Abstract
Piwi proteins and Piwi-interacting small RNAs (piRNAs) have known functions in transposon silencing in the male germline of fetal and newborn mice. Both are also present in adult testes; however, their function here remains a mystery. Here, we confirm that most piRNAs in meiotic spermatocytes originate from clusters in non-repeat intergenic regions of DNA. The regulation of these piRNA clusters, including the processing of the precursor transcripts into individual piRNAs, is accomplished through mostly unknown processes. We present a possible regulatory mechanism for one such cluster, named cluster 1082B, located on chromosome 7 in the mouse genome. The 1082B precursor transcript and its 788 unique piRNAs are repressed by the Alkbh1 dioxygenase and the testis-specific transcription repressor Tzfp. We observe a remarkable >1000-fold upregulation of individual piRNAs in pachytene spermatocytes isolated from Alkbh1- and Tzfp-deficient murine testes. Repression of cluster 1082B is further supported by the identification of a 10-bp Tzfp recognition sequence contained within the precursor transcript. Downregulation of LINE1 and IAP transcripts in the Alkbh1- and Tzfp-deficient mice leads us to propose a potential role for the 1082B-encoded piRNAs in transposon control.
INTRODUCTION
Spermatogenesis is a cyclic developmental process by which spermatogonia generate mature spermatozoa (1,2). The process takes place in the seminiferous tubules of the testis and involves three phases: mitotic proliferation of spermatogonia, meiosis and finally, spermiogenesis, which involves a stepwise maturation of the spermatids to mature spermatozoa. Meiosis involves two successive cell divisions of spermatocytes resulting in the formation of haploid spermatids. During the prolonged prophase I of meiosis, the primary spermatocytes go through the leptotene, zygotene, pachytene, diplotene and diakinesis stages in a highly organized, sequential manner that involves homologous chromosome pairing, synaptonemal complex formation and meiotic recombination.
Piwi-interacting small RNA (piRNA) is a distinct class of small non-coding RNAs. They are defined by their association with Piwi proteins, a subgroup of the Argonaute protein family (AGO), which are well-known mediators in small RNA-mediated silencing (3). The Piwi proteins were first identified in Drosophila melanogaster (4) but have now been characterized in many different organisms, including mammals. Three murine Piwi proteins have been identified: Miwi, Mili and Miwi2 (5–7). Unlike other small non-coding RNAs like microRNAs (miRNAs) and endo-small interfering RNAs (endo-siRNAs), piRNAs seem to be highly specific for the male germline in mammals (8–11). All three Piwi proteins are essential for spermatogenesis in mouse as deletion of all the genes renders male knockout mice infertile (5–7).
Each of the three mouse Piwi proteins associate with a specific subset of piRNAs and have different expression patterns (12). Mili and Miwi2 are expressed during embryogenesis and just after birth (7,8) and bind piRNAs called pre-pachytene piRNAs. Deep sequencing analyses have shown that these piRNAs often map to repeat-associated DNA sequences (13), and it is well established that these complexes are important for transposon silencing (7,14–16). Mili is also expressed in adults and can be detected in all spermatogenic cells until the round spermatid stage (8). Miwi is expressed in adult animals and can be detected in pachytene spermatocytes to elongating spermatids (5). Miwi-associated piRNAs are called pachytene piRNAs and are relatively depleted of repeat elements (13). Whether the Piwi–piRNA complexes in adult testes primarily control RNA stability, gene transcription, chromatin organization or protein synthesis is not well understood (17). However, both Mili and Miwi have been shown to be endonucleases (15,18,19), and their function is thought, at least in part, to be piRNA-guided degradation of target transcripts. In addition, some evidence suggest that they may be involved in translational control (20,21).
piRNAs are believed to derive from long single-stranded transcripts and processed either through a primary processing pathway or the so-called ping-pong amplification cycle (15,22,23). The production of pachytene piRNAs from long primary transcripts was recently confirmed by Ragan et al. using the novel NORAHDESK tool (24). The primary processing probably involves splicing of long transcripts from piRNA-rich genomic sequences called piRNA clusters (8,9). This process is thought to be independent of the endonuclease activity of Piwi proteins (25). piRNA clusters may extend for tens of thousands of bases, and each cluster encodes a precursor transcript that can generate many different piRNA sequences, some of which may be partially overlapping. No secondary structures for the primary transcripts have been determined. Many clusters give rise to piRNAs that map to both genomic strands, suggesting bidirectional transcription (13,15,22).
We previously characterized the Alkbh1−/− mouse, which display developmental defects and sex-ratio distortion. Mutant animals are viable and fertile, but have a decreased survival rate and display increased apoptosis in adult testes (26). Alkbh1 is a member of the mammalian AlkB family of dioxygenases and is proposed to be involved in epigenetic regulation (26–29). Alkbh1 localizes to nuclear euchromatin (27), and recent studies suggest that ALKBH1 is a histone H2A dioxygenase (Ougland et al., submitted for publication). Recently, we generated a Tzfp mouse model (hereafter referred to as the TzfpGTi/GTi mouse). Tzfp is a testis-specific transcription repressor belonging to the BTB/POZ Zn finger family. Mice lacking this protein are viable and fertile and have no obvious phenotype (unpublished data). The zinc finger domain of Tzfp binds to a specific genomic sequence, the Tzfp-binding site (tbs) (TGTACAGTGT) located upstream of the Aie1 gene (30). The interaction with the tbs has a repressive effect on the target gene. Expression analyses have shown that both Tzfp and Alkbh1 are highly expressed in adult testes and that the expression peaks at the pachytene stage during spermatogenesis (26) (Furu et al., in preparation).
In the present work, we present a possible regulatory mechanism of a non-repeat piRNA cluster located on chromosome 7, consisting of 788 unique anti-sense piRNAs. The precursor transcript and in-dividual piRNAs derived from this cluster are dramatically upregulated in the absence of Alkbh1 and Tzfp, as shown in the two mouse models TzfpGTi/GTi and Alkbh1−/−. The cluster contains the Tzfp-binding sequence, implying that the interaction partners Tzfp and Alkbh1 regulate the expression of the precursor by binding to the tbs motif. Further, we provide evidence for repression of long interspersed elements 1 (LINE1) and intracisternal A-particle (IAP) transcripts in these mouse models, indicating that upregulated piRNAs derived from cluster 1082B are involved in transposon control.
MATERIALS AND METHODS
Mouse handling and genotyping
All mice experiments were approved by the Norwegian Animal Research Authority (Ref. nr. 08/9940; 12/10-2388) and done in accordance with institutional guidelines at the Centre for Comparative Medicine at Oslo University Hospital. Animal work was conducted in accordance with the rules and regulations of the Federation of European Laboratory Animal Science Associations. Alkbh1-null mice were previously generated by targeted deletion of exon 6, and heterozygous were backcrossed onto a C57BL6/J background (26). Tzfp-targeted mice were designed by a gene-trap procedure using an embryonic stem (ES) cell clone containing a splice acceptor site from the gene trap vector (Omnibank Gene Trap Vector 76) upstream of exon 1 of the Tzfp gene (Furu and Klungland, in preparation). The clone was obtained from Texas Institute of Genomic Medicine C57BL6/J ES cell clone library. For genotyping, ear-clip samples were degraded by incubation in 75 µl Hot Shot Lysis Buffer (25 mM NaOH, 0.2 mM Na2EDTA, pH 12) at 95°C for 30 min and then cooled down to 4°C before adding 75 µl Hot Shot Neutralization Buffer (40 mM Tris–HCl, pH 5). Samples were PCR amplified for 35 cycles with an annealing temperature of 60°C for the Alkbh1 gene and 40 cycles with an annealing temperature of 58°C for the Tzfp gene. See primers 1–6 in Supplementary Table S1.
