4.40 (1H, dd, J = 5.7 and 3.6 Hz, H3′); 3.86–3.91 (1H, m, H4′); 3.63–3.69 (1H, dd, J = 11.9 and 4.9 Hz, H5′); 3.50–3.56 (1H, dd, J = 11.9 and 5.4 Hz, H5′′); 3.29–3.31 (3H, s, 2′-OMe). 13C NMR (100 MHz, D6-DMSO): δ (ppm) 156.23, 154.23, 152.43, 120.99, 117.87, 88.01, 86.67, 79.79, 69.27, 62.21, 57.91. HRMS (ES+) (m/z): Found, 398.0091 and 400.0061 [M+Na]+; C11H14N5O5Br.requires 398.0076 and 400.0056.
Oligonucleotide synthesis and purification
Standard 3′–5′ synthesis procedures were employed with extended coupling times (3 min) for 2′-O-Me phosphoramidites. Cleavage of the oligonucleotides from the linker and removal of base labile protecting groups was achieved by heating the support-bound oligomers at 55°C in 33.3% ammonium hydroxide for 8 h. The 5′-DMT group was retained to facilitate purification by preparative reverse-phase HPLC prior to lyophilization and DMT removal by treatment with 20% aqueous AcOH for 15 min at room temperature. Removal of the AcOH solution in vacuo was followed by partitioning between water and EtOAc to remove the DMT residues. Purity of the oligonucleotides was measured by HPLC with UV detection at 254 nm, quantification was achieved by determination of the concentration of aqueous solutions of the oligos from their UV absorbencies at 260 nm.
A list of all the ODNs prepared together with their retention times on HPLC and analysis by electrospray mass spectrometry is presented in the Supplementary Material.
PAGE analysis of primer extension reactions
Primer extension reactions were performed in a buffer containing 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 10 mM DTT, 250 μg ml-1 BSA. For gel-based analyses, the 16-mer primer (5′–ATGTCGACTCCCAATG–3′) was labelled at the 5′-end with [γ-32P]ATP (6000 Ci mmol-1) and T4 polynucleotide kinase. Unincorporated radionucleotide was removed by size-exclusion chromatography using G-25 Sephadex mini Quick Spin columns. 10 µl primer extension reactions were initiated by mixing 5 µl each of two solutions. One solution was prepared by incubating purified [γ-32P]-labelled primer and template DNA (5′-TCANCATTGGGAGTCGACATAGCGC–3′, N = 2′-O-methylguanosine or 8-nitro-2′-O-methylguanosine) in a 1:2 molar ratio in reaction buffer at 70°C for 20 min followed by cooling at room temperature for 15 min. AMV-RT or human DNA polymerase β was then added to the annealed primer-template solution. The second solution was made by heating various concentrations of single dNTPs or all four dNTPs in reaction buffer at 37°C for 10 min. The final concentrations of primer extension components were 20 nM primer, 40 nM template, and 2 U µl-1 AMV-RT or 750 nM human DNA polymerase β. Reactions were incubated at 37°C for 60 min then quenched with the addition of 10 µl gel loading dye (98% (v/v) formamide, 10 mM EDTA, 0.1% (w/v) bromophenol blue, 0.1% (w/v) xylene cyanol). Primer extension products were separated on an 8 M urea/18% (w/v) (acrylamide to bisacrylamide ratio 29:1) polyacrylamide gel (20 cm × 47 cm × 0.04 cm) that was electrophoresed at 2200 V until the bromophenol blue had migrated 35 cm. Gel bands were visualized by phosphorimaging and band intensities quantified with ImageQuant™ software (GE Healthcare). Tabulated band intensities derived from Figures 5 and 6, calculated as percentage of total band intensities in a gel lane, are presented in the Supplementary Material.
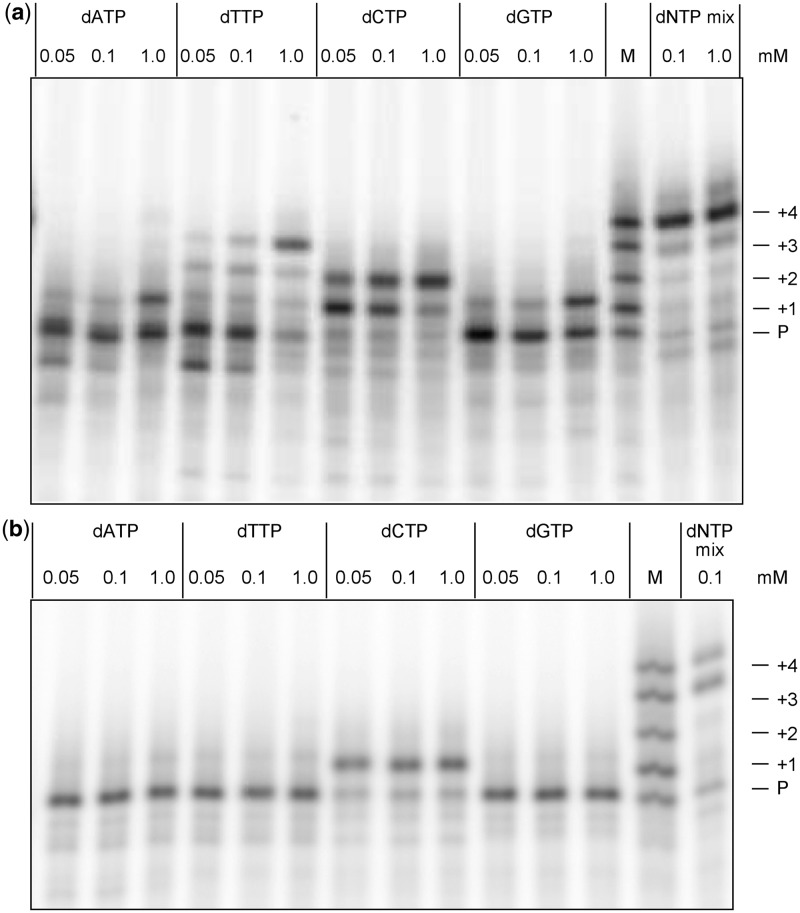
Figure 5.
Gel analysis of primer extension products synthesized by (a) AMV-RT and (b) human DNA polymerase β, from the control template containing 2′-O-methylguanosine. Varying concentrations of single deoxynucleoside triphosphates or an eqimolar mixture of all four dNTPs (dNTP mix) served as substrates for reactions as indicated. Lane M, 5′-[32P]-labelled oligonucleotide size standards consisting of primer (P; 16-mer) and synthetic oligonucleotides generated by incremental addition of nucleotides, forming a 17-mer (+1), 18-mer (+2), 19-mer (+3) and 20-mer (+4). The +4 band corresponds to a primer that has been extended to the end of the template. Some 21-mer is visible in the dNTP mix reactions of AMV-RT, and is likely to result from non-templated terminal transferase activity of that enzyme (41). Band intensities are reported in the Supplementary Material.
