Abstract
Bacterial RNA polymerases (RNAPs) are targets for antibiotics. Myxopyronin binds to the RNAP switch regions to block structural rearrangements needed for formation of open promoter complexes. Bacterial RNAPs containing the major variant σ54 factor are activated by enhancer-binding proteins (bEBPs) and transcribe genes whose products are needed in pathogenicity and stress responses. We show that (i) enhancer-dependent RNAPs help Escherichia coli to survive in the presence of myxopyronin, (ii) enhancer-dependent RNAPs partially resist inhibition by myxopyronin and (iii) ATP hydrolysis catalysed by bEBPs is obligatory for functional interaction of the RNAP switch regions with the transcription start site. We demonstrate that enhancer-dependent promoters contain two barriers to full DNA opening, allowing tight regulation of transcription initiation. bEBPs engage in a dual switch to (i) allow propagation of nucleated DNA melting from an upstream DNA fork junction and (ii) complete the formation of the transcription bubble and downstream DNA fork junction at the RNA synthesis start site, resulting in switch region-dependent RNAP clamp closure and open promoter complex formation.
INTRODUCTION
Changing environments, stresses and developmental programmes require regulated gene expression. RNA polymerases (RNAPs) catalyse the transcription of DNA into RNA coordinated by the actions of a range of transcription factors. Multisubunit RNAPs share a common crab-claw shape, two catalytic magnesium ions and flexible active centre domains that undergo structural rearrangements during the catalytic cycle (1,2). Bacterial RNAPs are sufficiently distinct from eukaryotic RNAPs to allow them to be used as targets for antibiotics. Emerging tolerances to antibiotics (3) have reignited interest in agents that specifically interfere with bacterial RNAPs (4).
Bacteria use two main classes of sigma (σ) factors, σ70 for the transcription of many house-keeping genes and σ54 as the major variant σ factor that regulates genes needed under wide-ranging stress conditions (5). These σ factors associate with core RNAP to produce two classes of holoenzymes (Eσ70 and Eσ54). In both cases, Eσ forms ‘closed’ promoter complexes (RPcs) where the DNA is fully duplexed. For Eσ70, isomerization to the ‘open’ complex (RPo) occurs often spontaneously whereas RPo formation in σ54-dependent transcription relies on adenosine triphosphate (ATP) hydrolysing bacterial enhancer-binding proteins (bEBPs) (6–9). Initially, the holoenzyme remains stalled at the upstream fork junction with the transcription start site DNA positioned outside of the RNAP, a first block to be overcome by activation (10). Upon binding of the bEBP and prior to ATP hydrolysis, the holoenzyme can engage with the non-template strand and capture the template strand in the active centre (11). These properties make the σ54-dependent system particularly interesting since it mimics key characteristics of the eukaryotic RNAP II including ATP hydrolysis and promoters that are regulated through enhancers and gene-specific transcriptional activators (12–14) and drives tightly regulated genes for a wide variety of functions (15,16).
RPo formation is accompanied by the structural rearrangements of a number of mobile RNAP elements. These include (i) the clamp, (ii) the jaw and (iii) the switch regions (17,18). The clamp has been proposed to be open in the RPc and closed in the RPo and to regulate the entry of the DNA template (19). The jaw domain rotates to ‘grip’ the downstream DNA duplex (20–24). The switch regions are flexible ‘hinges’ that connect the clamp to the body of the RNAP (Figure 1). In Eσ70, the switch regions have been proposed to ‘sense’ the presence of nucleic acids in the active centre and couple DNA binding with clamp closure by undergoing conformational changes (4,18,19,25–28). The structural rearrangements of all these elements accommodate the requirements of RPo formation. Evidence presented here suggests that Eσ54 uses the mobile elements differently in a bEBP-dependent manner: phage protein Gp2 binds the jaw domain of both Eσ classes but only inhibits Eσ70 (21). Certain σ54 mutants that bypass the activator dependence of Eσ54 become sensitive to Gp2 inhibition and similarly, replacing fast hydrolysing dATP with the slower hydrolysing analogue dATPγS leads to inhibition (29). These data suggest that Gp2 inhibition underlies kinetic restrictions that are weak in Eσ70 complexes but strong in activator-dependent Eσ54 complexes allowing Eσ54 holoenzymes to escape Gp2 inhibition. Gp2 and the downstream DNA duplex form interactions with multiple binding sites in the jaw domain that are individually weak but in combination provide a strong interface (30). In Eσ54, these interactions probably accumulate in a combinatorial fashion and Gp2 binding can compete against the interactions between the jaw domain and downstream DNA.
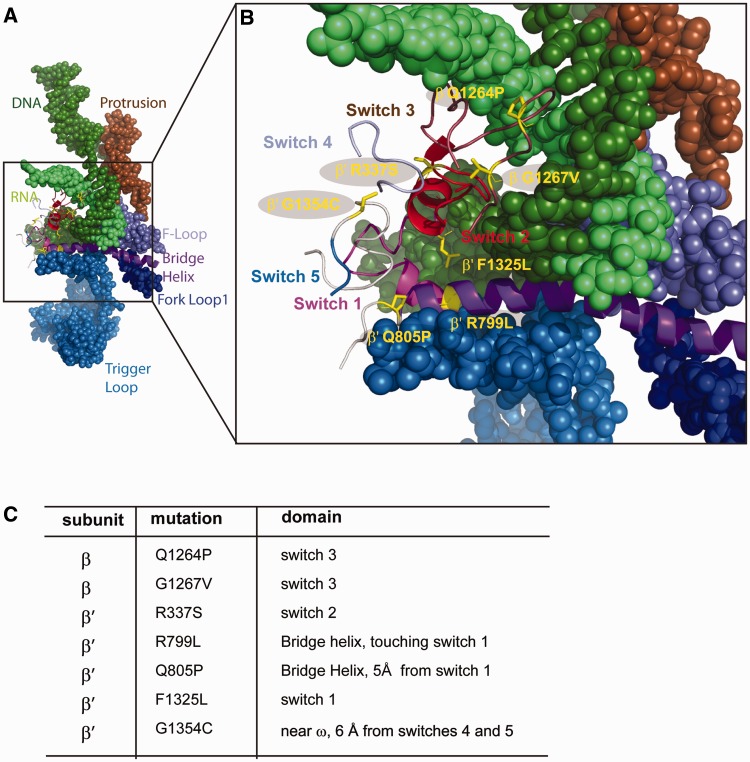
Figure 1.
DksA suppressor mutations are localized in or near the switch regions. (A) RNA–DNA hybrid and RNAP flexible elements that undergo structural rearrangements during the catalytic cycle (56). (B) Switch regions connect the clamp to the RNAP body. Highlighted are residues mutated in this study. (C) RNAP mutations investigated and positions.
The RNAP switch regions are the target of a novel class of antibiotics such as myxopyronin and lipiarmycin (4,31–35). Myxopyronin targets the switch regions of the bacterial Eσ70 and has been suggested to operate via a ‘hinge-jamming’ mechanism that prevents clamp closure (36). Accordingly, the DNA template cannot be loaded into the active centre resulting in a block to transcription initiation. We now show that while myxopyronin efficiently inhibits Eσ70-dependent transcription, Eσ54 transcription is less affected. The growth fitness of an Escherichia coli strain lacking σ54 decreases in the presence of myxopyronin suggesting that E. coli Eσ54 contributes to the natural antibiotic resistome. We provide evidence that bEBPs kinetically facilitate the necessary structural rearrangements of the switch regions. Upstream and downstream fork junction barriers for RPo formation are overcome by bEBPs, which establish the interactions between the switch regions and transcription start site that trigger start site melting.
MATERIALS AND METHODS
Knockout strains
ΔRpoN and ΔTolC ΔRpoN knockout strains were generated by P1 phage transduction of the ΦΔRpoN::KanR (Keio collection) into pop3 or pop3 ΔTolC::Tn TetR strains [gift from E. Koronakis and V. Koronakis (37)]. The strains were routinely grown on LB agar with 200 µg/ml l-glutamine to ensure growth of the ΔRpoN mutant.
Zone of inhibition assays
For zone of inhibition assays, bacteria were grown at 37°C as day cultures in Luria-Bertani (LB) medium to similar densities, pelleted, resuspended in minimal medium (40 mM K2HPO4, 15 mM KH2PO4, 2 mM Na3C6H5O7, 0.4 mM MgSO4·7H2O, 0.4% glucose, 15 µg/ml l-arginine, 2 mg/ml l-glutamine), mixed with minimal top agar (minimal medium, 0.75% agar) and plated on minimal agar (minimal medium, 1.5% agar). One microlitre of 25 mg/ml chloramphenicol (positive control), methanol (negative control) or 10 mM myxopyronin were spotted directly onto the agar. Cells were grown at 37°C and zone of inhibition diameters were analysed after 24 h. For the ΔTolC ΔRpoN strain, the zone of inhibition was defined as a clear zone with an absence of bacterial growth plus an outer ring where bacterial growth was significantly reduced in comparison to the control. The ΔTolC strain did not show a zone of reduced growth and the zone of inhibition was therefore defined as the absence of growth. The figures stated for the diameter of the zones of inhibition represent an average of four (myxopyronin) or three (chloramphenicol) zones (plus and minus standard deviations).