DNA microarray analysis
Total RNA was isolated from three Alkbh1+/+ and three Alkbh1−/− 12-week-old testes using the Fast RNA Pro Green Kit (MP Biomedicals) according to the manufacturer’s protocol. DNA remnants were removed using TURBO DNase (Ambion), and the RNA quality was checked using Agilent Bioanalyzer 2100 (RIN value between 9.8 and 10.0). Fifteen micrograms of biotinylated and fragmented cRNA was then hybridized onto the GeneChip Mouse Genome 430 2.0 Array (Affymetrix) according to the manufacturer’s protocol. Quality checks including scale factor, background, noise, spikes and RNA degradation were performed and validated using the yaqcaffy library (http://www.bioconductor.org/packages/2.3/bioc/%20vignettes/yaqcaffy/inst/doc/yaqcaffy.pdf). Affymetrix raw data were generated with GCOS 1.4 (GeneChip Operating Software, Affymetrix), and the signal intensities of each probe set were normalized with the robust microarray analysis (RMA) algorithm. To find differentially expressed genes, t-test with randomized variance was used as statistical test, and the cutoff (P value) was set to 0.05 with a FDR correction. Fold change for all genes that passed the above criteria was computed, and only the genes with ≥2-fold change were studied. A heatmap was generated using the GeneSpring GX 10 demoversion (Agilent). All data are MIAME compliant, and the raw data have been deposited in a MIAME compliant database under accession number GSE22073.
STAPUT isolation of pachytene cells
Pachytene cells were isolated from C57BL6/J, Alkbh1−/− and TzfpGTi/GTi 12-week-old testes using an adapted version of the STAPUT method (Bellvè, 1993). Each isolation required a total of six males, and 2× six mice for each genotype were used. The mice were killed using CO2 and testes were taken out and put in ice-cold DMEM medium containing antibiotics. Testes were detunicated and the tubules treated with DNaseΙ, collagenase, trypsin and hyaluronidase (all from Sigma-Aldrich) at 34°C to remove connective tissue and somatic cells, yielding a cell suspension of germinal cells in DMEM containing 0.5% BSA. The cell suspension was loaded into the cell loading chamber of the STAPUT apparatus and separated by sedimentation velocity at unit gravity in a 2–4% w/v BSA gradient in DMEM medium at 4°C for 2.5 h. After sedimentation, 10-ml fractions were collected and checked under the microscope. Fractions containing pachytene spermatocytes were pooled and the cell number was counted in a Countess® Automated Cell Counter (Invitrogen). Cells were spun down, and the pellet was snap frozen in liquid nitrogen before being placed in −70°C. An aliquot of purified cells was fixed on SuperFrost Plus slides (VWR) using cell adherence solution (Crystalgen, Lot no 425081) for microscopic analyses (Supplementary Figure S2). One isolation yielded ∼1.5 × 106 pachytene cells with an average size of 12.5 µm.
TaqMan® and SYBR® green gene expression analysis
Total RNA was isolated from C57BL6/J, TzfpGTi/GTi and Alkbh1−/− testes using the Fast RNA Pro Green Kit (MP Biomedicals) according to the manufacturer’s protocol. Twelve-week-old animals were used unless stated otherwise. DNA remnants were removed using TURBO DNase (Ambion), and complementary DNA (cDNA) was made using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The quantitative PCRs were carried out on a StepOnePlus instrument (Applied Biosystems).
TaqMan® analysis
Reactions were set up using 50 ng cDNA, TaqMan® Fast Universal PCR Master Mix and appropriate TaqMan primers and probes (all reagents from Applied Biosystems). Default PCR program settings were used. Pre-designed primers and probes were applied for target genes (Alkbh1, Tzfp, 4933440M02Rik) and endogenous control (Gapdh). See TaqMan probes 7–10 in Supplementary Table S1.
SYBR® green analysis
Reactions were set up using 5 ng cDNA, Power SYBR® Green PCR master mix and appropriate primers (300 nM) (all reagents from Applied Biosystems). Default PCR program and melt curve settings were used. Transposon primer sequences were similar to the ones used by Carmell et al. (7). Pachytene purity from STAPUT was verified using quantification of the pachytene marker Lcn2 on isolated large RNAs (>200 nt). SYBR Green analysis with appropriate primers designed in http://eu.idtdna.com/SCITOOLS/Applications/PrimerQuest/Default.aspx were used. GADPH was used as endogenous control. See primers 17–28 in Supplementary Table S1.
At least two biological parallels were used for each genotype. All samples were run in triplicates and with two technical parallels. The relative expression was calculated using the equation RQ = 2−ΔΔCT, where RQ is the relative quantity of the target gene. ΔΔCT is the difference in CT values between target gene and the endogenous control minus the difference in CT values between the reference gene and the endogenous control.
miScript PCR system for quantification of piRNAs
Small RNAs <200 nt were isolated from 2× C57BL6/J, Alkbh1−/− and TzfpGTi/GTi pachytene cells using the mirVana miRNA Isolation Kit (Ambion) in line with the manufacturer’s protocol. Any DNA remnants were removed using TURBO DNase (Ambion), and cDNA was made using miScript Reverse Transcription Kit (Qiagen). The quantitative PCRs were carried out on a StepOnePlus instrument using 1 ng cDNA, miScript SYBR Green PCR Kit (Qiagen) and Custom miScript Primer Assays (piR-19852, piR-12359, piR-103121, piR-17918 and piR-4749) (Qiagen). The RNU6B miScript PCR control (snRNA, 45 nt) was chosen as endogenous control (Qiagen). All samples were run in triplicates with two technical parallels. The relative expression was calculated as for the TaqMan® Gene Expression Analysis. Primers 11–15 are listed in Supplementary Table S1.
Sequencing and computational analysis of small RNAs
Small RNAs from pachytene cells used in the miScript PCR system were also run in a high-throughput sequencing pipeline. Of RNA (<200 nt), 50 ng was diluted to 5.7 µl, entering the Small RNA Sample Preparation Guide (Illumina Part # 1004239 Rev. B August 2009) at the 5′-adapter ligation step. Subsequently, 3′-adapter ligation and cDNA synthesis followed by PCR amplification (15 cycles) were performed using the Small RNA Sample Prep Kit (FC-102-1009, Illumina) according to the manufacturer’s recommendations. Finally, the small RNA library (105–110 bp) was sequenced (36-bp Single-End Read) using the Illumina Genome Analyzer ΙΙx (GAΙΙx).