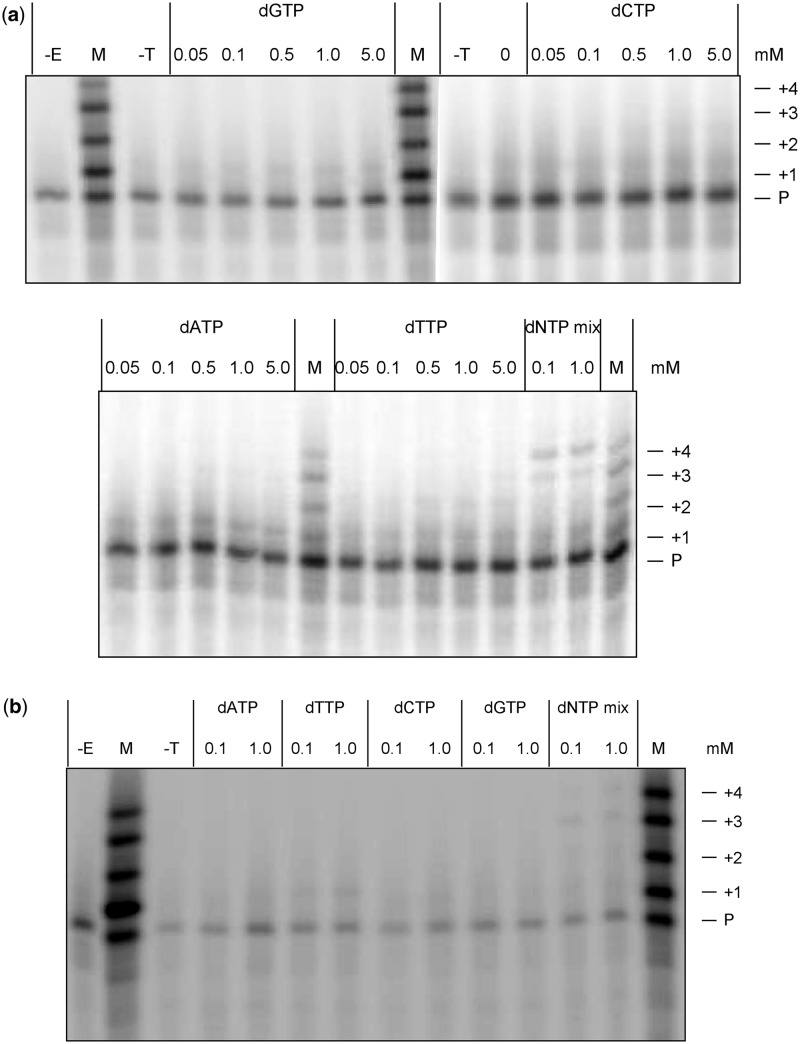
Figure 6.
Gel analysis of primer extension products synthesized by (a) AMV-RT and (b) human DNA polymerase β, from template containing 8-nitro-2′-O-methylguanosine. The experimental details are as described in Figure 5. The tracks marked –E and –T correspond to control reactions carried out without the addition of enzyme and template, respectively. Band intensities are reported in the Supplementary Material.
MALDI-TOF mass spectrometric analysis of primer extension reactions
Samples were prepared by carrying out primer extension reactions as described above with the following exceptions: (i) 10 pmol of unlabelled primer was annealed to 20 pmol template; (ii) reactions contained 1 mM each of the four deoxynucleoside triphosphates; and (iii) reactions were terminated by heating samples at 90°C for 5 min. To remove salt contaminants and concentrate extension products, reactions were processed through ZipTipC18 pipette tips. The desalting agent triethylammonium acetate, pH 7.0 was added to each primer extension reaction to give a final concentration of 0.1 M. Chromatographic tips were washed with 50% acetonitrile and equilibrated with 0.1 M triethylammonium acetate, pH 7.0. Primer extension samples were bound to the tip resin, and then washed sequentially with 0.1 M triethylammonium acetate, pH 7.0 and Milli-Q water. Oligonucleotides were eluted with 2 µl of the MALDI-TOF matrix 3-hydroxypicolinic acid (3-HPA) and spotted directly on stainless steel target plates. The matrix was prepared by vortexing a solution of 50 mg ml-1 3-HPA in 50% acetonitrile containing 10 mg ml-1 ammonium citrate for 1 min, followed by sonication for 15 min. Undissolved matrix was separated from the solution by centrifugation in a microcentrifuge at 13 000 rpm for 10 min.
Other techniques
Experimental details and data relating to the depurination studies, pKa measurements and thermal melting studies are all presented in the Supplementary Material.
RESULTS and DISCUSSION
Synthesis and hydrolytic stability of 8-nitro-2′-O-methylguanosine
8-Nitroguanine has previously been incorporated into ODNs in a site-specific manner through photochemical nitration of a single dG residue present in the sequence (12,19). This procedure gave extremely low yields (∼3% based on the starting ODNs) of the hydrolytically unstable nitrated oligonucleotides and the products were only available in nanomole quantities, which was insufficient to perform any physicochemical studies on this lesion (19). In addition, this method is not suited to the preparation of oligonucleotides containing multiple guanine bases. As described above, the approach adopted here was to prepare hydrolytically stable analogues of 8-nitrodG that could be incorporated into chemically synthesized ODNs.
Saito et al. (20) have previously described the synthesis of 8-nitroguanosine (6a, Scheme 2), in addition to its 2′-fluoro and 5′-azido derivatives, and established that these nucleosides are relatively stable to depurination (half-life of 72 and 4.5 days for the 2′-fluoro and 5′-azido derivatives, respectively, at pH 7.4). These results suggested that in order to stabilize the glycosidic bond, 8-nitroguanine could be incorporated into ODNs as its 2′-O-methyl riboside as this electronegative substituent stabilizes the glycosidic bond (21). The 2′-O-methyl nucleosides are widely available and have been extensively investigated with regard to antisense oligonucleotides and in comparison to the ribonucleoside, has the advantage that no protection of the 2′-position is necessary (22). While the 2′-fluoro modification could also be considered for this purpose, the greater electronegativity of the fluorine atom causes these analogues to adopt a sugar conformation that is even further removed from that of 2′-deoxynucleosides, when compared with ribonucleosides (23). Thus, Saito’s synthesis of 8-nitroguanosine was repeated as shown in Scheme 2 and also applied to the synthesis of 8-nitro-2′-O-methylguanosine (6b). This reaction scheme was also repeated starting with 2′-deoxyguanosine, but in this case only depurinated products were detected after nitration, confirming the extreme lability of 8-nitrodG.
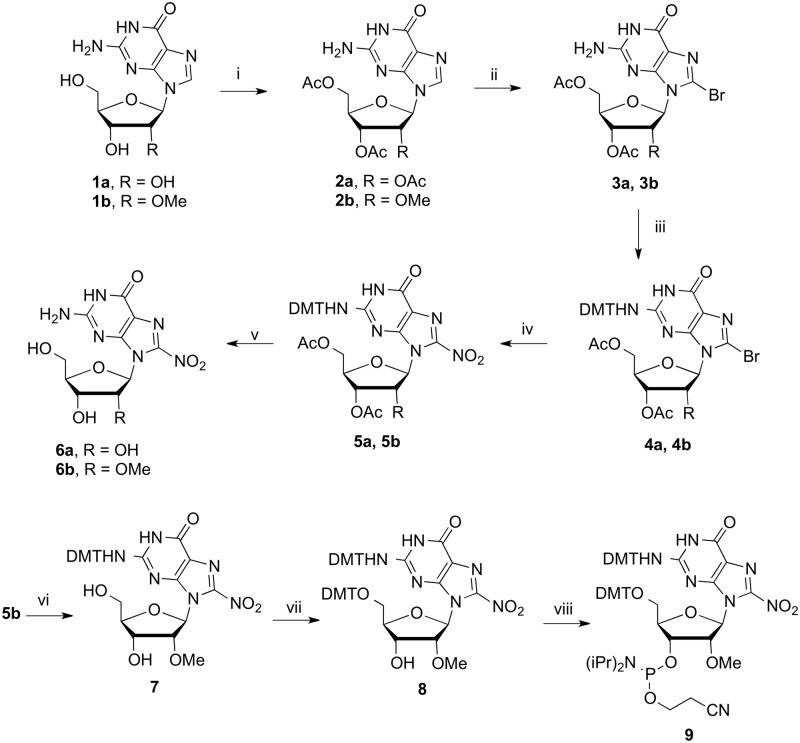
Scheme 2.