Constructs
Single amino acid substitutions of the E. coli rpoB and rpoC genes (pIA661, pIA458 and pIA545) were introduced by site-directed mutagenesis and reconstituted in pVS10 as described (38).
DNA probes and proteins
DNA templates representing the Sinorhizobium meliloti nifH promoter were 5′-32P-labelled (where appropriate) and annealed to the complementary strand as described (39). Escherichia coli RNAP (wild-type and switch region variants), PspF1–275 (Phage shock protein F) and Klebsiella pneumoniae σ54 were overexpressed in E. coli and purified as described (11,40,41). Escherichia coli σ70 was a gift from S. Wigneshweraraj.
Eσ54 formation assays
One micromolar 32P-HMK-σ54 (gift from N. Joly; HMK: heart muscle kinase) was incubated with 25–100 nM RNAP (wild-type or switch variants) and 500 µg/ml α-lactalbumin in STA buffer [25 mM Tris–acetate pH 8.0, 8 mM magnesium acetate, 10 mM KCl, 3.5% (w/v) polyethylene glycol (PEG) 6000] at 37°C for 15 min. Reactions were quenched with the addition of native buffer before products were analysed on 4.5% non-denaturing gels. Products were visualized and quantified using a Fuji FLA-5000 PhosphorImager. For quantitation purposes the photostimulated luminescence (PSL) transfer scale was adjusted using the AIDA software and for illustration purposes the Curves scale was adjusted using Photoshop CS3. These image manipulations were performed for all computer-generated images. All Eσ54 formation assays were performed as titrations with three different RNAP concentrations (25, 50 and 100 nM).
Transcription assays
Promoter-dependent spRNA (short-primed RNA) assays were performed as described (11) in a 10 -μl reaction volume containing 25–100 nM σ54/RNAP or σ70/RNAP (reconstituted using 40 µM of σ and 25–100 nM RNAP) and 25 nM promoter DNA probe. ADP·AlF (adenosine diphosphate - aluminium fluoride) trapped and open promoter complexes were formed as described (11) in the presence of 7 μM PspF1–275. All transcription assays were performed as titrations with three different RNAP concentrations (25, 50 and 100 nM).
Specific promoter-independent transcription assays (‘minimal scaffold assays’) were performed as described (38). These assays permit determination of the catalytic activity of the RNAP variants in the absence of any requirements for promoter recognition or interaction with σ factors. All minimal scaffold assays were performed as titrations with three different RNAP concentrations (25, 50 and 100 nM).
Gp2 or myxopyronin inhibition
Transcription reactions were carried out as described but in the presence of 100 nM Gp2 (gift from S. Wigneshweraraj) or 1 µM myxopyronin (Ascenion GmbH) for 10 min prior to adding the promoter DNA. To analyse the effect of myxopyronin on the switch variants, reactions were carried out in triplicates with 100 nM RNAP.
Inorganic pyrophosphate assay
In a post-extension state, RNAPs were capable of catalysing the cleavage of the terminal nucleotide from a transcript upon addition of inorganic pyrophosphate (PPi). This activity is pyrophosphorolysis and is one of the proofreading mechanisms of RNAP (42). Pyrophosphatase assays were performed as described (38) as titrations with three different RNAP concentrations (25, 50 and 100 nM).
Binding assays
Minimal scaffold and promoter binding assays were conducted as described (11,38,43). All reactions were performed as titrations with three different RNAP concentrations (25, 50 and 100 nM).
RESULTS
A σ54 knockout strain is more sensitive to myxopyronin than the parental strain
bEBPs trigger Eσ54 transcription in situations such as membrane stress, pathogenicity and nitrogen starvation. Recently, one signalling cascade has been shown to help bacteria to survive in the presence of antibiotics, the phage shock protein (psp) response that uses bEBP PspF to manage inner membrane stress (44). This prompted us to consider the potential role of Eσ54 in activating downstream mechanisms that result in increased tolerance towards antibiotics. Gram-negative bacteria can escape the toxic effects of certain antibiotics as the membrane transporter TolC pumps them out of the cell, thus depleting them intracellularly (37). In order to use E. coli to study the effect of myxopyronin on the Eσ54 system, the intracellular drug concentration had to be sufficiently maintained. We therefore generated an RpoN knockout in a ΔTolC background (gift from V. Koronakis and E. Koronakis). In zone of inhibition assays, a strain lacking σ54 (ΔTolC ΔRpoN, 28 ± 1.5 mm) was sensitive to chloramphenicol to a similar degree as the parental strain (ΔTolC, 29 ± 1.1 mm). On minimal media agar, the ΔTolC ΔRpoN strain showed a larger zone of inhibition around myxopyronin (29 ± 1.5 mm) in comparison to the parental ΔTolC strain (21 ± 1.4 mm) thus indicating a greater degree of acquired drug sensitivity (Figure 2A). In vitro, Eσ54-dependent promoter-specific transcription initiation was inhibited by ∼4-fold higher myxopyronin concentrations than Eσ70-dependent initiation (50% of maximum activity was reached with ∼3 µM (Eσ54) and ∼0.7 µM (Eσ70), Supplementary Figure S1). We conclude that Eσ54 may contribute to natural E. coli resistomes.
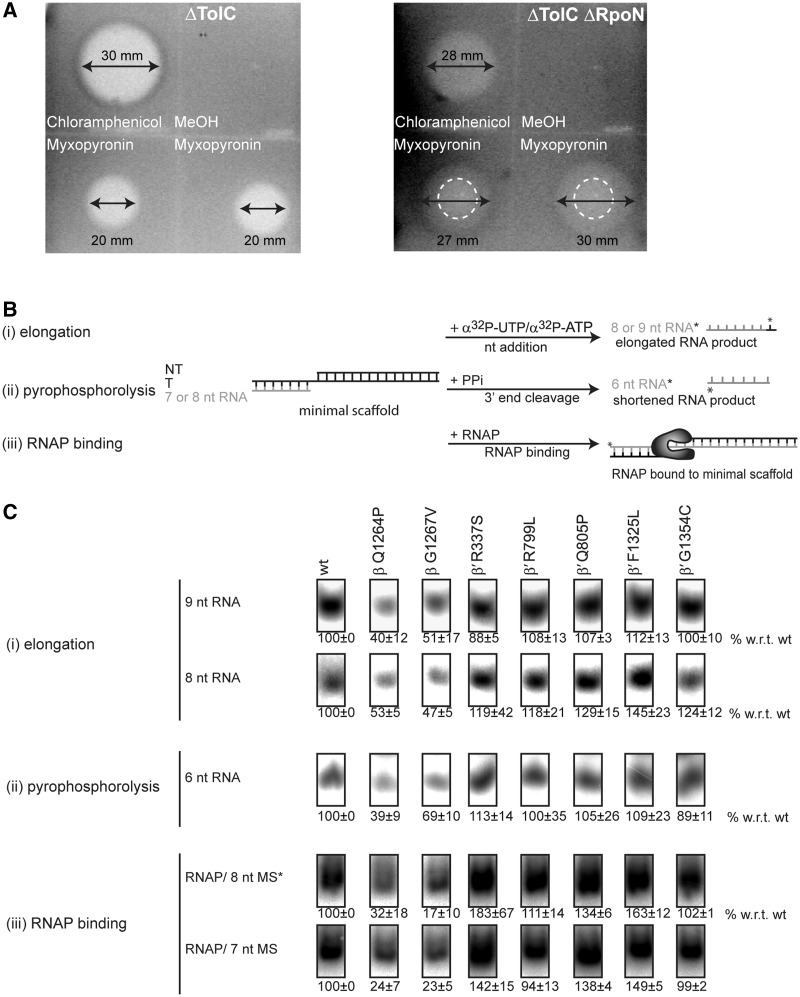
Figure 2.
Myxopyronin sensitivity in vivo and switch 3 mutants have reduced DNA template affinity. (A) Zone of inhibition assays. The loss of RpoN increases sensitivity to myxopyronin. Myxopyronin inhibits growth of the ΔTolC strain within a zone of 20 mm. The zone of inhibition (27–30 mm, indicated by arrows) for the ΔTolC ΔRpoN strain was defined as a combination of a central zone with no growth (15 mm, dashed circle) plus a ring of impaired growth. (B) Schematic of the (i) minimal scaffold (extension of an RNA hybridized to DNA by a single labelled nucleotide), (ii) pyrophosphatase (shortening of a 5′-end-labelled RNA upon addition of PPi) and (iii) minimal scaffold binding (binding of RNAP to the minimal scaffold containing a 5′-end-labelled DNA template strand) assay. (C) The switch 3 mutants, β-Q1264P and β-G1267V are significantly reduced in supporting RNA extension, (top), in cleaving the terminal nucleotide (middle) and in binding the minimal scaffold (bottom). Assays were titrations at three concentrations (25, 50 and 100 nM RNAP) and activity was calculated relative to the wild-type (wt) at the respective concentrations (100%). The numbers reflect the activity obtained as an average from the three concentrations and the error bars are standard deviations.