Prior to analyzing, the sequence reads consisting solely of adapter sequences were removed. The remaining reads were aligned to the reference genome (mm9) using Novoalign (http://www.novocraft.com/), with settings allowing maximum one mismatch and automatic adapter trimming. For reads that had a best hit to fewer than six positions in the genome, all positions were considered. The rest were considered to have an unknown location. Known piRNA clusters were downloaded from piRNABank (31) and piRNAdb (http://kbrb.ioz.ac.cn/piRNA). All non-overlapping clusters from these two sources were considered for annotation. Other non-coding RNAs (miRNA, small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), transfer RNA (tRNA), long intergenic non-coding RNA (lincRNA), ribosomal RNA (rRNA) and miscellaneous RNA miscRNA) were downloaded from Ensembl using the BioMart system (http://www.ensembl.org/biomart/). In addition, GtRNAdb (http://gtrnadb.ucsc.edu/) was used for annotated tRNAs. Annotated exons, introns, 5′-UTR and 3′-UTR, in addition to the RepeatMasker track, were downloaded from the UCSC table browser.
Annotation of the mapped reads to the various non-coding RNA classes, as well as the gene and repeat tracks, was done using the intersectBed script from the BedTools software suit (32). The intersections were initially done against the annotated piRNA clusters. The resulting uncharacterized reads were then mapped to miRNA, rRNA, tRNA, snoRNA, snRNA, lincRNA and finally, miscRNA. Similarly, all reads were annotated to the RepeatMasker classes. The reads were organized into LTR, LINE and SINE. These reads were considered repeat derived. Any read that mapped to a different repeat category was classified as other repeat.
Potential piRNAs included reads that overlapped with the annotated piRNA clusters. The expression of piRNAs was measured at the cluster level by summing the number of mapped reads for each cluster intersected with intersectBed. Reads that mapped to several positions were divided between them such that fractions were used instead of full counts. To avoid counting unique sequences resulting from sequencing errors as novel piRNAs, we required at least two reads for a novel piRNA sequence to be called.
In order to detect significantly differentially expressed clusters, the DESeq package for the statistical environment R was used (33). Significantly differentially expressed clusters were defined as all clusters below a 10% false discovery rate.
Search for potential piRNA targets
Potential piRNA targets in the July 2007 (NCBI37/mm9) genome assembly of the Mus musculus strain C57BL6/J were identified using BLAST version 2.2.26 (34) with options set to detect highly diverged sequences (-W 7 -F F -r 3 -q -2 -G 5 -E 5). SWIPE version 2.0.3 (35) was also used for this purpose. The complete 4227-bp sequence of cluster 1082B was used as the query. Sequences matching only in the 97-bp 5′-LINE1 region or the adenine/guanine-rich region located at 2180–2470 bp were ignored. Matching regions were inspected using the UCSC genome browser (36).
RESULTS
Increased expression of a piRNA precursor in Alkbh1−/− and TzfpGTi/GTi testes
Alkbh1 deficiency in mice leads to reduced survival rate in embryos and causes embryonic abnormalities, and surviving mice have a dramatic sex-ratio skewing (26). Although male mice homozygous for the mutant gene were fertile, the sex-ratio distortion may possibly be due to aberrant formation of male germ cells. The hypothesis that Alkbh1 has a role in germ cell formation was strengthened by the high Alkbh1 expression in testis.
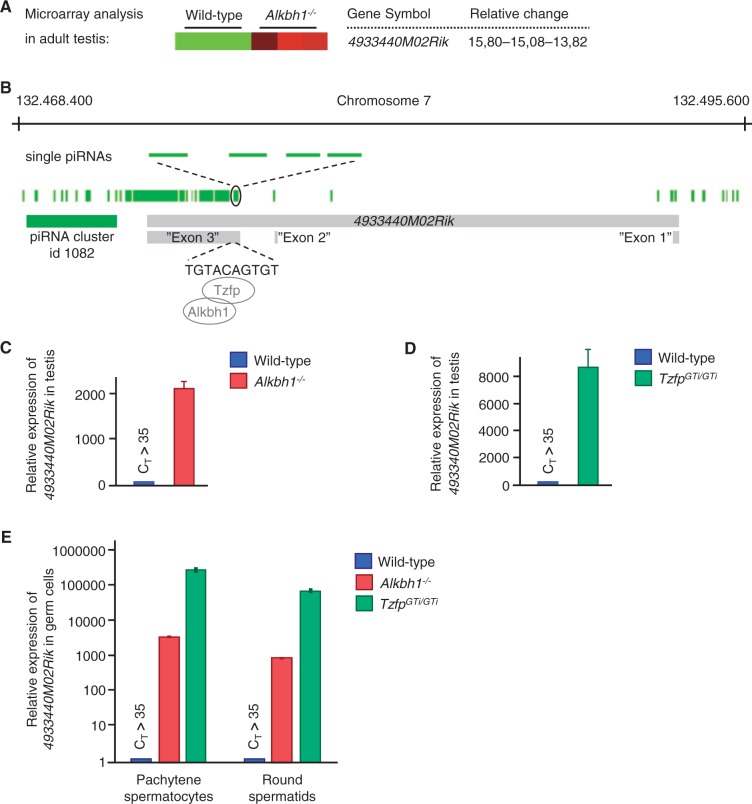
To gain more insight into the role of Alkbh1 in mouse spermatogenesis, we searched for Alkbh1-regulated genes in adult testes using microarray analysis. Using class comparison strategy, we identified 25 differentially expressed genes in Alkbh1−/− testes. Although most of the genes affected are likely indirect targets of Alkbh1, one of the genes, 4933440M02Rik, caught our interest due to its tremendous 15-fold upregulation (Figure 1A). The RefSeq for 4933440M02Rik is suppressed as no support for a protein encoded by this transcript has been found. Parallel sequence alignment of 4933440M02Rik in ParAlign 5.0 resulted in hits on 30 different mouse piRNAs. An expanded search in piRNABank (http://pirnabank.ibab.ac.in) revealed a cluster of 225 individual piRNAs containing the 4933440M02Rik transcript on chromosome 7 (Figure 1B). The piRNA cluster is defined with id 1082 and has a cluster length of 39.2 kb.
Figure 1.
The expression of 4933440M02Rik is upregulated in the Alkbh1−/− and TzfpGTi/GTi testes. (A) Microarray analysis performed on whole testes indicating a 15-fold upregulation of the 4933440M02Rik gene in Alkbh1−/− testes. (B) Schematic representation of the 4933440M02Rik gene and transcript, a likely precursor for a piRNA cluster with id 1082 (adopted from http://pirnabank.ibab.ac.in). Magnification of indicated part of the cluster shows single piRNAs (light green) spliced from the piRNA precursor. The 10-bp Tzfp binding sequence (tbs) is located within ‘Exon 3’. (C,D) qPCR analyses of the 4933440M02Rik transcript in Alkbh1−/− (red) (C) and TzfpGTi/GTi (green) (D) indicate a dramatic upregulation in the mutant testes when compared to wild type (blue). (E) qPCR analyses of the 4933440M02Rik transcript in wild type (blue), TzfpGTi/GTi (green) and Alkbh1−/− (red) pachytene spermatocytes (PC) and round spermatids (RS). Wild type is used as reference sample (RQ = 1).
The tremendous upregulation of the 4933440M02Rik transcript in the Alkbh1−/− testes indicates a possible role for Alkbh1 in the regulation of the piRNA cluster located within the 4933440M02Rik gene. This hypothesis was strengthened by the identification of a binding site for Tzfp, a possible Alkbh1 interaction partner, within the gene. Tzfp binds a specific genomic sequence, the tbs TGTACAGTGT (30), located within ‘Exon 3’ of the 4933440M02Rik gene (Figure 1B). Tzfp is a known transcription repressor with a remarkably high expression in testis (37,38).