Synthesis of 8-nitroguanosine nucleosides. Reagents and conditions: (i) Ac2O (2.5 eq), Et3N (2.6 eq), DMAP (0.1 eq), MeCN 0°C, r.t. 2 h (94%, 2b) (ii) Sat. bromine water (excess), H2O, r.t. (90%, 3b) (iii) DMTCl (1.2 eq), pyridine, r.t., 6 h (94% 4a, 70% 4b); (iv) KNO2 (10 eq), 18-C-6 (10 eq), DMF, 100°C, 6 h (36% 5a, 43% 5b); (v) TsOH (1.1 eq), CHCl3, r.t. 1 h then 7 M NH3 in MeOH, r.t., 48 h (73% 6a, 81% 6b); (vi) 7 M NH3 in MeOH, r.t., 48 h (94%) (vii) DMTCl (1.2 eq), pyridine, r.t., 16 h (80%) (viii) DIPEA (12.5 eq), 2-cyanoethydiisopropylchlorophosphoramidite (1.2 eq), DCM, 0°C to r.t. 3 h (67%).
In preparation for oligonucleotide synthesis, efforts were made to introduce the usual acyl or formamidine protection at the N2 position (24) of the nitrated guanine nucleosides. However the N2 amino group proved stubbornly unreactive and these N2 protected derivatives could not be prepared by routine procedures. In an alternative approach to N2 protection, the 8-bromo nucleoside (3a) for example, could be protected at the N2 position as the isobutyryl amide or at O6 as the 2-(4-nitrophenyl)ethyl ether (24), however these compounds could not then be converted through to the 8-nitro derivative and, so far, acceptable nitration yields have only been achieved with dimethoxytrityl (DMT) protection at N2. This failure to derivatize the N2 position demonstrates that the reactivity of the guanine base is significantly reduced by the nitro group, suggesting that it should be possible to introduce 8-nitro-2′-O-methylguanosine (6b) into DNA without permanent protection of the nucleobase. Thus, through manipulation of the protecting groups, nucleoside (6b) was converted through to its phosphoramidite (9) (Scheme 2). The N2-DMT group was retained, in order to aid solubility and purification of the intermediate products, even though it would undoubtedly be removed along with the 5′-O-DMT group during oligonucleotide synthesis.
Prior to incorporation into oligonucleotides it was important to establish that 8-nitro-2′-O-methylguanosine was sufficiently resistant to depurination in order to withstand the conditions of chemical DNA synthesis and to allow unambiguous biological studies. The half-lives of both 8-nitroguanosine (6a) and 8-nitro-2′-O-methylguanosine (6b) were measured over a range of pHs and are reported in Table 1. As can be seen, (6b) had a half-life of 66.7 h (pH 7.0 and 37°C), which dropped to 11.0 h at pH 2.0. Interestingly, the ribose derivative (6a) had a significantly shorter half-life at both pHs (22.1 h and 5.35 h at pHs 7.1 and 2.0, respectively). Prior to this work there was one published study on the hydrolytic stability of 8-nitroguanosine (6a), which reported a shorter half-life of 5 h (pH 7 and 37°C). The results presented here show a half-life for the 8-nitro-2′-O-Me-guanosine, which is closer to that recently published for the related 8-nitro-5′-azidoguanosine [half-life of 108 h at a slightly higher pH (pH 7.4 and 37°C)] (20).
Table 1.
Half-lives for the hydrolysis of 8-nitroguanosine nucleosides at pHs, 2.1, 5.0 and 7.1 and at 37°C
| Half-life (h) |
|||
|---|---|---|---|
| pH 2.1 | pH 5.0 | pH 7.1 | |
| 8-NO2-G (6a) | 5.35 | 15.1 | 22.1 |
| 8-NO2-2′-O-Me-G (6b) | 11.0 | 26.4 | 66.7 |
Experimental details and the plotted data are presented in the Supplementary Material.
It is useful to compare these results to studies previously conducted on the depurination of 2′-deoxy-8-nitroguanosine either in DNA or ODNs. For example, the half-life for the release of 8-nitroguanine from a 13-mer ODN was measured (25) to be 1 h at 37°C (pH 7.2), while for an 11-mer ODN the depurination half-life (19) was 20 h at 23°C (pH 7), which was extrapolated to a half-life of 2 h at 37°C. Interestingly, in double-stranded calf-thymus DNA, 8-nitro-dG appears to be more stable (half-life ∼4 h, 37°C, pH 7.4), (10) indicating that secondary structure and other factors such as sequence context are also likely to influence the rate of depurination. It is clear that the 2′-O-methyl group exploited here considerably stabilizes the glycosidic bond, as compared with 8-nitrodG.
Synthesis and physiochemical studies on ODNs containing 8-NO2-2′-O-Me-G
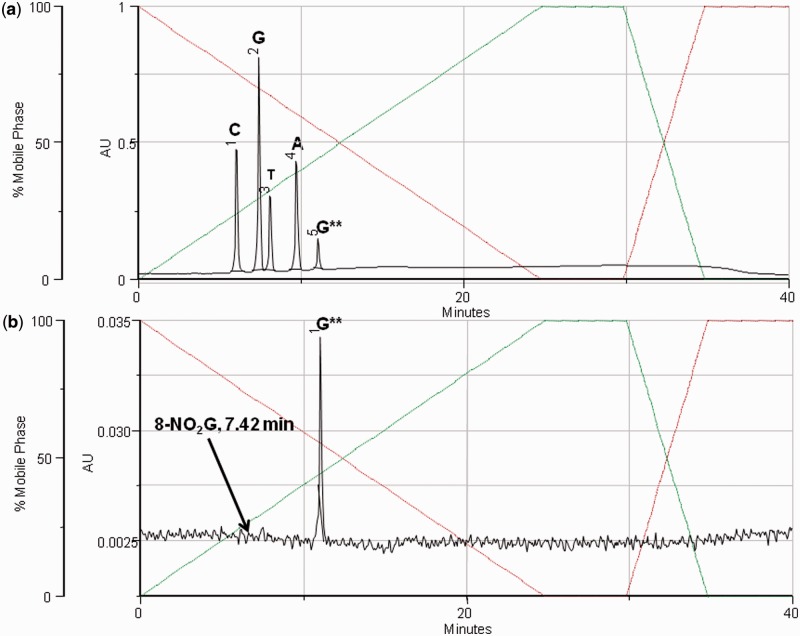
ODNs containing 8-nitro-2′-O-methylguanosine were prepared using phosphoramidite (9) (Scheme 1) in a standard coupling procedure. ODNs were purified as their 5′-DMT derivatives by reverse-phase HPLC and the DMT group finally removed by treatment with acetic acid:water (20:80, 15 min). This deprotection step was deliberately kept short to minimize the possibility of depurination. If necessary oligonucleotides were repurified to ∼98–99% purity, as determined by HPLC and characterized by mass spectrometry (Supplementary Material). The presence of 8-nitro-2′-O-methylguanosine within the oligomer could also be established by enzymatic hydrolysis to the constituent nucleosides and analysis of the hydrolysate by HPLC (Figure 1). The HPLC analysis was also monitored at 350 nm (Figure 1), which specifically reveals 8-nitro-2′-O-methylguanosine. The chromatogram shows no observable peak corresponding to the depurination product 8-nitroguanine, which also absorbs at this wavelength.