Myxopyronin binds to residues in or very close to all five switch regions (36). A potential explanation for the decreased sensitivity of Eσ54 towards myxopyronin thus was that Eσ70 and Eσ54 may use the switch regions in different ways. To investigate the roles of the RNAP switch regions in the enhancer-dependent transcription system, we generated seven RNAPs with substitutions at residues of which the potentially relevant biological functions had previously been established (45). These residues had been identified within the switch 1, 2 and 3 regions or in close proximity to the switch 1, 4 and 5 regions and are likely to interact with the switches (45) (Figure 1). The switch mutants we selected had been identified as DksA suppressor mutants (45). DksA is a transcriptional regulator that binds in the secondary channel of RNAP and is a well-recognized inhibitor of rRNA gene transcription. Depending on the promoter sequence, DksA can also activate the transcription of certain genes of which the products predominantly serve a metabolic function. In a ΔDksA background, suppressor mutants are those which restore the transcription of DksA activated genes to wild-type levels and repress the transcription of DksA repressed promoters. In vitro, they shift the RPc <-> RPo equilibrium in the direction of dissociation (45).
Switch 3 mutants are defective in minimal scaffold binding
The mutant RNAPs were generated by site-directed mutagenesis, overexpression and affinity purification. The biochemical integrity of their active centres was confirmed by performing minimal scaffold assays measuring the extension of a short RNA species hybridized to a DNA template by a single nucleotide (Figure 2B). This minimal scaffold lacks σ-mediated promoter recognition sequences and supports RNA-primed transcription from a single-stranded DNA template. The switch 1 and 2 mutants, as well as those which have been proposed to interact with the switch 1, 4 and 5 regions (hereafter referred to as switch 1 or switch 4 and 5 mutants, respectively), showed a catalytic activity that was greater or equal to wild-type levels. The switch 3 mutants, by contrast, only reached 40–50% of the activity levels of the wild-type (Figure 2B and C and Supplementary Figure S2A and B). These observations were mirrored by a pyrophosphorolysis assay (an alternative to test the catalytic activity of the active site) where the switch 3 mutants displayed 40–70% of wild-type activity and the switch 1, 2, 4 and 5 mutants were at least wild-type-like activity (Figure 2B and C). Finally, gel mobility shift assays illustrated that the binding of the switch 3 mutants to the minimal scaffold was reduced to 15–35% in comparison to wild-type RNAP, thus explaining their apparent defects in catalytic activity (Figure 2B and C).
Switch 3 mutants are defective in holoenzyme formation
Next, we turned to investigating the requirements of promoter-directed transcription and assessed the capability of the RNAP variants to form Eσ54. Again, the switch 1, 2, 4 and 5 mutants showed at least wild-type-like activity in forming Eσ54 whereas the switch 3 mutants only formed 30–40% Eσ54 compared to the wild-type (Figure 3A). We infer that the switch 3 mutants represent a population of RNAPs that exclude both the DNA template and σ54 from their active centre, presumably due to the closed clamp conformation. Our results also suggest that the DNA template and σ54 normally interact with the same feature of the RNAP active centre and or DNA-binding clefts and so might compete with each other for binding at some point during transcription initiation.
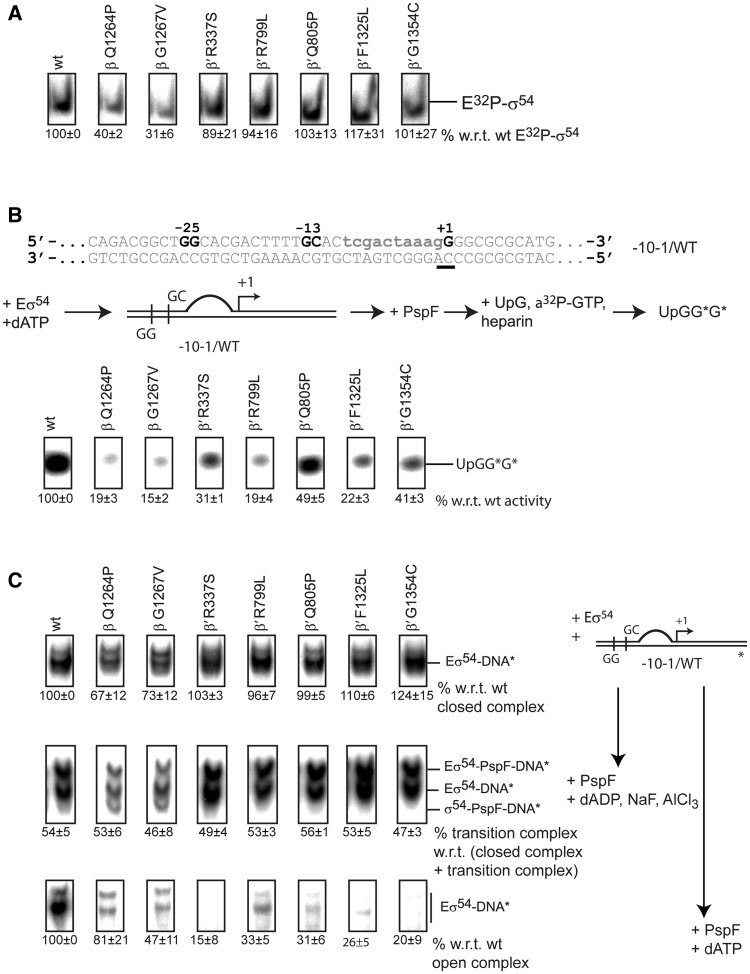
Figure 3.
The switch 1, 2, 4 and 5 mutants are defective in RPo formation. (A) EMSA with 32P-σ54. Complex formation of E32P-σ54 was measured as a supershift with respect to unbound 32P-σ54. The switch 3 mutants, β Q1264P and β G1267V, are defective in forming E32P-σ54. (B) Top: spRNA assay using a linear template representing the nifH promoter with a preformed transcription bubble (mismatch indicated in lower case). The start site is underlined. Middle: The spRNA assay: the RPc was incubated with PspF + dATP followed by an extension mix of UpG primer, heparin and α-32P-GTP (guanosine triphosphate). Labelled (UpGGG*) was measured. Bottom: Reduced accumulation of extended UpG species indicated significant defects in supporting promoter-directed transcription for all mutants. (C) Right: Schematic of the three binding reactions. Eσ54 was incubated with DNA only to form RPcs (top), with DNA, PspF and ADP·AlF to form RPis (middle) or with DNA, PspF and ATP to form RPos. Left: Switch 3 mutants, β-Q1264P and β-G1267V, are reduced in RPc formation (top). Approximately 50% of all RPcs shift into the trapped RPi state confirming that all variants interact with PspF (middle). RPo formation is severely affected in all mutants (bottom). For open and trapped complexes, 200 µg/ml heparin was added for 5 min to outcompete non-specific binding interactions.
Defects in the switch 1, 2, 4 and 5 regions interfere with open complex formation
We next looked at the ability of the RNAP variants to support activator-dependent transcription initiation in spRNA assays. In these assays, a specific dinucleotide primer hybridizes at the start site of a known promoter sequence and is extended by one or two radiolabelled nucleotides (depending on the primer and the promoter sequence) to form a short RNA (11). We used a linear template representing the nifH promoter. The non-template strand was mismatched from −10 to −1 (−10−1/WT template) to create a pre-formed transcription bubble that favours transcription, thereby resulting in higher nucleotide incorporation compared to a fully duplexed template (Figure 3B, Supplementary Figure S3A). Transcription from this template still relies on activator-driven initiation (Supplementary Figure S4A). We measured the accumulation of the tetramer to evaluate the capacity of the holoenzymes to initiate from the nifH promoter. As expected, the switch 3 mutants are impaired in this assay and only produced 15–20% of the wild-type activity. The activity of the remaining mutants was drastically reduced as well, reaching 20–50% of the wild-type activity (Figure 3B). We reasoned that these mutants failed in one or more of the following steps: (i) interaction with the −10−1/WT template, (ii) interaction with the activator or (iii) formation of open promoter complexes.