The ALKBH1–TZFP interaction was identified by screening a testis library by yeast two-hybrid analysis with full-length human ALKBH1 as bait (Supplementary figure S1A). The protein–protein interaction was verified using a second reporter gene (HTX assay, Supplementary figure S1A) as well as plasmid control digest (Supplementary figure S1B) and was further verified by dot blot analysis (Supplementary figure S1C). Although we have not been able to verify the ALKBH1-TZFP interaction through Co-IP analysis, the results indicate that these two proteins may be binding partners in vivo.
The strong upregulation of the tbs-containing 4933440M02Rik gene could imply that Tzfp, in cooperation with Alkbh1, represses the piRNA precursor (Figure 1B). To test this, we used the two mouse models Alkbh1−/− and TzfpGTi/GTi to quantitate the 4933440M02Rik transcript in Alkbh1−/− and TzfpGTi/GTi testes. Indeed, quantitative PCR (qPCR) revealed a remarkable 2000-fold upregulation of the 4933440M02Rik piRNA precursor in Alkbh1−/− testes (Figure 1C) and an 8000-fold upregulation in TzfpGTi/GTi testes (Figure 1D). To determine whether this transcript is normally expressed during certain stages of spermatogenesis, male pups were killed at specific time points corresponding to the appearance of different spermatocytes and spermatids in the first wave of spermatogenesis in the juvenile mouse (day 10–26). In addition, pachytene spermatocytes and round spermatids were isolated from adult male mice. qPCR analyses revealed low abundance of the 4933440M02Rik transcript at all stages in juvenile mice (data not shown). In adult mutant cells, this transcript was detected at remarkably high levels in both pachytene spermatocytes and round spermatids, although a lower level was seen in round spermatids (Figure 1E, note the logarithmic scale on the y-axis). Since repression of the piRNA transcript requires both Alkbh1 and Tzfp, it seems likely that the regulation is caused by a stable or transient Alkbh1-Tzfp interaction at a specific DNA sequence.
Alkbh1 and Tzfp repress specific piRNAs in the pachytene stage of meiosis
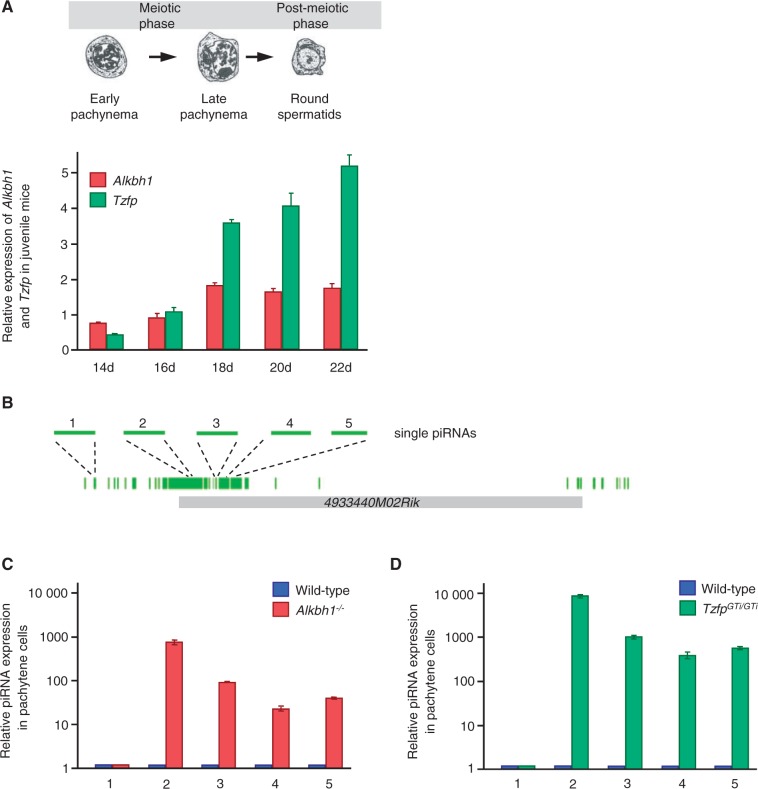
It is previously shown that Alkbh1 expression is particularly high in adult testis (26). For Tzfp, the testis-specific expression is especially pronounced (37,38). By analyzing RNA from juvenile testes, we find that both Alkbh1 and Tzfp expression increases during pachynema (Figure 2A). The upregulation in the pachytene stage is more dramatic for Tzfp than for Alkbh1. Pachynema is the third stage of prophase I of meiosis, in which synapsis is completed and homologous recombination occurs.
Figure 2.
Individual piRNAs derived from parts of cluster 1082 are upregulated in Alkbh1−/− and TzfpGTi/GTi pachytene cells. (A) The upper panel shows a schematic drawing of the meiotic and post-meiotic phases during spermatogenesis (adopted from San Agustin and Witman, 2001). The appearance of early and late pachytene spermatocytes and round spermatids is indicated. The lower panel shows qPCR analyses of Alkbh1 (red) and Tzfp (green) in male juvenile testes at 14–22 days after birth (d). Day 16, which is when mid-pachytene spermatocytes appear, is used as reference sample (RQ = 1). (B) Map showing the location of single piRNAs (light green) investigated by qPCR (numbered 1–5). The indicated piRNAs are distributed along the length of cluster 1082. (C, D) miScript qPCR analyses of the expression of piRNA 1–5 in isolated pachytene cells from wild type (blue), Alkbh1−/− (red) (C) and TzfpGTi/GTi (green) (D). Wild type is used as reference sample (RQ = 1).
Next, we wanted to investigate the expression profile of mature piRNAs derived from the area containing the 4933440M02Rik gene during the pachytene stage. To address this question, we performed qPCR analysis on individual piRNAs (piRNA 1–5) from cluster 1082 on purified pachytene spermatocytes (Figure 2B). A dramatic increase in expression of piRNA 2–5 was found in both mutants (Figure 2C and D) and correlated very well with the expression of the 4933440M02Rik gene described above (Figure 1E). Surprisingly, piRNA 1 expression was not affected by Alkbh1 deletion and was slightly downregulated in the Tzfp mutant (Figure 2C and D). This piRNA is positioned furthest away from the tbs site and was found to be located outside the 4933440M02Rik transcript, but within cluster 1082 as defined by piRNABank (http://pirnabank.ibab.ac.in).
Based on these results, Alkbh1 and Tzfp evidently regulate parts of cluster 1082 during pachynema of meiosis. The repression, however, does not comprise the whole length of this cluster. To get more detailed information on which area of cluster 1082 that is regulated, we performed high-throughput sequencing on small RNAs from Alkbh1−/−, TzfpGTi/GTi and wild-type pachytene cells. These studies will also uncover the possible regulation of other piRNA clusters by Alkbh1 and Tzfp.
