Abstract
Hfq is a post-transcriptional regulator that plays a key role in bacterial gene expression by binding AU-rich sequences and A-tracts to facilitate the annealing of sRNAs to target mRNAs and to affect RNA stability. To understand how Hfq from the Gram-positive bacterium Staphylococcus aureus (Sa) binds A-tract RNA, we determined the crystal structure of an Sa Hfq–adenine oligoribonucleotide complex. The structure reveals a bipartite RNA-binding motif on the distal face that is composed of a purine nucleotide-specificity site (R-site) and a non-discriminating linker site (L-site). The (R–L)-binding motif, which is also utilized by Bacillus subtilis Hfq to bind (AG)3A, differs from the (A–R–N) tripartite poly(A) RNA-binding motif of Escherichia coli Hfq whereby the Sa Hfq R-site strongly prefers adenosine, is more aromatic and permits deeper insertion of the adenine ring. R-site adenine-stacking residue Phe30, which is conserved among Gram-positive bacterial Hfqs, and an altered conformation about β3 and β4 eliminate the adenosine-specificity site (A-site) and create the L-site. Binding studies show that Sa Hfq binds (AU)3A ≈ (AG)3A ≥ (AC)3A > (AA)3A and L-site residue Lys33 plays a significant role. The (R–L) motif is likely utilized by Hfqs from most Gram-positive bacteria to bind alternating (A–N)n RNA.
INTRODUCTION
Small RNAs (sRNAs) are non-coding RNAs that play a pivotal role in gene regulation in response to a variety of stresses as well as in the coordinated expression of virulence factors in pathogenic bacteria (1–4). The large and growing number of sRNAs found in both Gram-negative and Gram-positive bacteria, including Staphylococcus aureus (5,6) further underscores their importance in many diverse regulatory processes (1). Adaptation to environmental signals is achieved through modulation of mRNA stability and translation by the base pairing of sRNAs with their target mRNAs. Thus, sRNA–mRNA complex formation can facilitate either activation or repression of translation in a complex-specific manner (7–12). However, as sRNAs are often encoded in trans and therefore exhibit imperfect complementarity to their target mRNAs, effective sRNA–mRNA annealing frequently requires the presence of auxiliary factors. In many bacteria, this role is executed by the RNA-binding protein Hfq (also called HF-1) (13,14).
Hfq is an abundant, heat-stabile protein that is highly conserved throughout the bacterial domain (15). Hfq displays similarity to Sm and Lsm proteins in eukaryotes and archea and forms doughnut shaped, hexameric rings. First identified as an Escherichia coli host factor involved in Qβ-phage replication (16,17), Hfq later emerged as a key player in post-transcriptional gene regulation mediated by bacterial sRNAs. Consequently, an hfq deletion entails pleiotropic effects such as growth defects, diminished stress tolerance and quorum sensing and impaired virulence in numerous pathogens (18–24). Experimental data currently support two non-exclusive models of Hfq function in riboregulation (14). The first model emphasizes the chaperone activity of Hfq requiring that Hfq alters the conformation of the sRNA or its target mRNA or both by partially unfolding the RNA structure to enable sRNA–mRNA hybridization or to alter access to the ribosome-binding site. The second model assumes that Hfq binds sRNA and mRNA simultaneously thereby favoring hybridization by increasing the local concentration of sRNA and mRNA.
Hfq preferentially binds single-stranded AU-rich RNA sequences in the proximity of double-stranded regions (25–28). Recently, the polyU tail of Rho-independent terminators of sRNAs from Gram-negative bacteria has been identified as important Hfq target site (29–31). Hfq-binding sites also exist along target mRNAs, whereby an (ARN)x motif in the upstream mRNA leader region of E. coli rpoS constitutes a high-affinity Hfq-binding site essential for gene regulation (32–35). In the presence of Ec Hfq sRNA stability is remarkably increased (36), probably due to protection from nuclease cleavage. However, once Hfq-assisted base pairing with the cognate mRNA has occurred within a ternary complex, either one or both RNAs may be degraded through Hfq-mediated recruitment of components of the degradosome, specifically RNase E (37,38).
To date only a few crystal structures of Hfq–RNA complexes have been published. These have confirmed the presence of several non-overlapping RNA-binding sites on the Hfq protein and revealed the roles of several key residues in RNA recognition. In the crystal structures of S. aureus Hfq (Sa Hfq) in complex with the oligoribonucleotide AU5G (39) and Salmonella typhimurium Hfq (St Hfq) in complex with U6 RNA (31), the RNA is bound to a circular rim around the central pore on the ‘proximal’ side of the Hfq hexamer. The proximal side or face of Hfq is defined as that on which the lone α-helix of the Hfq core lays. In these structures the uracil bases are fixed by similar nucleobase-aromatic stacking interactions in repeated binding pockets between neighboring protomers. However, in the St Hfq–U6 complex structure the RNA sugar–phosphate backbone adopts a strikingly different, highly constrained conformation. The resulting position of the ribose allows the specific recognition of the free terminal hydroxyl group by residue His57 thereby explaining the preferential binding of U-rich 3′-ends. Interestingly, a newly reported structure of Ec Hfq bound to AU6A shows a hybrid-binding mode in which three of the six uridines and one adenosine are bound in the proximal pore as seen in the Sa Hfq–AU5G structure and two uridines above the pore (40). The remaining uridine binds in a novel manner by lying in a pocket near the N-terminus of the lone α helix. The other adenosine is found in the R-site of a second Hfq hexamer. Hence, these structures reveal that U-rich sequences can bind to the proximal face of Hfq in multiple, but related conformations and provide a rationale for the finding that this face is the preferred binding site for U-rich sRNAs (41–43), although recent work implicates involvement of an additional, lateral surface-binding site for RybB (44).