Figure 1.
Top trace shows the nucleoside composition analysis of GCGTACG**CATGCG obtained by enzymatic digestion with SVPD and alkaline phosphatase, followed by analysis by RP-HPLC at 260 nm. The bottom trace was recorded at 350 nm, where only 8-nitroguanine derivatives have a significant absorbance. The elution time for 8-nitroguanine is marked on the chromatogram. (G** = 8-nitro-2′-O-methylguanosine).
To analyse the ODNs for contamination with abasic sites resulting from loss of 8-nitroguanine, GCGTACG**CATGCG (G** = 8-nitro-2′-O-methylguanosine) was treated at pH 5 and analysed by HPLC. The chromatogram (Supplementary Figure S2b) revealed a new peak with the expected slightly shorter retention time corresponding to the ODN product containing the abasic site (26). The 8-nitro-2′-O-methylguanosine appeared to be considerably more resistant to hydrolysis when incorporated into ODNs and the HPLC peak attributed to depurination was only observed in significant quantities at pH 2. However, at this pH the ODNs also began to decompose further and thus the depurinated ODN could not be isolated and characterized and the half-life of depurination within the ODN could not be measured. Prior to treatment at pH 5 the extent of depurination in the purified ODN sample was estimated to be less than 2%.
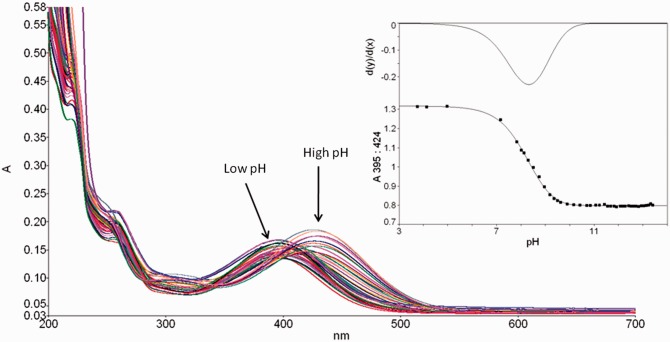
pKa is an important property of nucleosides influencing their base pairing properties, since the strength of the hydrogen-bonding interactions is correlated with the relative pKa values of the donor and acceptor nucleobases (27,28). 8-Nitroguanine nucleosides have previously been shown to have a maximum absorption at around 390 nm (29). The pH-dependent shift in the absorption spectrum was used to determine the pKa for deprotonation at N1 (Figure 2). Thus, 8-nitroguanosine (6a) and 8-nitro-2′-O-methylguanosine (6b) were shown to have pKa values of 8.4 and 8.3, respectively, and hence are more acidic than guanosine (30) (pKa = 9.4) and 2′-O-methylguanosine (pKa = 9.5), due to the electron withdrawing nitro group. Due to the wavelength of absorption, the pKa of 8-nitro-2′-O-methylguanosine could also determined within the single-stranded oligonucleotide (GCGTACG**CATGCG, where G** = 8-nitro-2′-O-methylguanosine) and in this situation the pKa was increased to 9.1. The decrease in acidity of 8-nitro-2′-O-methylguanosine within the ODN is probably due to deprotonation being suppressed by the negatively charged phosphodiester backbone; the pKa will also be modulated by the sequence context (31). However, it should be noted that the conformational studies presented below largely rule out base pairing schemes involving the N1 position of the 8-nitroguanine base.
Figure 2.
pH-dependent shift in spectrophotometric absorption of 8-nitro-2′-O-methylguanosine. Insert shows the plot of pH versus absorption at 395 nm for 8-nitro-2′-O-methylguanosine and the first derivative of the curve.
To assess the thermal stability of DNA containing the 8-nitroguanine lesion, ODN duplexes were prepared in which either 2′-O-methylguanosine or its 8-nitro derivative was paired in turn with each of the DNA nucleosides and spectrophotometric thermal melting curves were recorded; the melting temperatures (Tm) are shown in Table 2. The DNA sequences used were based on those previously studied in the analysis of the base pairing preferences of 8-oxo-2′-deoxyguanosine (32). Duplexes containing the control 2′-O-methylguanosine all showed a slight destabilization (∼1-3°C) when compared with the Tm data for the corresponding duplexes containing dG (32), but importantly revealed the same trends for the melting temperatures (C >> T > G > A). These data established that 2′-O-methyl ribose is a sufficiently good mimic of 2′-deoxyribose for the purpose of these thermal melting studies. Tm data obtained for the duplexes containing 8-nitro-2′-O-methylguanosine showed a considerable lowering of the Tm for the 8-nitroG·C pairing, relative to the 2′-O-methylguanosine control (52.4°C versus 61.0°C, Table 2). While the nitro group also significantly destabilized the G·T pair, it had no effect on G·A, and it actually stabilized the G·G pair (54.2°C versus 51.0°C). Comparing the thermal stabilities of the 8-nitroG·C and 8-nitroG·G base pairings, (52.4°C and 54.2°C) the data are consistent with a greater stability for the G.G pairing (see Supplementary Material for statistical analysis).
Table 2.
Melting temperatures (average of three determinations) of a family of DNA duplexes with and without the 8-nitroG modification
| 5′-GCGTACXCATGCG | |||
|---|---|---|---|
| 3′-CGCATGYGTACGC | |||
| Y | X |
||
| dG (°C) | 2′-OMeG (°C) | 8-NO2-2′-OMeG (°C) | |
| C | 63.6 | 61.0 | 52.4 |
| T | 55.3a | 54.2 | 48.7 |
| A | 51.5a | 50.0 | 49.9 |
| G | 54.9a | 51.0 | 54.2 |
Conditions: 1 M NaCl, 0.1 M MgCl2, 60 mM sodium cacodylate (pH 7), 3µM duplex. aTaken from reference (31).
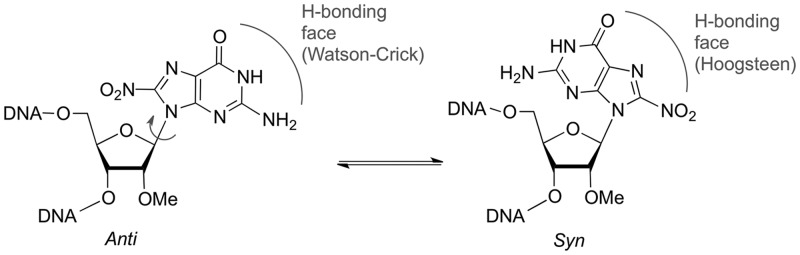
To provide further insight to the base pairing behaviour of this lesion we examined the conformation about the glycosidic bond for 8-nitro-2′-O-methylguanosine, as it determines which hydrogen bonding face is presented to the complementary nucleobase (Figure 3). It is well known that the introduction of sterically demanding groups at the 8-position of purine nucleosides favours a conformational change from anti to syn (33). NMR shifts are diagnostic of conformation and a change from anti to syn is characterized by a downfield shift of the H2′, C1′, C3′ and C4′ signals and an upfield shift of C2′ (34,35). A comparison of the NMR spectra for 2′-O-methylguanosine and its 8-nitro derivative (Table 3) showed that all of the chemical shifts indicative of a syn conformation were observed for the nitrated nucleoside. Data are also presented for 8-Br-2′-OMeG as 8-bromoguanosine nucleosides are established to adopt the syn conformation (33). Interestingly, a large downfield shift is observed for H1′ of 8-nitro-2′-O-methylguanosine and is probably due to the close proximity of the nitro group to H1′ in the syn conformation.