The small RNA population is similar in wild-type and mutant pachytene spermatocytes
High-throughput sequencing of small RNAs isolated from purified pachytene spermatocytes was performed for the two mutants utilizing the 36-bp single-read sequencing on the Illumina Genome Analyzer ΙΙx (GAΙΙx). Each mutant sample was analyzed along with a wild-type sample, resulting in the analysis of 2× six C57BL6/J adult mice and 1× six Alkbh1 and Tzfp mutant mice. After adapter trimming and mapping of the sequences to the reference genome, the Alkbh1−/− sample yielded about 10 million reads, whereas the TzfpGTi/GTi sample generated >15 million reads per sample (Supplementary Table S2). The number of reads is appreciably high and makes a promising basis for analysis with high statistical strength (13,39).
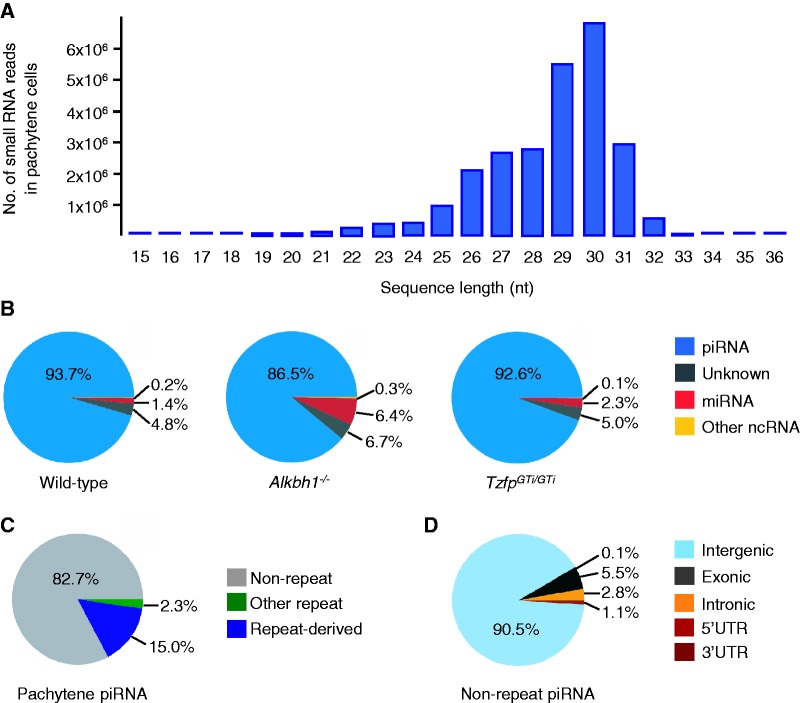
Small RNA reads were annotated as piRNAs if they mapped to a known piRNA cluster (see ‘Materials and Methods’ section). Our analysis demonstrated that pachytene piRNAs have a peak length of 29–30 nt (Figure 3A). Strikingly, as much as 93.7% of all sequence reads were piRNAs, whereas miRNAs only made up 1.4% of the matched reads (Figure 3B, left). The distribution of piRNAs and miRNAs was roughly the same in wild-type and TzfpGTi/GTi pachytene spermatocytes (Figure 3B, right), whereas the miRNA population was slightly higher in Alkbh1−/− pachytene spermatocytes (Figure 3B, middle).
Figure 3.
Small RNA sequencing analysis reveals a majority of 29–30 nt long non-repeat intergenic piRNAs in pachytene spermatocytes. (A) Sequence length distribution of small RNAs in wild-type pachytene cells after high-throughput sequencing, indicating peak expression of 29–30 nt long small RNAs. (B) Distribution of small RNA classes in isolated pachytene cells from wild type (left), Alkbh1−/− (middle) and TzfpGTi/GTi (right). (C) Distribution of pachytene piRNAs into non-repeat associated, repeat associated (LTR, LINE and SINE) and other repeat sequences (analyses performed on the sample from (A)). (D) Distribution of non-repeat pachytene piRNAs into intergenic, exonic, intronic and UTR sequences (analyses performed on the sample from (A)).
The sequencing data revealed a 5′-U bias and a variable 10th position in the pachytene piRNAs (data not shown), suggesting a lack of amplification by the ‘ping-pong’ mechanism. This is in accordance with previous findings and indicates that these piRNAs are generated through primary processing of precursor transcripts only (13,18).
Pachytene piRNAs can be divided into non-repeat, repeat-derived (LTR, LINE, SINE) and other repeat sequences (Figure 3C). Non-repeat piRNAs can be further subdivided into intergenic, exonic, intronic, 5′-UTR and 3′-UTR sequences (Figure 3D). We identified 82.7% of the wild-type piRNAs to be unique, i.e. non-repeat derived (Figure 3C) and 90.5% of the non-repeat piRNAs to be intergenic piRNAs (Figure 3D).
Based on the Illumina reads, we can conclude that the major portion of small RNAs in the meiotic pachytene stage is non-repeat intergenic piRNAs with a peak length of 29–30 nt. These findings are similar to other studies in adult testes from mouse and rat (9–11,13,14). In general, the profile of small RNAs was similar in wild-type and mutant pachytene spermatocytes, suggesting that Alkbh1 and Tzfp are not engaged in global piRNA regulation or biogenesis.
Cluster 1082B is the sole piRNA cluster regulated by Alkbh1 and Tzfp
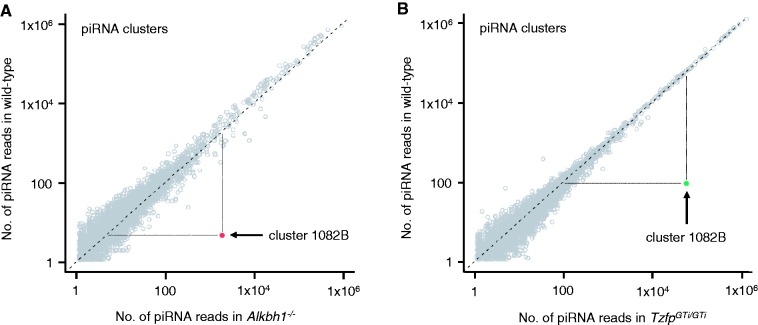
Despite the strong upregulation seen for some of the piRNAs located within cluster 1082, deep sequencing of small RNAs from isolated pachytene spermatocytes revealed that the global small RNA profile was unchanged in mutant pachytene cells when compared to wild type. To investigate whether more piRNA clusters were differentially expressed, we first determined the expression profile of clusters by summing the number of mapped reads for each cluster intersected with intersectBed (see ‘Materials and Methods’ section). By mapping the sequence reads against known piRNA clusters from piRNABank (http://pirnabank.ibab.ac.in) and piRNAdb (http://kbrb.ioz.ac.cn/piRNA), a total of 4944 piRNA clusters were identified. Next, we established if any of the clusters were differentially expressed using the DESeq package for statistical testing (see ‘Materials and Methods’ section). Even though several piRNA clusters showed an increased or decreased expression in mutants compared to wild type (Supplementary Figure S3 and Supplementary Table S3), a part of cluster 1082 was the only region displaying a considerable upregulation in both Alkbh1−/− and TzfpGTi/GTi (Figure 4, Supplementary Table S3). This region was defined as cluster 1082B.
Figure 4.