These three possibilities were addressed by analysing the ability of the Eσ54 to form RPcs, the trapped transition state (RPi) and RPos. When we incubated the Eσ54 with the promoter template in the absence of PspF activation, the switch 1, 2, 4 and 5 mutants formed a similar number of RPcs as wild-type Eσ54. As expected, the efficiency of the switch 3 mutants in RPc formation was lower reaching only ∼70% in comparison to wild-type Eσ54. We therefore concluded that the switch 1, 2, 4 and 5 mutants are not affected in their ability to interact with the −10−1/WT template and to form RPcs [we define RPc as the promoter complex in which the start site is closed and outside of the RNAP (10,46)]. Next, PspF and ADP·AlF were added to drive the reaction into the ‘trapped state’ (RPi). ADP·AlF has been suggested to mimic the conformation of ATP immediately prior to hydrolysis (47). Hence, the Eσ54–PspF–ADP·AlF–DNA complex is believed to assume a conformation that represents an intermediate en route to open complex formation (10). To analyse the ability of the RNAP variants to reach the RPi, we measured the proportion of complexes that succeeded in progressing from the RPc to the RPi. The proportion was constant across all the variants with ∼50% of all RPcs reaching the RPi state, suggesting that in each mutant tested interactions with the bEBP occur in a wild-type-like fashion and are unlikely to be affected by any of the switch mutations studied. However, adding PspF and hydrolysable dATP revealed that all the mutants were impaired in forming RPos. Surprisingly, in comparison with the switch 1, 2, 4 and 5 mutants, the switch 3 mutants seemed less affected in their ability to form RPos despite their defects in allowing DNA and σ54 to access their active centre, suggesting that the switch 3 region is not greatly impaired in the later steps of the pathway from RPi to RPo (Figure 3C and Supplementary Figure S3B and C).
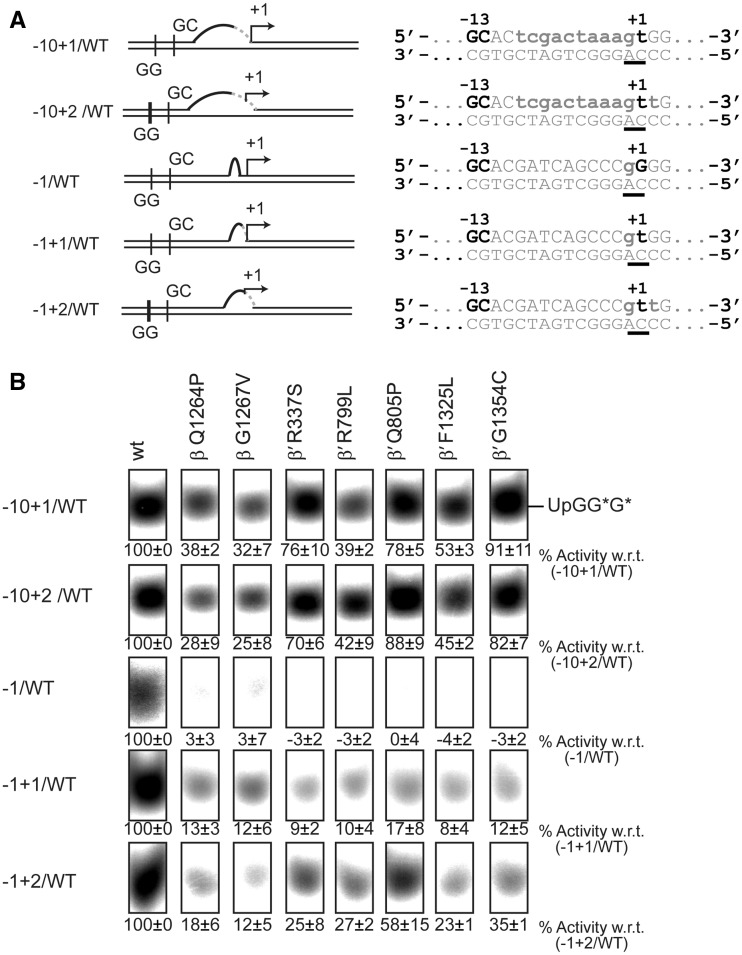
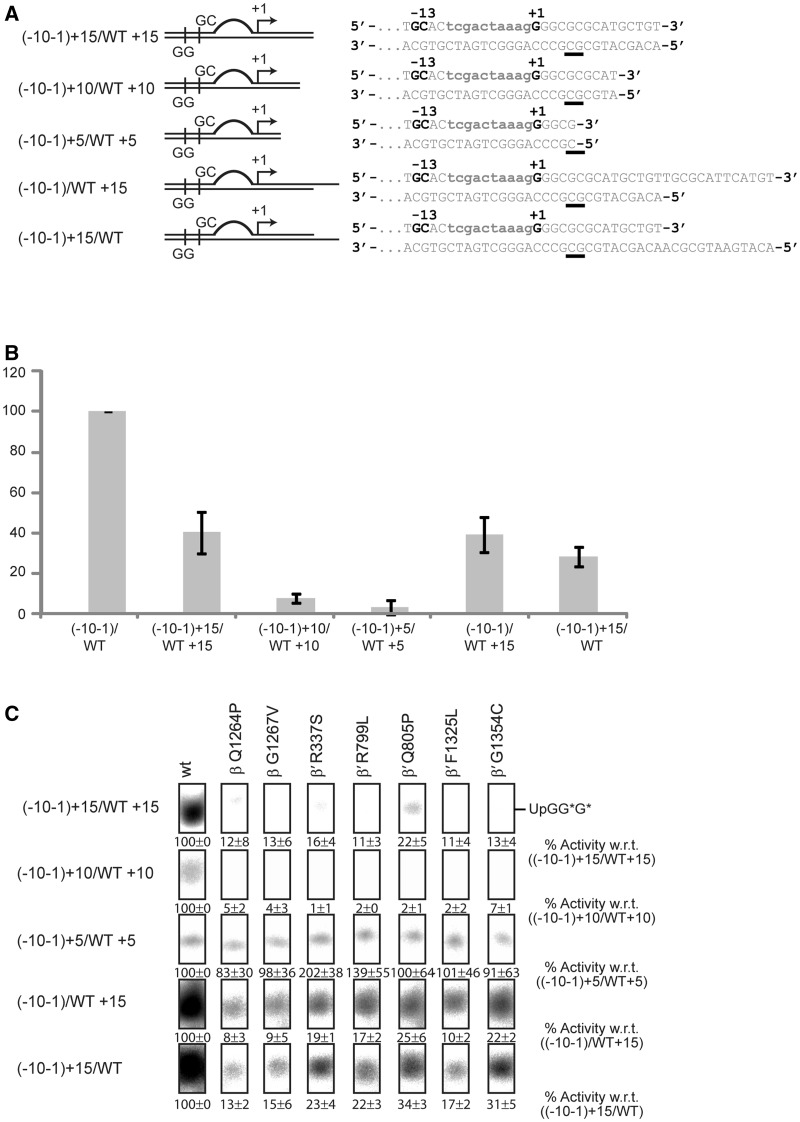
Precise start site pre-opening suppresses defects in transcription initiation
RPo formation comprises two steps: (i) melting of the DNA template to form the transcription bubble and (ii) closing of the RNAP clamp. Essentially, melting of the DNA template must include the opening of the start site for the incoming +1 nucleotide. To determine which of these steps is predominantly affected by the mutations in the switch regions, we bypassed the start site melting requirement of the RNAP by extending the artificial transcription bubble to include the start site (−10+1/WT) and the second nucleotide (−10+2/WT) as unpaired DNA bases (Figure 4A). Strikingly, the switch mutants partially recovered from their defects in spRNA assays when these templates were used and their activity approximately doubled as soon as the +1 site became available. Activity did not further increase when the +2 site was melted as well (Figure 4B). The melting of the start site did not bypass the requirement of the bEBP and the activity observed in the absence of PspF was negligible (Supplementary Figure S4B and C). Also, melting out of the base pairs around the start site alone resulted in a similar recovery effect (Figure 4A and B) confirming that start site melting specifically and not the overall size of the transcription bubble was the crucial parameter that led to the recovery of the mutants. These observations showed that the active start site melting requirement is bypassed and a transcriptionally productive state is reached by shifting the equilibrium towards the RPo. We therefore propose that start site melting is a crucial step in the formation of the RPo and dependent on the conformation of the switch 1, 2, 4 and 5 regions. To establish if (i) the switch mutants still contain an intact binding pocket for myxopyronin and if (ii) the start site melting step can still be bypassed when myxopyronin is bound to Eσ54, 1 µM myxopyronin was added to Eσ54 prior to adding the DNA template and PspF. The capability of wild-type Eσ54 to support transcription from the −10−1/WT template was to reduced ∼60% in comparison with the Eσ54 uninhibited by myxopyronin. The switch mutants did not show any activity on this template. A similar outcome was observed when the WT/WT template was used. Using the −10+1/WT template we discovered that none of the mutants recovered (Supplementary Figure S5). This established that, despite the mutations, myxopyronin is still capable of binding and inhibiting Eσ54. Since myxopyronin interacts with RNAP via a number of residues in the switch regions, it is conceivable that the binding pocket may remain largely intact if only single residues are mutated (36). Also, start site pre-opening fails to compensate for the inhibitory effect of myxopyronin. This is in contrast to observations predicting that myxopyronin interferes with start site melting (35,48). Presumably, myxopyronin acts at a step prior to start site opening in the enhancer-dependent system, consistent with the proposed hinge-jamming mechanism (36).
Figure 4.
Bypassing start site melting partially restores the activity of the mutants. (A) Promoter templates with an opened start site. (B) Extending the −10−1/WT transcription bubble to include the +1 or the +1 and +2 sites led to partial recovery of the mutants. Opening of the start site alone produced a similar recovery.