Figure 3.
The anti and syn conformations of 8-nitro-2′-O-methylguanosine showing the change in hydrogen bonding face.
Table 3.
Selected NMR data for studies on the syn/anti conformation
| Nucleoside | H1′ | H2′ | C1′ | C2′ | C3′ | C4′ |
|---|---|---|---|---|---|---|
| 2′-OMeG | 5.82 | 4.28 | 85.85 | 82.73 | 68.66 | 84.32 |
| 8-Br-2′-OMeG | 5.79 | 4.78 | 88.01 | 79.79 | 69.27 | 86.67 |
| 8-NO2-2′-OMeG | 6.32 | 4.68 | 89.12 | 80.88 | 69.55 | 86.02 |
1H and 13C NMR shifts of sugar protons and carbons of the 2′-O-methyl derivatives of guanosine (2′-OMeG), 8-bromoguanosine (8-Br-2′-OMeG) and 8-nitroguanosine (8-NO2-2′-OMeG), measured in D6-DMSO. Downfield and upfield chemical shifts for the syn conformation are indicated by turquoise and yellow shading, respectively.
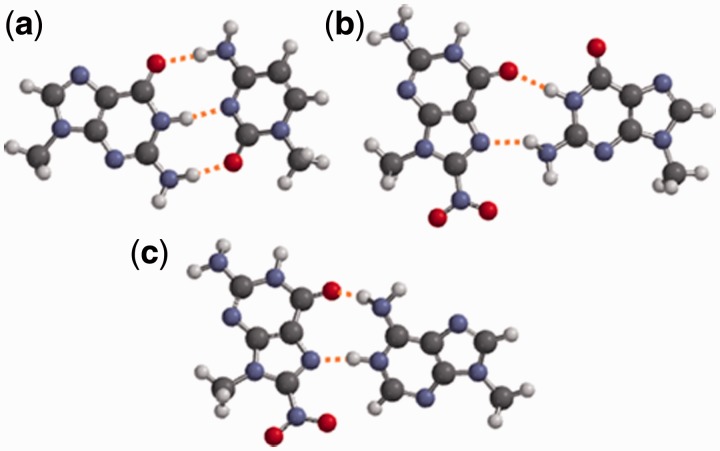
The NMR data, taken in conjunction with the results of the thermal melting studies are consistent with the 8-nitroG·C base pair being destabilized by a change from the anti to the syn conformation on introduction of the nitro group. This conformational change presents the Hoogsteen face of 8-nitroG, which can form two hydrogen bonds to a paired guanine base (Figure 4). Molecular modelling calculations performed on the N9 methylated bases, show that the syn-8-nitroG·G base pair, with its two hydrogen bonds is considerably weaker than the regular G·C pair. Anti-G·syn-A and anti-G·anti-A base pairs are both known from crystal structures (36) of oligonucleotides and anti-8-nitroG·anti-A base pairing was also proposed by Suzuki et al. (12) to explain their observed incorporation of dA opposite this lesion in their primer extension reactions. While analogous structures with 8-nitroG may form, they would be inconsistent with the preferred syn conformation suggested here by NMR. It is also of interest to compare 8-nitroG to the behaviour of the notionally similar 8-oxoG, which is established to adopt a syn-8-oxoG·anti-A base pair in crystal structures (37). However, the equivalent syn-8-nitroG·A structure is less plausible, since formation of two hydrogen bonds requires protonation (Figure 4c) and the calculated energy for this protonated syn-8-nitroG·A pair was considerably greater (−166 kJmol−1) than the aforementioned syn-8-nitroG·G pairing.
Figure 4.
Computational modelling of N9-methylated bases. Calculated using Spartan 08 v1.2.0 at AM1 level: (a) Watson–Crick G·C base pair (−701 kJ mol−1) (b) syn-8-nitroG·G (−354 kJ mol−1) (c) protonated syn-8-nitroG·A base pair (−166 kJ mol−1).
DNA synthesis directed by a template containing 8-nitroG
As discussed previously, Suzuki et al. have investigated the mutagenic behaviour of 8-nitroguanine in primer extension reactions performed using an ODN template containing the natural 8-nitrodG lesion, which was produced photochemically. Despite being derived from a ribonucleoside, 8-nitro-2′-O-methylguanosine has potential advantages over the natural lesion in that it can be incorporated into ODNs by conventional chemical synthesis and the 2′-O-methyl substituent stabilizes the glycosidic bond. It was therefore of interest to compare the results of Suzuki’s primer extension studies to those obtained with our stabilized template. To conduct these studies, 8-nitro-2′-O-methylguanosine was incorporated into an ODN template, and primer extension experiments were carried out (Scheme 3). A 16-mer primer was designed to anneal to a template such that the first nucleotide to be added would be paired with 8-nitroG, with the unmodified 2′-O-methylguanosine as a control. Two polymerases were investigated: firstly the DNA polymerase AMV-RT, which naturally uses both DNA and RNA as a template and is particularly suited to our lesion analogue as it is known to extend past 2′-O-methyl nucleotides on the template (38,39). Secondly, and to allow direct comparison with Suzuki’s work, the higher fidelity human DNA polymerase β (pol β), was also selected (40).
Scheme 3.
Oligonucleotides used in primer extension. The X indicates the position of 2′-O-methyl G or 8-nitro-2′-O-methyl G in the template.
Primer extension reactions using single deoxynucleoside triphosphates as well as mixtures of the four dNTPs were analyzed by polyacrylamide gel electrophoresis (Figure 5 and 6). In reactions containing single nucleoside triphosphates and the control 2′-O-methyl G template, AMV-RT showed selectivity in pairing dCTP opposite the target site at low nucleotide concentrations (50–100 µM), showing that fidelity was not seriously compromised by the 2′-O-methyl modification. At significantly higher concentrations (1 mM), the enzyme incorporated each of the other three dNTPs with high efficiency (Figure 5a). The predominant products in Figure 5a are consistent with the addition of any single nucleotide after the last correctly templated addition at high dNTP concentration. With dCTP, a second nucleotide is added, and with dTTP, a third, after the ‘correct’ T in the second position (Scheme 3). In contrast, pol β efficiently incorporated only dCTP, even at high concentrations (Figure 5b), again suggesting that the 2′-O-methyl analogue is appropriately discriminated by the enzyme. With a mixture of all four dNTPs, both enzymes could efficiently extend past 2′-O-methylG to give 20-mer products corresponding to extension to the end of the template (Scheme 3). Pol β also gave a significant fraction of 19-mer product (Figure 5b).