Cluster 1082B is the sole piRNA cluster upregulated in pachytene cells lacking Alkbh1 and Tzfp. Scatterplot analysis of piRNA cluster reads annotated by small RNA sequencing. Expression of piRNA clusters in pachytene cells is plotted on a log2 scale in a pairwise comparison of wild type (y-axis) and Alkbh1−/− (A) or TzfpGTi/GTi (B) (x-axis). Clusters with a similar number of reads in the two samples line up along the center line (gray, open circles). Cluster 1082B, with a significantly higher number of reads in mutant samples than wild type, deviates noticeably from the center line (red, filled circle in (A); green, filled circle in (B)). The number of clusters presented is 4944. Dashed lines indicate the expression level in each genotype, showing an average expression level for cluster 1082B in wild type and a highly increased expression in the two mutants.
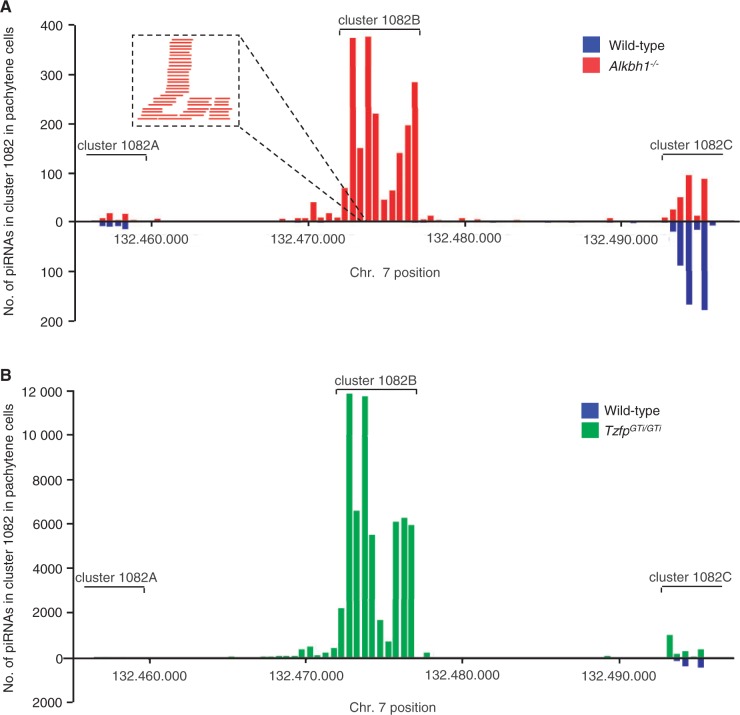
The sequence analysis showed that cluster 1082B covers a 4.2-kb sequence in chromosomal position 132 472 367–132 476 593 on chromosome 7 (Figure 5). Cluster 1082B is a relatively small piRNA cluster, both compared to cluster 1082, which originally had a cluster length of 39.2 kb, and to the average cluster length of 32 kb presented in a similar study (13). In our study, cluster 1082 is redefined into three separate clusters, called cluster 1082A, 1082B and 1082C (Figure 5). Cluster 1082A is a sense-strand cluster, whereas cluster 1082B and 1082C both consist of anti-sense piRNAs. This is in accordance with what Gan et al. found in their deep sequencing analyses (13).
Figure 5.
A 4.2-kb segment of piRNA cluster 1082 makes up cluster 1082B and is regulated by Alkbh1 and Tzfp. Chromosomal view of cluster 1082 on chromosome 7 with back-to-back histograms illustrating piRNA reads in wild type (blue bars) compared to Alkbh1−/− (red bars) (A) and TzfpGTi/GTi (green bars) (B). After deep sequencing, cluster 1082 was redefined into the three separate clusters 1082A, 1082B and 1082C as indicated. Cluster 1082B is dramatically upregulated in mutant pachytene cells and corresponds to chromosomal position 132 472 367–132 476 593 containing 788 unique anti-sense piRNAs. A high-resolution view of a small segment of the upregulated region is given in (A) to illustrate individual, partially overlapping piRNAs.
The precursor sequence for cluster 1082B is partially located within the 4933440M02Rik gene (‘Exon 3’) and contains the Tzfp recognition sequence. An 80% identical 1082B sequence is found on chromosome 1 in the rat genome, and the Tzfp recognition sequence is also conserved (data not shown). None of the other clusters investigated contained the tbs sequence (data not shown). The two mutants show an extreme upregulation in the expression of cluster 1082B, with a logfold change of 8.86 in Alkbh1−/− and 9.25 in TzfpGTi/GTi compared to wild type (Figure 4 and Supplementary Table S3). Cluster 1082B contains 788 unique non-repeat piRNAs, and the distribution of the piRNAs along the cluster shows an almost identical distribution in Alkbh1−/− and TzfpGTi/GTi during pachynema (Figure 5). In wild-type pachytene spermatocytes, the expression of cluster 1082B is comparable to the expression level of other clusters investigated (Figure 4). In the mutant samples, however, cluster 1082B expression is highly above the expression level of most clusters. This means that many of the piRNA clusters are expressed at low levels and are only detectable with highly sensitive methods, e.g. deep sequencing.
We conclude that Alkbh1 and Tzfp specifically repress a 4.2-kb intergenic piRNA cluster on chromosome 7 during the pachytene stage of meiosis. The location of the tbs within this cluster indicates that the upregulated sequence represents a piRNA precursor transcript regulated by Alkbh1 and Tzfp.
Removal of Tzfp and Alkbh1 leads to LINE1 and IAP transcript downregulation
The detailed role of Miwi and its associated piRNAs have, until recently, been largely unknown, although a role in mRNA target silencing has been suggested (20). Reuter et al. recently showed that Miwi binds repeat-derived pachytene piRNAs in adult testes and cleaves target LINE1 RNA complementary to the piRNA sequence (18). This silencing was found to be ping-pong independent and not related to DNA methylation of LINE1 elements. The function of piRNAs derived from unannotated genomic regions, however, still remains a mystery. These piRNAs are not complementary to any other parts of the genome.
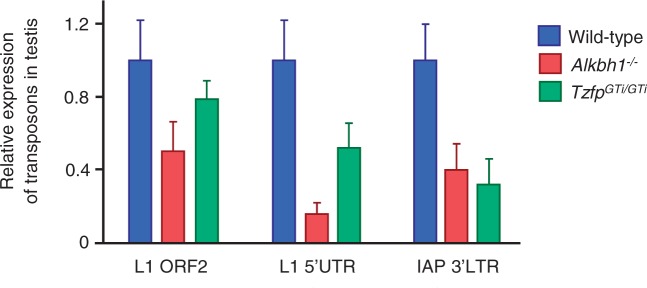
To investigate whether the 1082B-derived piRNAs might affect mRNA stability, we performed mRNA sequencing on wild-type and Alkbh1−/− pachytene spermatocytes. Even though five genes showed a more than 4-fold enrichment in the mutant preparation (Supplementary Figure S4 and Supplementary Table S4), none of the differentially expressed mRNAs are likely targets of the piRNAs from cluster 1082B. Next, we investigated whether LINE1 and IAP transcripts were affected in our two mouse models. Surprisingly, qPCR analysis of IAP (3′-LTR) and LINE1 (5′-UTR and ORF2) elements revealed significant downregulation in the TzfpGTi/GTi and Alkbh1−/− testes, with an average reduction of ∼50% (P < 0.001) (Figure 6). This could indicate that the piRNAs regulated by Alkbh1 and Tzfp in pachytene spermatocytes have the potential to silence LINE1 and IAP sequences and that piRNA upregulation seen in the TzfpGTi/GTi and Alkbh1−/− mice thus causes a reduction in transposon transcripts.