Bypassing start site melting restores inhibition by Gp2
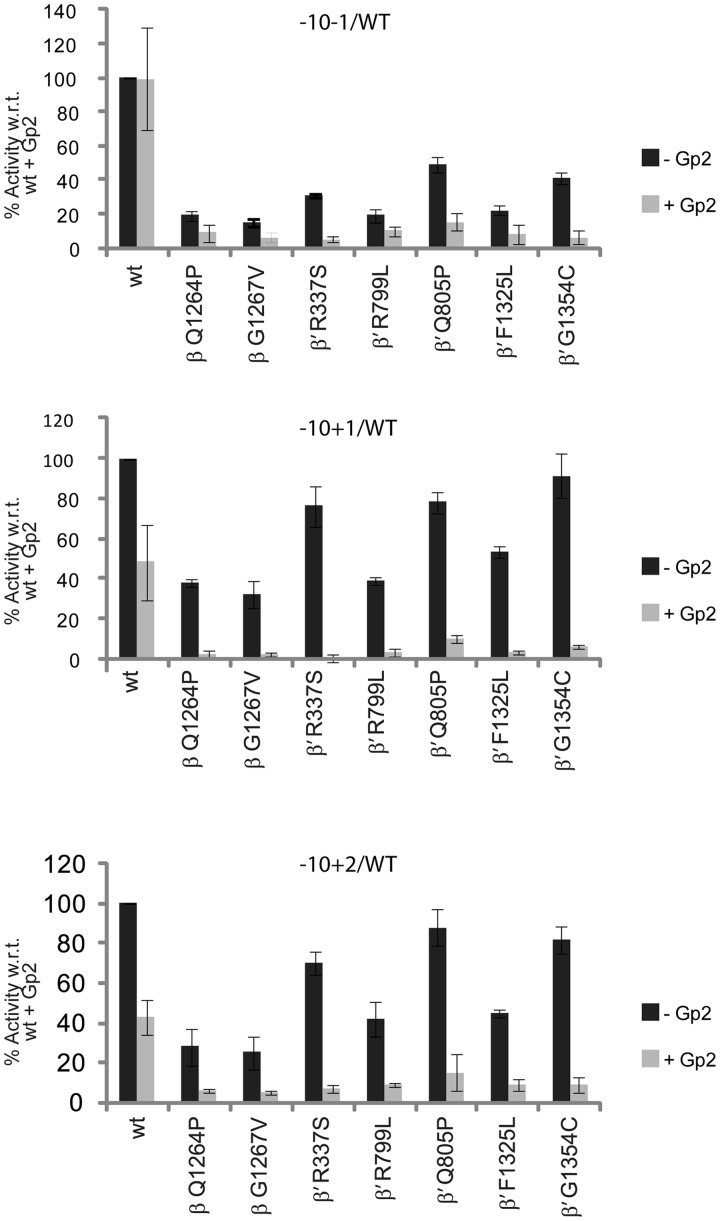
We next asked how start site melting relates to clamp closure. Phage protein Gp2 is a transcriptional regulator that interacts with the RNAP jaw domain, competing with the downstream DNA duplex for interaction sites in the jaw domain (21). In Eσ54 complexes, the reaction equilibrium is predicted to be in favour of the formation of interactions between the jaw domain and DNA when the bEBP is present driving RPo formation (11,12,49) and excluding Gp2 from forming interactions with the jaw domain. Using the −10−1/WT template, we confirmed that wild-type Eσ54 could not be inhibited by Gp2. The switch mutants, on the other hand, were inhibited by Gp2 indicating that their structural defects hinder the normally fast transition into the RPo that would preclude inhibitory Gp2 interactions with the jaw domain (Figure 5 and Supplementary Figure S6). When templates including the pre-melted start site were used, the switch mutants were unable to recover their activity in the presence of Gp2. Additionally, the activity of the wild-type was reduced to ∼50%. We reasoned that templates with a pre-melted start site bypassed the formation of interactions required for start site melting. Albeit sufficient to partially restore the activity of the switch mutants, start site opening did not compensate for a mechanism that establishes interactions with the downstream duplex before Gp2 can inhibit.
Figure 5.
Gp2 inhibition. Measuring accumulation of a four base spRNA product in the presence of Gp2. Top: Gp2 inhibits mutant but not wild-type (wt) Eσ54. Middle and bottom: Bypassing start site melting leads to inhibition of wt Eσ54.
Positioning of the downstream duplex DNA is not affected by mutations in the switch regions
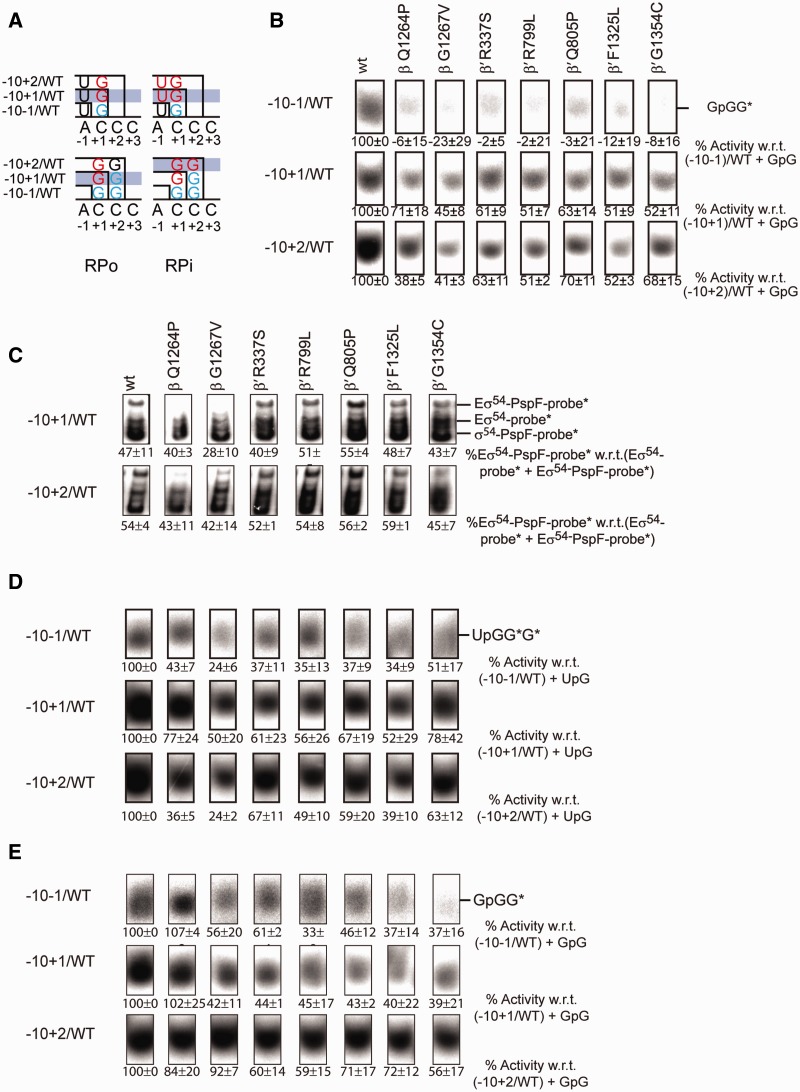
An alternative explanation for the inhibition effect observed in the presence of Gp2 is that in Eσ54 mutants the downstream DNA might fail to establish contacts with the downstream DNA-binding channel (dwDBC) that forms with the clamp, the jaw domain and other downstream mobile elements (22–24). Both, the correct positioning of the downstream DNA and the presence of a specific length of DNA for interaction with the jaw domain are crucial for the formation of productive open complexes (50). Defects in the switch regions, however, might lead to an altered configuration of the DNA in the active centre where it (i) cannot interact with the dwDBC in its usual way and/or (ii) might interfere with the position of the dwDBC in the RPo. Shortening the downstream DNA might remove any interfering effects or, in the case of misalignment of the jaw domain, would at least not increase the suppression effect observed with the switch mutants. The −10−1/WT template represents the nifH promoter from −60 to +28. Shortening the downstream end of both DNA strands to +15, +10 and +5 (Figure 6A) and comparing these templates in spRNA assays using wild-type Eσ54 showed that they are less favoured with hardly any transcription occurring from the shortest template (Figure 6B). This loss of activity strongly indicates that the presence and appropriate length of downstream DNA are both essential factors for the jaw domain to interact and to stabilize the open complex. The switch mutants displayed an additive effect on these templates, reflecting both the defects of the mutants in supporting transcription from the nifH promoter as well as the downstream DNA requirements and resulted in an almost total loss of transcription (Figure 6C). Clearly, the switch mutants still depend on a sufficient length of downstream DNA to retain some activity. We inferred that the configuration of the downstream DNA duplex in the active centre of these mutants is still intact and next investigated whether the template or the non-template strands are required for the interactions with the jaw domain, each being shortened to +15 (Figure 6A). These two templates performed similarly to the template with two shortened strands, indicating that both DNA strands are required to maintain the stability of the complex (Figure 6B and C). To stabilize the contacts between the jaw and the DNA, the DNA duplex must be ‘sandwiched’ between the closed clamp and the jaw (48). Clamp closure seems to be delayed in those switch mutants that are defective for RPo formation. Taken together, our results suggest that the switch regions act in a relay mechanism that is triggered by interactions with the start site, leads to template melting and is further propagated to the clamp which results in clamp closure.