In contrast, a template containing 8-nitro-2′-O-methylguanosine resulted in a substantial reduction in both the level and specificity of nucleotide insertion opposite this lesion (Figure 6). AMV-RT inserted single dNTPs at levels 10–60% of that measured for 2′-O-methylG, with dATP apparently most preferentially inserted, whereas pol β incorporated dCTP at a level <20% of the control, although dTTP incorporation was actually increased around 2-fold (from a low level). Consistent with this, the formation of extended 19- and 20-mer products in the presence of all four dNTPs was reduced by 70–90% with AMV-RT and over 90% with pol β, even at high (1 mM) dNTP concentrations.
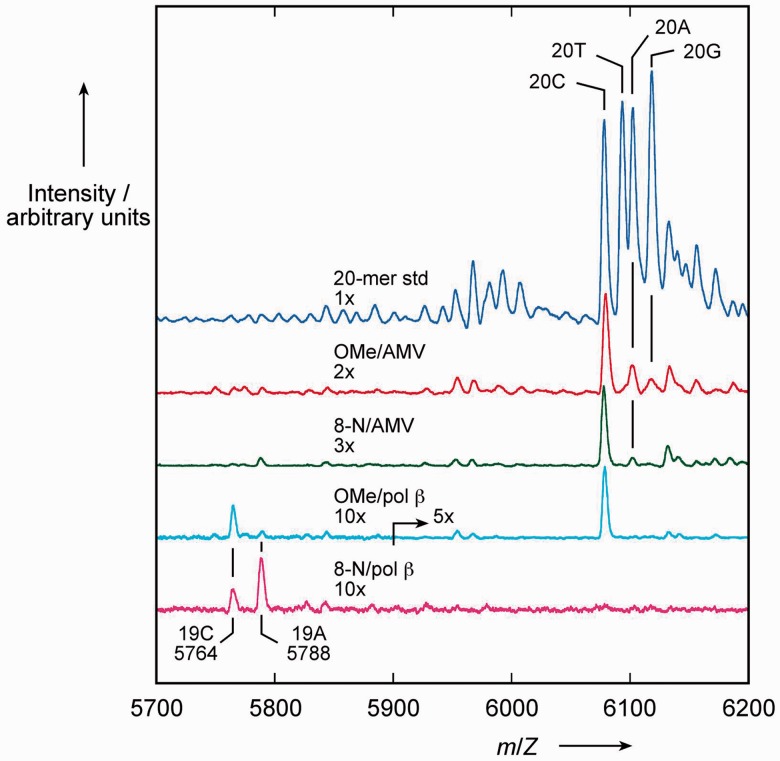
The composition of these extended products was analysed by MALDI-TOF mass spectrometry (Figure 7), using synthetic 20-mer products with the 4 alternative nucleotides at position 17 as standards. MALDI-TOF is widely used for the detection and quantitation of single nucleotide polymorphisms and allele frequencies using similar primer extension reactions; see for example (42). Consistent with the gel results, AMV-RT gave 20-mer products containing predominantly dC (with minor fractions containing dA and dG) with the control template, whereas pol β incorporated only dC into 19-mer and 20-mer products. With the 8-nitroG template, AMV-RT incorporated predominantly dC (with a small fraction of dA) into fully-extended product, whereas pol β preferentially incorporated dA over dC in a ratio of 2:1—only 19-mers were detected in this experiment. There was no evidence for incorporation of dTTP in the extended 19-mer products with pol β. The products of single dTTP incorporation visible in Figure 6b may not serve as a competent substrate for further extension.
Figure 7.
MALDI-TOF mass spectrometric analysis of primer extension reactions. Reactions contained 1 mM each of dATP, dTTP, dCTP and dGTP and either a 2′-O-methylguanosine (OMe) or 8-nitro-2′-O-methylguanosine (8-N) template with AMV reverse transcriptase (AMV) or human DNA polymerase β (pol β). The mass spectra of these reactions are aligned with the spectrum of a mixture of oligonucleotides (20-mer std) representative of the fully-extended primers that would result from incorporation of one of the four dNTPs opposite 2′-OMeG or 8-nitro-2′-OMeG (20C, 20T etc.). 19-mer pol β products incorporating a C or an A opposite the modified nucleotide (19C, 19A) are identified by their calculated m/Z values. For clarity, the spectra have been expanded along the arbitrary y-axis by the factors indicated (2x, 3x etc.).
The experiments with DNA polymerase β show a strong tendency towards the incorporation of dA opposite the lesion and in this respect they agree qualitatively, but not quantitatively with those of Suzuki et al. performed using a related ‘running start’ primer extension reaction with a template containing an unstable 8-nitrodG produced photochemically (a 1:18 dA:dC ratio) (12). There are possible reasons for this difference from the 2:1 dA:dC ratio that we see. Our use of the more stable 2′-O-methyl derivative may have perturbed the incorporations, although our control experiments suggest that the non-nitrated 2′-O-methyl G template does not significantly affect the specificity of the enzyme. Another possible cause is a pH difference. Suzuki et al. (12) carried out their pol β primer extensions at pH 8.8, which we now know from our experiments is rather close to the pKa of 8-nitroG in an oligonucleotide (9.1); deprotonation of the 8-nitroG may have a significant effect on specificity. In contrast, we used pH 8.0 based on previous literature, specifically the work of Kosa and Sweasy (43), from whom we obtained our enzyme. In addition, we did not observe a significant level of incorporation of nucleotides opposite the 8-nitroG lesion, whereas Suzuki et al. report retardation of the extension one base prior to and opposite the lesion, with human pol α and β (12). The lower level of nucleotide incorporation opposite the 8-nitroG lesion seen in the current study could be due to the 2′-O-methyl derivative or the pH used, as discussed above, or may be due to the difference in the primer template systems; the construct used here does not provide a ‘running start’ (44).
Once again it is of interest to compare the behaviour of 8-nitroG to that of 8-oxoG, where primer extension is not markedly impaired, but incorporation of dA through pairing with the syn conformation of the modified base is common (45,46). With human pol β, there is a reported 2:1–4:1 preference for incorporation of dC over dA opposite 8-oxodG (46,47), compared with the 1:2 preference seen here opposite 8-nitro-2′-O-methylG. Thus based on the present experiments, the 8-nitroG lesion appears to have more mutagenic potential than 8-oxoG.
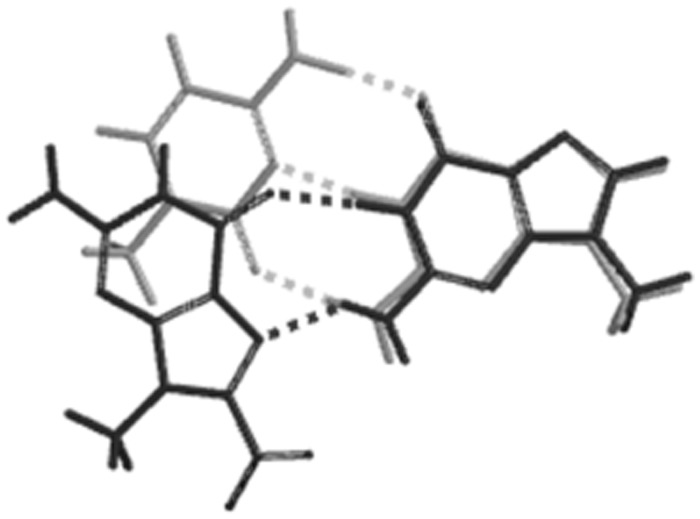
With regard to the 8-nitroG·G base pairing, there is little evidence for this arrangement in the presence of either of the polymerases. This is not altogether unexpected; for example, O6-methylguanine forms its most stable base pair with cytosine, but DNA polymerase ι prefers to incorporate T opposite this lesion because of better shape complementarity to a Watson–Crick pair (48). Related to this, studies on non-polar isosteres of DNA bases, has also revealed that shape complementarity between the incoming nucleotide and template is an important factor (49). A comparison of the G·C base pair with the syn-8-nitroG·G base pair shows that the G·G pairing has a poor match to the Watson–Crick G·C pair, particularly with regard to the position of the methyl carbon atoms, which represent C1′ of the sugar (Figure 8). Thus, while the syn-8-nitroG·G base pair emerges as the most stable pairing in the duplex, this arrangement shows a poor superpositional fit to a G·C base pair and the polymerases are likely to discriminate against it on this basis. Molecular dynamics studies of conformational changes in the pol β active site both before and after the incorporation of mismatched nucleotides have suggested that G·G mispairs are particular disfavoured both in initial incorporation and subsequent extension (50,51). These results are also consistent with kinetic studies on the extension of mutagenic intermediates by pol β (52).