Figure 6.
LINE1 and IAP transcripts are downregulated in the TzfpGTi/GTi and Alkbh1−/− testis. qPCR analyses of transposable elements in adult testes from wild type, TzfpGTi/GTi and Alkbh1−/− samples. The expressions of LINE1 (5′UTR and ORF2) and IAP (3′LTR) transcripts were tested. The two-tailed Student’s t-test was used to check for significant difference in expression between wild type and mutants. A P value <0.001 was found for all three transcripts in both mouse models. Wild type is used as reference sample (RQ = 1).
Search for potential piRNA targets
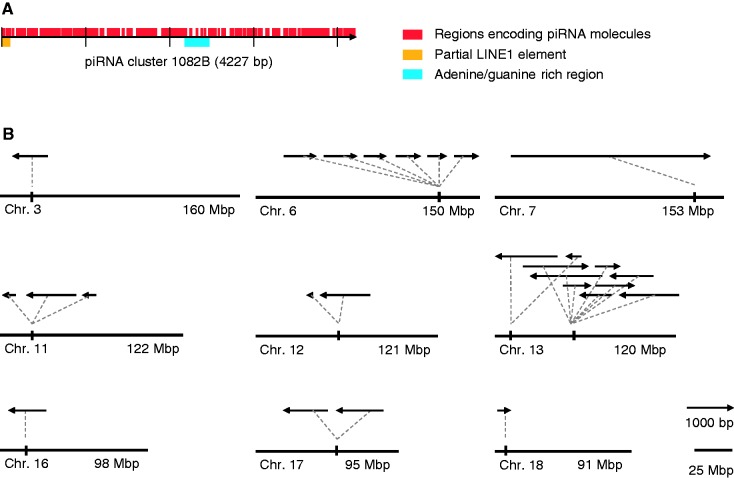
To further investigate the possible function of the piRNAs derived from cluster 1082B, a homology search was performed to see whether these piRNAs can bind to other transcripts or genomic regions. Similarity searches using the 4227-bp sequence of cluster 1082B as query identified several potential piRNA target regions in the mouse genome. Potential targets with significant sequence similarity were identified on chromosomes 3, 6, 11, 12, 13, 16, 17 and 18 (Figure 7 and Supplementary Table S5). The best match was to a region on chromosome 13 that includes the gene 4921525O09Rik (E = 10−45). Interestingly, Girard et al. and Lau et al. have previously mapped several piRNAs to 4921525O09Rik (9,10), indicating that it probably contains a piRNA precursor transcript. The regions on chromosome 11 and 17 include the pseudogene 4932414J04Rik and the gene gm324, respectively, both encoding putative serine/threonine protein kinases. The region on chromosome 16 includes a gene (4933404G15Rik) with unknown function.
Figure 7.
Potential target regions in the mouse genome of piRNAs from cluster 1082B identified by sequence similarity. (A) Location of regions encoding piRNAs (red), a partial LINE1 element (1–97 bp) (yellow) and an adenine/guanine-rich region (2180–2470 bp) (blue) in the sequence of cluster 1082B. (B) The chromosomal location, direction and length of potential piRNA target regions identified by homology search using the 1082B sequence as query. Chromosome 7 with cluster 1082B is included for reference. Distances between target regions are not drawn to scale.
DISCUSSION
The high abundance of piRNAs in immature male germ cells indicates that they have a vital biological role. To study the possible regulation of piRNAs by Alkbh1 and Tzfp, we performed small RNA sequencing on pachytene spermatocytes isolated from mutant and wild-type testes. We obtained a notable ∼15 million reads and ∼10 million reads per sample after adapter trimming and mapping of the sequences to the reference genome, allowing high-quality analysis.
The mouse Piwi protein Miwi is detected from the mid-pachytene stage to elongating spermatids (8). Mili is expressed from primordial germ cells until the round spermatid stage (8). piRNAs expressed during the pachytene stage of meiosis can thus bind both Miwi and Mili, resulting in at least two subpopulations of piRNAs in adult testes (8,9,20). IP analysis has shown that Mili-interacting piRNAs peak at 26–28 nt, whereas Miwi bind 29–31 nt long piRNAs (8). Using deep sequencing analyses, we could confirm that pachytene piRNAs have a peak length of 29–30 nt, indicating that they primarily interact with Miwi (40). In accordance with other studies, we found the majority of small RNAs in pachytene spermatocytes to be piRNAs derived from non-repeat associated, intergenic sequences (13).
ALKBH1 localizes to nuclear euchromatin and recent evidence shows that ALKBH1 is a dioxygenase targeting a novel methylated lysine in histone H2A (27) (Ougland et al., submitted for publication), suggesting a role for the protein in gene regulation through an epigenetic mechanism (26–28). Tzfp, a member of the C2H2-type zinc finger family, binds to the tbs, leading to repression of target genes (30). Here, we show that both proteins are highly expressed during pachynema in spermatogenesis. Using deep sequencing analyses, we found that piRNAs derived from a non-repeat cluster on chromosome 7, called cluster 1082B, are highly upregulated in Alkbh1−/− and TzfpGTi/GTi pachytene spermatocytes. The non-coding RNA transcript covering cluster 1082B was also greatly upregulated in mutant testes, indicating a role for Tzfp and Alkbh1 in the regulation of the precursor transcript encoding the piRNAs within this region. This hypothesis is further substantiated by the identification of the 10-bp Tzfp recognition sequence contained within the cluster. No other piRNA cluster was found to be clearly differentially expressed, indicating that Tzfp and Alkbh1 are not involved in global piRNA regulation.
Combined, these results indicate that Alkbh1 and Tzfp together regulate cluster 1082 in testis. We believe that the repression of the piRNA precursor occurs by Tzfp binding to the tbs sequence, while Alkbh1 changes the surrounding chromatin environment by histone H2A demethylation. During the progress of this project, we also generated the Alkbh1−/−/TzfpGTi/GTi double knockout mice. These mice had reduced fertility and viability comparable to the single Alkbh1−/− knockout mouse (data not shown). This suggests an epistatic relationship between Alkbh1 and Tzfp. The regulatory mechanism of pachytene piRNA clusters has previously been unknown, but our results indicate that piRNA precursor transcripts are individually regulated by stage-specific proteins determining the timing and amount of piRNA expression. Gan et al. found that a large number of transcripts expressed during spermatogenesis, including pseudogenes, are piRNA precursors and suggest that these genes are regulated by alternative splicing and anti-sense transcripts (13). Our findings invite us to speculate whether some of the long ESTs with unknown functions in mouse testes are single-stranded piRNA precursors regulated by members of the C2H2-type zinc finger transcription factors.
Despite intense research over the past few years, the precise function of pachytene piRNAs is still largely unknown. As these piRNAs originate from clusters spread throughout the genome, one of the functions may be patronage of their respective loci, as seen for scnRNAs in Tetrahymena (41), but other functions such as post-transcriptional regulation and roles in epigenetic programming cannot be ruled out. It is now well established that Mili and Miwi2 are essential for de novo DNA methylation and repression of LINE1 in fetal and newborn mice (7,14,16), but as only ∼17% of adult piRNAs map to annotated repeats in mammals (11,14), it has been unclear whether this occurs in adult testes. Interestingly, Grivna et al. observed that Miwi is associated with polysomes throughout spermatogenesis and that it associates with several mRNAs through the cap binding complex (11,20). This would suggest that Miwi-associated piRNAs may be capable of silencing target mRNA transcripts. Other studies have shown that some piRNAs are located in the meiotic nucleus of the male mouse, indicating that these piRNAs could be involved in chromosome remodeling during meiosis and post-meiotic stages (42,43).