Figure 6.
The downstream DNA duplex is required for the activity of the mutants. (A) Shortened templates based on the −10−1/WT template. (B) Activity of the wild-type (wt) Eσ54 decreases when the downstream DNA duplex is removed. (C) Shortened downstream DNA duplexes increase the intrinsic defects of the mutants.
bEBP ATP hydrolysis elicits start site melting
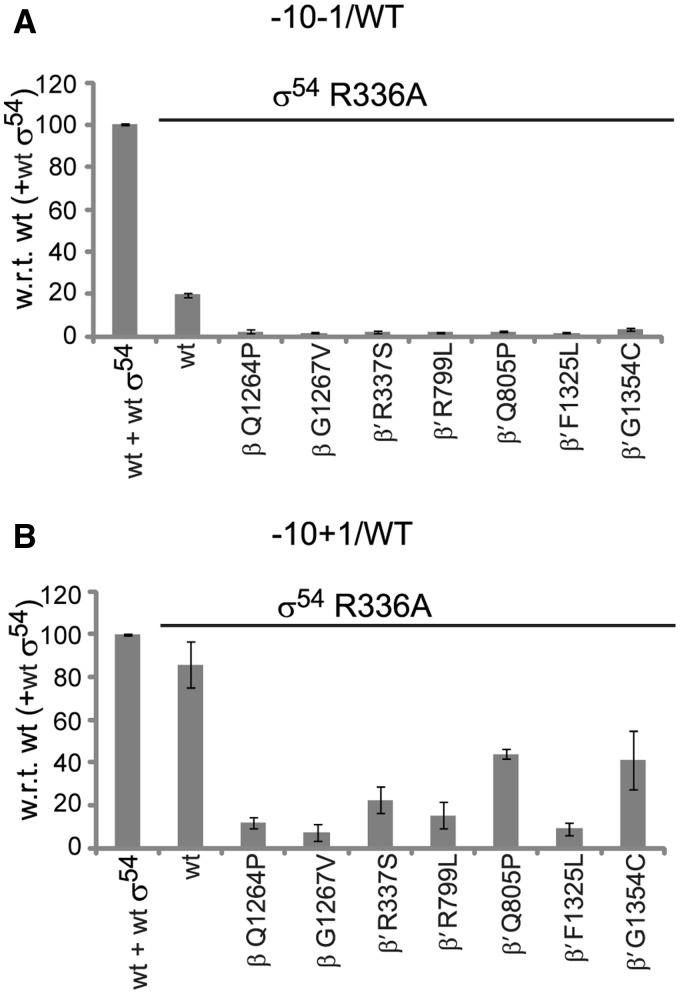
σ54-R336A is referred to as the ‘activator bypass mutant’ because it forms an Eσ54 which requires neither the bEBP nor dATP in order to form transcriptionally competent RPos. Eσ54-R336A probably assumes a conformation that mimics that of a post-hydrolysis state of Eσ54. When forming holoenzymes with σ54-R336A, the switch mutants failed to support transcription from the −10−1/WT template (Figure 7A). They did, however, recover when the −10+1/WT template was used illustrating that ATP hydrolysis catalysed by bEBP is the key trigger for start site melting (Figure 7B), supporting the proposal that the start site opening contributes a thermodynamic barrier.
Figure 7.
ATP hydrolysis is required for start site melting. (A) Bypassing the ATP hydrolysis step by using σ54 variant R336A, the switch mutants fail in supporting transcription from the −10−1/WT template. (B) If the start site melting step is bypassed as well, the switch mutants support transcription.
When the UpG dinucleotide primer was utilized in spRNA assays, base-pairing interactions occurred with the −1 and +1 bases (A,C). Nucleotides that extend this primer recognize the +2 (C) and +3 (C) sites. In order to investigate the accessibility of the start site in the open complex, we performed spRNA assays on the −10−1/WT template using the dinucleotide primer GpG, which interacts with the +1+2 (CC) site and is extended by a single guanosine triphosphate (GTP) to form a labelled trimer. Dependent on the accessibility of the +2 site, this reaction can be predicted to become more or less productive. Overall, the signals obtained with GpG were substantially weaker than those with UpG, probably reflecting both the requirements for primer base pairing in the absence of at least one unpaired base binding site and the necessity to melt the site for the incoming dinucleotide primer. However, the signal intensities increased if templates with the pre-melted start site (−10+1/WT or −10+2/WT) were used (Figure 8B, Supplementary Figure S7). Again, the mutants partially recovered from their defects when the +1 site was pre-melted. The activity did not significantly increase when the +2 site was exposed as well (Figure 8B). This suggests that in the RPo the accessibility of the +1 site is the crucial determinant to allow further melting and transcription to occur, a proposal explored below.
Figure 8.
The dinucleotide binding requirements differ in the trapped and open complex. (A) Schematic of the start site requirements in the RPo (left) and RPi (right) state. In RPo, the biggest increase in activity occurs as the start site becomes available. In RPi the biggest increase in activity occurs as the dinucleotide can hybridize to the template. (B) Extension of GpG established the requirement for the availability of the +1 site. (C) If the −10+1/WT or the −10+2/WT templates are used, the proportion of RPos reaching the RPi state is constant and still reaches ∼50%. (D) Extension of UpG in RPi demonstrates a recovery effect when the +1 site becomes available. (E) Extension of GpG in RPi demonstrates a recovery effect when the +2 site becomes available.
Assuming that template melting follows σ54 activation by PspF, we predicted that in the RPi, the accessibility of the binding site for the dinucleotide primer should be an important and dominant determinant of transcription levels. Evaluating the extension of the UpG or GpG dinucleotide primer, respectively, from the RPi, we observed an overall increase in activity when templates with the pre-melted start site were used. All the switch mutants gained activity as soon as the +1 site became exposed, with no further increase, and in some cases (β-Q1264P, β-G1267V and β′-F1325L) even a decline in activity, when the +2 site was opened (Figure 8C and D). With GpG instead of UpG, the mutants became gradually more active in correlation with template opening. Exceptions were β-Q1264P, which experienced a mild decline in activity suggesting a saturation effect, and β-G1267V and β′-R337S, which displayed an initial drop of activity when the −10+1/WT template was used and an increase in activity when the −10+2/WT template was used (Figure 8C and E). The necessity for the GpG dinucleotide to establish base-pairing interactions was in line with our expectations. It demonstrated that prior to the start site melting step, the accessibility of the primer binding site in RPi is sufficiently high to allow primer extension to then occur. In the RPo, on the other hand, the capacity to melt the template overcomes the thermodynamic requirements and base-pairing interactions with a single nucleotide are sufficient (Figure 8A). The switch 3 mutants are more active under trapping conditions when compared to the remaining mutants. Their performance was similar or better than that of other switch mutants and even reached wild-type levels when GpG was used.
DISCUSSION
Eσ54-dependent transcription contributes to the bacterial resistome
Effective antibiotics target processes that are essential for the survival of a bacterial cell. Signalling cascades triggering persister cell formation (44) act via the Eσ54-dependent transcription system, suggesting it has a key role in contributing to the natural bacterial resistome. Eσ54 is tolerant to myxopyronin and we propose that this is due to the action of the bEBP, which kinetically facilitates the transition from the RPc to the RPo and thereby shortens the time-window during which the switch regions are in the appropriate conformation for the antibiotic to bind and act.
The switch regions act in a pathway that controls start site melting and clamp closure
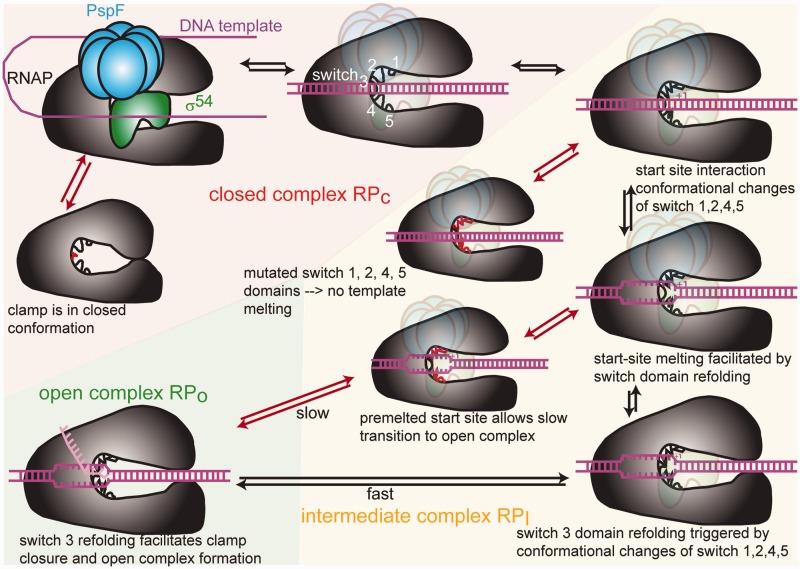
The switch regions control the same key steps during both Eσ70 and the Eσ54-dependent transcription initiation i.e. template melting and clamp closure. However, the pathway by which DNA is delivered into Eσ54 to form the RPo is so far unknown. Our experimental system provided us with a toolset to shed new light on the impacts of the switch region movements and the domain movements they control in relation to the enhancer-dependent system. We were able to follow the importance of the switch regions through steps leading to RPo formation from RPi and RPc. We propose that the switch 1, 2, 4 and 5 regions act in a step that is distinct from that controlled by the switch 3 regions and that all five switch regions together may cooperate in a relay mechanism (Figure 9). Our data were consistent with observations made by others and give novel insight into the sequence of events and kinetic relationships. We are now able to propose an extended view on the mechanism of bEBP-dependent transcription that relies on a dual switch whereby the kinetic restrictions preventing switch region refolding are lifted and, by inference, start site melting and clamp closure occur.