Figure 8.
Overlay of N9-methylated base pairs modelled using Spartan. Watson–Crick G–C (light grey) and syn-8-nitroG·G (dark grey).
CONCLUSIONS
8-Nitro-2′-O-methylguanosine has been incorporated into DNA as a stabilized, ribose derivative of 8-nitrodG. Thermal melting studies conducted on DNA duplexes containing this modification show that this lesion forms an apparently more stable base pair with dG than with dC. This is also consistent with 8-nitro-2′-O-methylguanosine preferentially adopting the syn conformation in which it can form a syn-8-nitroG·G base pair with two hydrogen bonds. In studies with DNA polymerases, the 8-nitroG lesion induces very significant stalling with both a reverse transcriptase and a high fidelity DNA polymerase, with a tendency towards the incorporation of dA opposite the lesion in the latter case, leading to the possibility of G-T transversion mutations. It is possible that the polymerases select against a syn-8-nitroG·G due to a poor geometric match with the natural Watson–Crick pairings.
As previously discussed, the 8-nitroG lesion has been proposed to cause G to T mutations through two mechanisms; either mispairing with A or through depurination to yield apurinic sites. It is clear from these studies that this mutational effect cannot entirely be attributed to apurinic sites and in this respect our results support the work of Suzuki (12). Which of these two mutational mechanisms are more important in vivo is unclear, as there is little relevant information such as the steady state levels of this lesion in cellular DNA and if or how the lesion is repaired. The use of this and other stabilized analogues of this lesion will enable us to perform structural studies on the 8-nitroG·G and 8-nitroG·A base pairs in order to understand why polymerases may discriminate between these pairings and also enable the study of potential repair pathways that might remove the lesion. Of particular interest for future studies will be the use of 8-nitroguanine nucleoside analogues in which the sugar moiety is a better mimic of deoxyribose, such as the cyclohexenyl nucleosides (53). To date the synthesis of these analogues has eluded us due to the difficulties in preparing suitable derivatives/precursors of the 8-nitroguanine nucleobase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–8, Supplementary Figures 1–7, Supplementary Methods and Supplementary Data.
FUNDING
Engineering and Physical Sciences Research Council (EPSRC) (research grant EP/002464/1); Albertus Magnus College Faculty Welfare and Development Committee (sabbatical fellowship to P.C.-P.). Funding for open access charge: University of Liverpool and EPSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Yvonne Woolerton and Rob Beynon of the University of Liverpool Proteomics Group for advice and access to mass spectrometry, Jean Wood and Paul Loughnane for technical assistance, and Marta Veses-Garcia for providing oligonucleotide standards.
REFERENCES
- 1.Hiraku Y. Formation of 8-nitroguanine, a nitrative DNA lesion, in inflammation-related carcinogenesis and its significance. Environ. Health Prev. Med. 2010;15:63–72. doi: 10.1007/s12199-009-0118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 5.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawa T, Ohshima H. Nitrative DNA damage in inflammation and its possible role in carcinogenesis. Nitric Oxide-biol. ch. 2006;14:91–100. doi: 10.1016/j.niox.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch. Biochem. Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 9.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 10.Yermilov V, Rubio J, Ohshima H. Formation of 8-nitroguanine in DNA treated with peroxynitrite in vitro and its rapid removal from DNA by depurination. Febs. Lett. 1995;376:207–210. doi: 10.1016/0014-5793(95)01281-6. [DOI] [PubMed] [Google Scholar]
- 11.Sawa T, Tatemichi M, Akaike T, Barbin A, Ohshima H. Analysis of urinary 8-nitroguanine, a marker of nitrative nucleic acid damage, by high-performance liquid chromatography-electrochemical detection coupled with immunoaffinity purification: association with cigarette smoking. Free Radic. Biol. Med. 2006;40:711–720. doi: 10.1016/j.freeradbiomed.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki N, Yasui M, Geacintov NE, Shafirovich V, Shibutani S. Miscoding events during DNA synthesis past the nitration-damaged base 8-nitroguanine. Biochemistry. 2005;44:9238–9245. doi: 10.1021/bi050276p. [DOI] [PubMed] [Google Scholar]
- 13.Schaaper RM, Kunkel TA, Loeb LA. Infidelity of DNA-synthesis associated with bypass of apurinic sites. Proc. Natl Acad. Sci. USA. 1983;80:487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, Marx A. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO J. 2010;29:1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko K, Akuta T, Sawa T, Kim HW, Fujii S, Okamoto T, Nakayama H, Ohigashi H, Murakami A, Akaike T. Mutagenicity of 8-nitroguanosine, a product of nitrative nucleoside modification by reactive nitrogen oxides, in mammalian cells. Cancer Lett. 2008;262:239–247. doi: 10.1016/j.canlet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Garrett ER, Mehta PJ. Solvolysis of adenine nucleosides. 1. Effects of sugars and adenine substituents on acid solvolyses. J. Am. Chem. Soc. 1972;94:8532–8541. doi: 10.1021/ja00779a040. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda A, Shinozaki M, Suzuki M, Watanabe K, Miyasaka T. A convenient method for the selective acylation of guanine nucleosides. Synthesis. 1986:385–386. [Google Scholar]
- 18.Sheu C, Foote CS. Reactivity toward singlet oxygen of a 7,8-dihydro-8-oxoguanosine (8-hydroxyguanosine) formed by photooxidation of a guanosine derivative. J. Am. Chem. Soc. 1995;117:6439–6442. [Google Scholar]
- 19.Shafirovich V, Mock S, Kolbanovskiy A, Geacintov NE. Photochemically catalyzed generation of site-specific 8-nitroguanine adducts in DNA by the reaction of long-lived neutral guanine radicals with nitrogen dioxide. Chem. Res. Toxicol. 2002;15:591–597. doi: 10.1021/tx015593l. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Taguchi H, Fujii S, Sawa T, Kida E, Kabuto C, Akaike T, Arimoto H. 8-Nitroguanosines as chemical probes of the protein S-guanylation. Chem. Commun. 2008:5984–5986. doi: 10.1039/b810771h. [DOI] [PubMed] [Google Scholar]
- 21.York JL. Effect of the structure of the glycon on the acid-catalysed hydrolysis of adenine nucleosides. J. Org. Chem. 1981;46:2171–2173. [Google Scholar]
- 22.Cummins LL, Owens SR, Risen LM, Lesnik EA, Freier SM, McGee D, Guinosso CJ, Cook PD. Characterization of fully 2′-modified oligoribonucleotide heteroduplex and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guschlbauer W, Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980;8:1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaucage SL, Iyer RP. Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron. 1992;48:2223–2311. [Google Scholar]