The present study can give new insight into the mechanisms related to pachytene piRNAs, as our results indicate that pachytene piRNAs derived from non-repeat sequences may have a role in transposon control. This hypothesis is based on the downregulation of LINE1 and IAP transcripts seen in the Alkbh1−/− and TzfpGTi/GTi testes. Although it cannot be ruled out that the downregulation is a secondary effect, it is tempting to speculate that these piRNAs target LINE1 and IAP transcripts, leading to reduced expression. Reuter et al. recently showed that Miwi and its associated repeat-derived piRNAs target LINE1 RNAs in adult testes, resulting in transcript cleavage by the endonuclease activity of Miwi (18). The cleavage was found to be dependent upon high degree of complementarity between piRNA and target sequence. Yet, the 1082B-derived piRNAs, like the majority of pachytene piRNAs, are devoid of sequences complementary to active transposons, indicating a slicer-independent mechanism. Further studies are needed to elucidate the mechanism by which these piRNAs work, but a miRNA-like mechanism is possible. Upon miRNA binding to target, only a small part of the 5′-end, called the miRNA ‘seed’, requires Watson–Crick pairing (44,45). Binding of the miRNA to the target transcript leads to repression rather than direct degradation.
As silencing of transposons is regarded important for the maintenance of a cell’s integrity, it is somewhat surprising that a cluster of piRNAs, with the potential to inactivate transposons, is repressed in wild-type pachytene cells. LINE1 is, however, known to be expressed in wild-type germ cells (46), and it is possible that a certain level of LINE1 expression during spermatogenesis is evolutionary beneficial. It has been speculated that LINE1 insertions may structurally modify endogenous genes and regulate gene expression as they move and duplicate over evolutionary time (47).
A homology search performed to find putative targets of the piRNAs derived from cluster 1082B yielded highly significant hits in several regions of the genome. None of the regions included LINE1 or IAP sequences and did not overlap with any of the piRNA clusters identified in our deep sequencing analysis. The region with highest degree of homology overlapped with the gene 4921525O09Rik, which has previously been shown to contain several piRNAs (9,10). Some of these piRNAs are anti-sense to the cluster 1082B primary transcript, indicating that 4921525O09Rik- and 1082B-derived piRNAs could bind to sense and anti-sense transcripts in target regions. So far, the biological relevance of non-repeat piRNAs is unclear, and more research is needed to identify the mechanism through which they work. Studies have revealed that the genomes of mammals are almost entirely transcribed (48,49), giving rise to a wide range of intergenic, anti-sense, overlapping and intronic non-protein-coding RNAs. Some of these transcripts could be regulatory RNAs, taking part in the networked interactions between transcription initiation and elongation, modulation of chromatin architecture, RNA editing, alternative splicing, RNA and protein signaling. Our findings emphasize the complexities of gene and chromatin regulation during differentiation in mammals.
In summary, we identified a piRNA cluster located in a non-repeat intergenic region of chromosome 7 that encodes a unidirectional piRNA precursor transcript giving rise to 788 unique anti-sense piRNAs. The cluster is transcriptionally repressed in wild type but was found to be the only cluster highly upregulated in TzfpGTi/GTi and Alkbh1−/− pachytene spermatocytes, indicating a role for Tzfp and Alkbh1 in the regulation of this cluster. The dramatic upregulation of the 1082B-derived piRNAs was followed by a downregulation of LINE1 and IAP transcripts in testes from both mutants. It is thus tempting to speculate that the piRNAs derived from cluster 1082B are involved in transposon control during the pachytene stage of meiosis in male germ cells.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–4, Supplementary Methods and Supplementary Reference [50].
AVAILABILITY
The raw sequence data from this study have been submitted to the NCBI Gene Expression Omnibus under accession number GSE37150.
FUNDING
Funding for open access charge: The Norwegian Research Council and the Norwegian Cancer Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Merete Worren from the Bioinformatics Core Facility at Oslo University Hospital for helping out with the mRNA sequencing analysis. We also thank Linda Ellevog for excellent technical assistance with the germ cell isolations and qPCR analysis. We thank the Centre for Comparative Medicine at Oslo University Hospital for the excellent service they provided.
REFERENCES
- 1.Hess RA, Renato de FL. Spermatogenesis and cycle of the seminiferous epithelium. Adv. Exp. Med. Biol. 2008;636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- 2.Russell LB. Patterns of mutational sensitivity to chemicals in poststem-cell stages of mouse spermatogenesis. Prog. Clin. Biol. Res. 1990;340C:101–113. [PubMed] [Google Scholar]
- 3.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 5.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 6.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 7.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 9.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 10.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 11.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gan H, Lin X, Zhang Z, Zhang W, Liao S, Wang L, Han C. piRNA profiling during specific stages of mouse spermatogenesis. RNA. 2011;17:1191–1203. doi: 10.1261/rna.2648411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 15.Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 18.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 19.De FS, Bartonicek N, Di GM, breu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 20.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl Acad. Sci. USA. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 2009;284:6507–6519. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 24.Ragan C, Mowry BJ, Bauer DC. Hybridization-based reconstruction of small non-coding RNA transcripts from deep sequencing data. Nucleic Acids Res. 2012;40:7633–7643. doi: 10.1093/nar/gks505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 26.Nordstrand LM, Svard J, Larsen E, Nilsen A, Ougland R, Furu K, Lien GF, Rognes T, Namekawa SH, Lee JT, et al. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS One. 2010;5:e13827. doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Z, Sikandar S, Witherspoon M, Dizon D, Nguyen T, Benirschke K, Wiley C, Vrana P, Lipkin SM. Impaired placental trophoblast lineage differentiation in Alkbh1(−/−) mice. Dev. Dyn. 2008;237:316–327. doi: 10.1002/dvdy.21418. [DOI] [PubMed] [Google Scholar]
- 28.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 30.Tang CJ, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev. Biol. 2006;290:398–410. doi: 10.1016/j.ydbio.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 31.Sai LS, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rognes T. Faster Smith-Waterman database searches with inter-sequence SIMD parallelisation. BMC Bioinformatics. 2011;12:221. doi: 10.1186/1471-2105-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin W, Lai CH, Tang CJ, Huang CJ, Tang TK. Identification and gene structure of a novel human PLZF-related transcription factor gene, TZFP. Biochem. Biophys. Res. Commun. 1999;264:789–795. doi: 10.1006/bbrc.1999.1594. [DOI] [PubMed] [Google Scholar]
- 38.Hoatlin ME, Zhi Y, Ball H, Silvey K, Melnick A, Stone S, Arai S, Hawe N, Owen G, Zelent A, et al. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood. 1999;94:3737–3747. [PubMed] [Google Scholar]
- 39.Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS. Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 41.Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcon E, Babak T, Chua G, Hughes T, Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- 43.Beyret E, Lin H. Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev. Biol. 2011;355:215–226. doi: 10.1016/j.ydbio.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 45.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 48.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 49.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.