Figure 9.
Model of the structural rearrangements occurring en route to open complex formation. When the clamp is open the DNA duplex can enter Eσ54. The switch regions are then in a conformation where they can interact with the start site. This leads to structural rearrangements of the switch 1, 2, 4 and 5 regions that then facilitate template melting and refolding of the switch 3 region. Switch 3 region refolding leads to clamp closure and the formation of RPo. If the switch 3 region is mutated, the clamp is closed, so the DNA duplex and σ54 remain excluded from the active centre. If the switch 1, 2, 4 and 5 regions are affected by mutations, start site melting cannot occur. If the start site is pre-melted, RPo formation is rescued but occurs at a much slower rate because crucial interactions between the DNA template and the switch regions are bypassed and the DNA template is not stabilized in the active centre.
Two promoter checkpoints control Eσ54-dependent initiation via a dual switch
We suggest that the two DNA fork junctions at the upstream (−12) and downstream (−1) end of the transcription bubble are targets for control of Eσ54 RPo formation by the ATPase of bEBPs and form two checkpoints that act as kinetic barriers to Eσ54-dependent transcription. These barriers are overcome in a dual switch consisting of (i) binding of the activator to allow the nucleated transcription bubble to spread from −12 and (ii) ATP hydrolysis to complete the transcription bubble by melting the start site. For Eσ70, the upstream fork junction does not completely stall RNAP (51). Eσ70 binding to a downstream fork junction positioned around +2 is much stronger than to a fork junction at −1 (50). The propagation of the fork junction to the +2 site is predicted to be coupled to a shift in the equilibrium between the duplexed and melted conformations of the DNA segment around the start site, allowing the transcription bubble to stabilize (50). In Eσ54-dependent initiation, on the other hand, the DNA duplex upstream of the start site is not destabilized because the +1 site is kept in check by a lack of interactions with the switch regions prior to ATP hydrolysis. Given the prior absence of the tools required to study the impacts of the switch mutants on any form of RPi with major or variant RNAP holoenzymes, it was not possible to establish a link between activation pathways (e.g. with ATP hydrolysis), interaction of the switch regions with the transcription start site and start site melting. With the use of specific switch region mutants, a range of promoter templates representing different DNA states en route to RPi and RPo formation and nucleotide analogues we have finally been able to bridge this gap. Our results show that upon ATP hydrolysis, interactions between the start site and the switch 1, 2, 4 and 5 regions are established, which allow the holoenzyme to overcome a second block in activation. As a consequence of interacting with the start site, the switch 1, 2, 4 and 5 regions are refolded and achieve start site melting. As a result, the second block is lifted and the downstream fork junction progresses along the template to the +2 site where binding interactions are stronger and shift the equilibrium towards a stable melted state of the DNA duplex.
The switch 1, 2, 4 and 5 regions control start site melting at the downstream fork junction
The switch 2 region is suggested to control the fitting of the template DNA into the active centre (34). Residue β′-R339 has been shown to directly contact the −1 to +2 phosphate backbone of the DNA template strands via electrostatic interactions (17,52,53) and mutation of this residue blocks DNA melting upstream of +1 (26). Substitution of R337 by serine has been demonstrated to change the contacts with the DNA template (35,54) and the switch 2 region has been proposed to undergo conformational changes during which contacts with the template DNA change to stabilize a conformation where melting around the start site can occur. All these observations are in line with our results showing that the switch 2 mutant is severely impaired in RPo formation due to its defects in melting the start site. The fact that promoter-directed transcription initiation can be partially restored if the start site melting requirement is bypassed confirms the key contribution of the switch 2 region to this particular step. Additionally, our results indicate that the switch 1, 4 and 5 regions cooperate with the switch 2 region in controlling start site melting. All these mutants are rescued in a very similar manner in the presence of the template with a pre-melted start site.
The switch 3 region controls clamp closure
The switch 3 region has been predicted to form a hydrophobic cleft for the first unpaired RNA base (−12), after it separates from the RNA–DNA hybrid, and subsequently bind to each RNA base in a nascent transcript as it dissociates from the DNA template (54,55) and seems to predominantly function in a clamp opening and closing mechanism. We observe that the switch 3 mutants have defects distinct from the switch 1, 2, 4 and 5 mutants, although we cannot exclude a contribution of the switch 3 region to the start site melting mechanism. Indeed, the switch 3 mutants show enhanced activity in the presence of a pre-melted start site. Both mutations in the switch 3 region generated RNAPs that are dysfunctional in both interacting with a DNA template and binding σ. Eσ forms an open clamp state where the DNA duplex can be loaded into the active centre and a closed clamp state. Mutations in the switch 3 region may then shift RNAP towards clamp closure and prevent access of the DNA template and the sigma factor to the active centre. We further argue that this state is not irreversible. The enzymes recovered their activity when they were trapped with a bEBP suggesting an equilibrium shift towards a more open clamp state by creating a longer time window for the clamp to swing from the closed to the open conformation. X-ray structures show the switch 3 region as a particularly long and flexible loop (56) and its mutation might lock it in a conformation that is predominant in a closed clamp state. We therefore propose that apart from its function in dissociating the transcript, the switch 3 region is key in coupling start site melting to clamp closure by undergoing conformational changes. The switch 3 regions seem to act at the end of the pathway that is initiated by the switch 1, 2, 4 and 5 regions. The mechanism of open complex formation is triggered by contacts between the switch 1, 2, 4 and 5 regions and the start site that lead to conformational changes of the switch 1, 2, 4 and 5 regions that, in turn, facilitate start site melting. The structural rearrangements are then relayed to the switch 3 domain, which also undergoes conformational changes that lead to clamp closure and RPo formation (Figure 9). One of the mutations in the switch 3 region, β-G1267V, was not only identified as a DksA suppressor mutant but was also shown to escape inhibition by DksA. For this reason, β-G1267 was postulated to act at the end of the signal transmission pathway that leads to inhibition by DksA (45). This view is consistent with our observations and supports the view that DksA interferes with the transmission of the structural rearrangements that the switch regions undergo en route to RPo formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–7.
FUNDING
The Biotechnology and Biological Sciences Research Council [BB/H012249/1 to M.B. supporting S.C.W.]; the Wellcome Trust [WT093044MA to M.B. supporting P.C.B.]. Funding for open access charge: Imperial College London.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Jovanovic and N. Zhang for help with protocols; S. Wigneshweraraj, V. Koronakis, E. Koronakis, R. Jansen and S. Heim for reagents and R. Weinzierl, E. James, C. Waite and S. Wigneshweraraj for useful comments on this article.