- 25.Tretyakova NY, Burney S, Pamir B, Wishnok JS, Dedon PC, Wogan GN, Tannenbaum SR. Peroxynitrite-induced DNA damage in the supF gene: correlation with the mutational spectrum. Mutat. Res. 2000;447:287–303. doi: 10.1016/s0027-5107(99)00221-3. [DOI] [PubMed] [Google Scholar]
- 26.Laayoun A, Décout JL, Defrancq E, Lhomme J. Hydrolysis of oligonucleotides containing 8-substituted purine nucleosides. A new route for preparing abasic oligodeoxynucleotides. Tet. Lett. 1994;35:4991–4994. [Google Scholar]
- 27.Acharya P, Cheruku P, Chatterjee S, Acharya S, Chattopadhyaya J. Measurement of nucleobase pK(a) values in model mononucleotides shows RNA-RNA duplexes to be more stable than DNA-DNA duplexes. J. Am. Chem. Soc. 2004;126:2862–2869. doi: 10.1021/ja0386546. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, McAllister MA, Lee JK, Houk KN. Short, strong hydrogen bonds in the gas phase and in solution: theoretical exploration of pKa matching and environmental effects on the strengths of hydrogen bonds and their potential roles in enzymatic catalysis. J. Org. Chem. 1998;63:4611–4619. [Google Scholar]
- 29.Byun J, Henderson JP, Mueller DM, Heinecke JW. 8-Nitro-2′-deoxyguanosine, a specific marker of oxidation by reactive nitrogen species, is generated by the myeloperoxidase hydrogen peroxide nitrite system of activated human phagocytes. Biochemistry. 1999;38:2590–2600. doi: 10.1021/bi9822980. [DOI] [PubMed] [Google Scholar]
- 30.Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. pp. 201–219. [Google Scholar]
- 31.Acharya S, Barman J, Cheruku P, Chatterjee S, Acharya P, Isaksson J, Chattopadhyaya J. Significant pK(a) perturbation of nucleobases is an intrinsic property of the sequence context in DNA and RNA. J. Am. Chem. Soc. 2004;126:8674–8681. doi: 10.1021/ja048484c. [DOI] [PubMed] [Google Scholar]
- 32.Plum GE, Grollman AP, Johnson F, Breslauer KJ. Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry. 1995;34:16148–16160. doi: 10.1021/bi00049a030. [DOI] [PubMed] [Google Scholar]
- 33.Hamm ML, Rajguru S, Downs AM, Cholera R. Base pair stability of 8-chloro- and 8-iodo-2′-deoxyguanosine opposite 2′-deoxycytidine: implications regarding the bioactivity of 8-oxo-2′-deoxyguanosine. J. Am. Chem. Soc. 2005;127:12220–12221. doi: 10.1021/ja052578k. [DOI] [PubMed] [Google Scholar]
- 34.Gannett PM, Sura TP. Base-pairing of 8-oxoguanosine and 8-oxo-2′-deoxyguanosine with 2′-deoxyadenosine, 2′-deoxycytosine, 2′-deoxyguanosine, and thymidine. Chem. Res. Toxicol. 1993;6:690–700. doi: 10.1021/tx00035a015. [DOI] [PubMed] [Google Scholar]
- 35.Uesugi S, Ikehara M. Carbon-13 magnetic resonance spectra of 8-substituted purine nucleosides. Characteristic shifts for the syn conformation. J. Am. Chem. Soc. 1977;99:3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- 36.Prive GG, Heinemann U, Chandrasegaran S, Kan LS, Kopka ML, Dickerson RE. Helix geometry, hydration, and G.A mismatch in a B-DNA decamer. Science. 1987;238:498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- 37.McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal-structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base-pairs. Biochemistry. 1994;33:10266–10270. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- 38.Maden BEH, Corbett ME, Heeney P, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal-RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 39.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid-determination of 16s ribosomal-RNA sequences for phylogenetic analyses. Proc. Natl Acad. Sci. USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad R, Beard WA, Strauss PR, Wilson SH. Human DNA polymerase beta deoxyribose phosphate lyase – substrate specificity and catalytic mechanism. J. Biol. Chem. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 41.Clark JM. Novel non-templated nucleotide addition-reactions catalyzed by procaryotic and eukaryotic DNA-polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohlke KL, Erdos MR, Scott LJ, Fingerlin TE, Jackson AU, Silander K, Hollstein P, Boehnke M, Collins FS. High-throughput screening for evidence of association by using mass spectrometry genotyping on DNA pools. Proc. Natl Acad. Sci. USA. 2002;99:16928–16933. doi: 10.1073/pnas.262661399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosa JL, Sweasy JB. The E249K mutator mutant of DNA polymerase β extends mispaired termini. J. Biol. Chem. 1999;285:40666–40672. doi: 10.1074/jbc.274.50.35866. [DOI] [PubMed] [Google Scholar]
- 44.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 45.Kim SK, Lee SH, Kwon O-S, Moon BJ. DNA.RNA heteroduplex containing 8-oxo-7,8-dihydroguanosine: base pairing, structures, and thermodynamic stability. J. Biochem. Mol. Biol. 2004;37:657–662. doi: 10.5483/bmbrep.2004.37.6.657. [DOI] [PubMed] [Google Scholar]
- 46.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA-synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 47.Miller H, Prasad R, Wilson SH, Johnson F, Grollman AP. 8-OxodGTP incorporation by DNA polymerase beta is modified by active-site residue Asn279. Biochemistry. 2000;39:1029–1033. doi: 10.1021/bi991789x. [DOI] [PubMed] [Google Scholar]
- 48.Pence MG, Choi J-Y, Egli M, Guengerich FP. Structural basis for proficient incorporation of dTTP opposite O-6-methylguanine by human DNA polymerase ι. J. Biol. Chem. 2010;285:40666–40672. doi: 10.1074/jbc.M110.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran S, Ren RX-F, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity, Proc. Natl Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, Beard WA, Wilson SH, Roux B, Broyde S, Schlick T. Local deformations revealed by dynamics simulations of DNA polymerase β with DNA mismatches at the primer terminus. J. Mol. Biol. 2002;321:459–478. doi: 10.1016/s0022-2836(02)00617-4. [DOI] [PubMed] [Google Scholar]
- 51.Arora K, Beard WA, Wilson SH, Schlick T. Mismatch-induced conformational distortions in polymerase β support an induced-fit mechanism for fidelity. Biochemistry. 2002;44:13328–13341. doi: 10.1021/bi0507682. [DOI] [PubMed] [Google Scholar]
- 52.Beard WA, Shock DD, Wilson SH. Influence of DNA structure on DNA polymerase β active site function: extension of mutagenic DNA intermediates. J. Biol. Chem. 2004;279:31921–31929. doi: 10.1074/jbc.M404016200. [DOI] [PubMed] [Google Scholar]
- 53.Veerle Kempeneers V, Renders M, Froeyen M, Herdewijn P. Investigation of the DNA-dependent cyclohexenyl nucleic acid polymerization and the cyclohexenyl nucleic acid-dependent DNA polymerization. Nucleic Acids Res. 2005;33:3828–3836. doi: 10.1093/nar/gki695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.