REFERENCES
- 1.Werner F. Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 2008;16:247–250. doi: 10.1016/j.tim.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 3.Rice LB. Mechanisms of resistance and clinical relevance of resistance to beta-lactams, glycopeptides, and fluoroquinolones. Mayo Clin. Proc. 2012;87:198–208. doi: 10.1016/j.mayocp.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srivastava A, Talaue M, Liu S, Degen D, Ebright RY, Sineva E, Chakraborty A, Druzhinin SY, Chatterjee S, Mukhopadhyay J, et al. New target for inhibition of bacterial RNA polymerase: 'switch region'. Curr. Opin. Microbiol. 2011;14:532–543. doi: 10.1016/j.mib.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck M, Bose D, Burrows P, Cannon W, Joly N, Pape T, Rappas M, Schumacher J, Wigneshweraraj S, Zhang X. A second paradigm for gene activation in bacteria. Biochem. Soc. Trans. 2006;34:1067–1071. doi: 10.1042/BST0341067. [DOI] [PubMed] [Google Scholar]
- 7.Wigneshweraraj SR, Burrows PC, Bordes P, Schumacher J, Rappas M, Finn RD, Cannon WV, Zhang X, Buck M. The second paradigm for activation of transcription. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:339–369. doi: 10.1016/S0079-6603(04)79007-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Chaney M, Wigneshweraraj SR, Schumacher J, Bordes P, Cannon W, Buck M. Mechanochemical ATPases and transcriptional activation. Mol. Microbiol. 2002;45:895–903. doi: 10.1046/j.1365-2958.2002.03065.x. [DOI] [PubMed] [Google Scholar]
- 9.Joly N, Zhang N, Buck M, Zhang X. Coupling AAA protein function to regulated gene expression. Biochim. Biophys. Acta. 2012;1823:108–116. doi: 10.1016/j.bbamcr.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Bose D, Pape T, Burrows PC, Rappas M, Wigneshweraraj SR, Buck M, Zhang X. Organization of an activator-bound RNA polymerase holoenzyme. Mol. Cell. 2008;32:337–346. doi: 10.1016/j.molcel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrows PC, Joly N, Buck M. A prehydrolysis state of an AAA+ ATPase supports transcription activation of an enhancer-dependent RNA polymerase. Proc. Natl Acad. Sci. USA. 2010;107:9376–9381. doi: 10.1073/pnas.1001188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman LJ, Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasse-Dwight S, Gralla JD. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 14.Lin L, Qian Y, Shi X, Chen Y. Induction of a cell stress response gene RTP801 by DNA damaging agent methyl methanesulfonate through CCAAT/enhancer binding protein. Biochemistry. 2005;44:3909–3914. doi: 10.1021/bi047574r. [DOI] [PubMed] [Google Scholar]
- 15.Reitzer L, Schneider BL. Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 2001;65:422–444. doi: 10.1128/MMBR.65.3.422-444.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, et al. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol. Microbiol. 2008;68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 17.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 18.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 19.Landick R. RNA polymerase clamps down. Cell. 2001;105:567–570. doi: 10.1016/s0092-8674(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 20.Wigneshweraraj SR, Savalia D, Severinov K, Buck M. Interplay between the beta' clamp and the beta' jaw domains during DNA opening by the bacterial RNA polymerase at sigma54-dependent promoters. J. Mol. Biol. 2006;359:1182–1195. doi: 10.1016/j.jmb.2006.04.063. [DOI] [PubMed] [Google Scholar]
- 21.Wigneshweraraj SR, Burrows PC, Nechaev S, Zenkin N, Severinov K, Buck M. Regulated communication between the upstream face of RNA polymerase and the beta' subunit jaw domain. EMBO J. 2004;23:4264–4274. doi: 10.1038/sj.emboj.7600407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gries TJ, Kontur WS, Capp MW, Saecker RM, Record MT., Jr One-step DNA melting in the RNA polymerase cleft opens the initiation bubble to form an unstable open complex. Proc. Natl Acad. Sci. USA. 2010;107:10418–10423. doi: 10.1073/pnas.1000967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kontur WS, Capp MW, Gries TJ, Saecker RM, Record MT., Jr Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry. 2010;49:4361–4373. doi: 10.1021/bi100092a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontur WS, Saecker RM, Capp MW, Record MT., Jr Late steps in the formation of E. coli RNA polymerase-lambda P R promoter open complexes: characterization of conformational changes by rapid [perturbant] upshift experiments. J. Mol. Biol. 2008;376:1034–1047. doi: 10.1016/j.jmb.2007.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santangelo TJ, Reeve JN. Deletion of switch 3 results in an archaeal RNA polymerase that is defective in transcript elongation. J. Biol. Chem. 2010;285:23908–23915. doi: 10.1074/jbc.M109.094565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pupov D, Miropolskaya N, Sevostyanova A, Bass I, Artsimovitch I, Kulbachinskiy A. Multiple roles of the RNA polymerase beta' SW2 region in transcription initiation, promoter escape, and RNA elongation. Nucleic Acids Res. 2010;38:5784–5796. doi: 10.1093/nar/gkq355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M. Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 2008;36:676–687. doi: 10.1093/nar/gkm1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majovski RC, Khaperskyy DA, Ghazy MA, Ponticelli AS. A functional role for the switch 2 region of yeast RNA polymerase II in transcription start site utilization and abortive initiation. J. Biol. Chem. 2005;280:34917–34923. doi: 10.1074/jbc.M502932200. [DOI] [PubMed] [Google Scholar]
- 29.Burrows PC, Joly N, Cannon WV, Camara BP, Rappas M, Zhang X, Dawes K, Nixon BT, Wigneshweraraj SR, Buck M. Coupling sigma factor conformation to RNA polymerase reorganisation for DNA melting. J. Mol. Biol. 2009;387:306–319. doi: 10.1016/j.jmb.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camara B, Liu M, Reynolds J, Shadrin A, Liu B, Kwok K, Simpson P, Weinzierl R, Severinov K, Cota E, et al. T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc. Natl Acad. Sci. USA. 2010;107:2247–2252. doi: 10.1073/pnas.0907908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho MX, Hudson BP, Das K, Arnold E, Ebright RH. Structures of RNA polymerase-antibiotic complexes. Curr. Opin. Struc. Biol. 2009;19:715–723. doi: 10.1016/j.sbi.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haebich D, von Nussbaum F. Lost in transcription–inhibition of RNA polymerase. Angew. Chem. Int. Ed. Engl. 2009;48:3397–3400. doi: 10.1002/anie.200900338. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol. Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tupin A, Gualtieri M, Leonetti JP, Brodolin K. The transcription inhibitor lipiarmycin blocks DNA fitting into the RNA polymerase catalytic site. EMBO J. 2010;29:2527–2537. doi: 10.1038/emboj.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belogurov GA, Vassylyeva MN, Sevostyanova A, Appleman JR, Xiang AX, Lira R, Webber SE, Klyuyev S, Nudler E, Artsimovitch I, et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;457:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, et al. The RNA polymerase "switch region" is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koronakis V. TolC–the bacterial exit duct for proteins and drugs. FEBS Lett. 2003;555:66–71. doi: 10.1016/s0014-5793(03)01125-6. [DOI] [PubMed] [Google Scholar]
- 38.Jovanovic M, Burrows PC, Bose D, Camara B, Wiesler S, Weinzierl RO, Zhang X, Wigneshweraraj S, Buck M. An activity map of the Escherichia coli RNA polymerase bridge helix. J. Biol. Chem. 2011;286:14469–14479. doi: 10.1074/jbc.M110.212902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joly N, Rappas M, Wigneshweraraj SR, Zhang X, Buck M. Coupling nucleotide hydrolysis to transcription activation performance in a bacterial enhancer binding protein. Mol. Microbiol. 2007;66:583–595. doi: 10.1111/j.1365-2958.2007.05901.x. [DOI] [PubMed] [Google Scholar]
- 40.Wigneshweraraj SR, Nechaev S, Bordes P, Jones S, Cannon W, Severinov K, Buck M. Enhancer-dependent transcription by bacterial RNA polymerase: the beta subunit downstream lobe is used by sigma 54 during open promoter complex formation. Methods Enzymol. 2003;370:646–657. doi: 10.1016/S0076-6879(03)70053-6. [DOI] [PubMed] [Google Scholar]
- 41.Joly N, Burrows PC, Buck M. An intramolecular route for coupling ATPase activity in AAA+ proteins for transcription activation. J. Biol. Chem. 2008;283:13725–13735. doi: 10.1074/jbc.M800801200. [DOI] [PubMed] [Google Scholar]
- 42.Rozovskaya TA, Rechinsky VO, Bibilashvili RS, Karpeisky M, Tarusova NB, Khomutov RM, Dixon HB. The mechanism of pyrophosphorolysis of RNA by RNA polymerase. Endowment of RNA polymerase with artificial exonuclease activity. Biochem. J. 1984;224:645–650. doi: 10.1042/bj2240645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang N, Joly N, Burrows PC, Jovanovic M, Wigneshweraraj SR, Buck M. The role of the conserved phenylalanine in the sigma54-interacting GAFTGA motif of bacterial enhancer binding proteins. Nucleic Acids Res. 2009;37:5981–5992. doi: 10.1093/nar/gkp658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popham DL, Szeto D, Keener J, Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 47.Chaney M, Grande R, Wigneshweraraj SR, Cannon W, Casaz P, Gallegos MT, Schumacher J, Jones S, Elderkin S, Dago AE, et al. Binding of transcriptional activators to σ 54 in the presence of the transition state analog ADP-aluminum fluoride: Insights into activator mechanochemical action. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saecker RM, Record MT, Jr, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wedel A, Kustu S. The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 50.Mekler V, Minakhin L, Severinov K. A critical role of downstream RNA polymerase-promoter interactions in the formation of initiation complex. J. Biol. Chem. 2011;286:22600–22608. doi: 10.1074/jbc.M111.247080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y, Lew CM, Gralla JD. Promoter opening by sigma(54) and sigma(70) RNA polymerases: sigma factor-directed alterations in the mechanism and tightness of control. Genes Dev. 2000;14:2242–2255. doi: 10.1101/gad.794800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struc. Mol. Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 54.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 55.Kent T, Kashkina E, Anikin M, Temiakov D. Maintenance of RNA-DNA hybrid length in bacterial RNA polymerases. J. Biol. Chem. 2009;284:13497–13504. doi: 10.1074/jbc.M901898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, Darst SA. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8:e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.