The structure of E. coli Hfq (Ec Hfq) bound to A15 RNA provided the mechanistic rationale for the binding of poly(A) tails by Ec Hfq (35,45). The RNA is bound on the opposite ‘distal’ face of Hfq and reveals a weaving, repeating tripartite (A–R–N)n RNA-binding motif, wherein an exposed adenosine specificity site (A-site) is created by two hydrogen bonds emanating from the peptide backbone, a purine nucleotide specificity site (R-site) is formed by a mostly hydrophobic crevice found between two β-strands and the third non-discriminatory ribonucleotide exit or entry site (E-site, now renamed the N-site to highlight its ability to be any nucleotide) extends into the solvent. More recently the structure of Bacillus subtilis Hfq (Bs Hfq) bound to the RNA aptamer (AG)3A was determined (46). Although the RNA binds to the distal face and uses the R-site to bind the adenosine moieties, there is no A or N site. Rather the guanine bases stack over the side chain of a glutamine and often make hydrogen bonds from their O6 or N7 atoms to the guanidinium side chain of a nearby arginine.
Investigations on Hfq function and mechanism have focused mainly on Hfq from enteric bacteria, notably E. coli and Salmonella (14). In these organisms the pivotal role Hfq plays in riboregulation and stress adaptation is indisputable. By contrast, the significance of Hfq on riboregulation in Gram-positive bacteria is controversial and still poorly understood and despite the structures of the Sa Hfq–AU5G and Bs Hfq–(AG)3A complexes, the role of RNA binding to Hfq is uncertain. Moreover, some Gram-positive bacteria, including Lactococci and Streptococci, lack an obvious hfq gene in their genome while some strains of S. aureus do not appear to express the protein leading to the hypothesis that Hfq plays only a minor role in these organisms and might be superseded by other RNA-binding molecules (47,48). Moreover, the deletion of the hfq gene from B. subtilis affected neither the growth nor sporulation of this strain (49) and an hfq deletion in the Newman, COL and RN6390 strains of S. aureus did not show any phenotype (48). By contrast Liu et al. have demonstrated that Hfq expression in S. aureus is highly strain dependent and in certain strains, including MRSA (methicillin-resistant S. aureus) strains, Hfq is expressed and contributes significantly to stress resistance and pathogenicity (50). Furthermore, many other Gram-positive bacteria encode one or more hfq genes in their genomes and these sequences contain key conserved residues. Indeed, the first Hfq-dependent sRNAs involved in stress regulation in a Gram-positive bacterium have been identified and characterized in Listeria monocytogenes (23,51).
To gain deeper biochemical and structural insight into the A-tract RNA-binding mechanism of Hfq from the pathogenic, low G+C Gram-positive bacterium, S. aureus, we determined the crystal structure of Sa Hfq in complex with the hepta-ribonucleotide A7. The structure of the complex reveals a fundamentally different mode of A-tract RNA binding from that utilized by Hfq from E. coli (35) but a highly similar mode to that used by Bs Hfq to bind the (AG)3A aptamer (46). Furthermore, binding studies on Sa Hfq and (AG)3A, (AC)3A, (AU)3A and (AA)3A RNA strongly point towards a conserved distal-face-binding mechanism for alternating (A–N) tracts among Gram-positive bacteria.
MATERIALS AND METHODS
Protein overexpression, purification and site-directed mutagenesis
Hfq proteins from S. aureus and E. coli were overexpressed in the E. coli strain ER3566 Δhfq using plasmid pTYB11 (NEB) and purified using the IMPACT-CN system as described (27,39). In order to remove any contaminating RNA, protein samples were treated with 20 µg/ml RNase A for 30 min, which was followed by gel filtration on Superdex G75. The purified protein was concentrated to a final concentration of 50–70 mg/ml and stored in 25 mM Tris–HCl pH 8.0, 150 mM NaCl and 0.5 mM EDTA. The protein concentration was determined by UV absorbance measurements.
The K33A mutant was introduced via site-directed mutagenesis according to the protocol of the manufacturer (Quick-Change Site-Directed Mutagenesis Kit, Stratagene) using the following primers: forward, 5′-CTAAACGGTTTCCAAATGGCGGGTGTTATTGAAGAATACGAC-3′; reverse, 5′-GTCGTATTCTTCAATAACACCCGCCATTTGGAAACCGTTTAG-3′. The mutant was expressed and purified as described for the wild-type protein.
Crystallization and data collection
Oligoribonucleotides (Oligos etc., Wilsonville, OR, USA) were dissolved in 10 mM sodium cacodylate pH 6.5 to a final concentration of 1 mM and used without further purification. The Hfq–RNA complex was formed by mixing the protein and the RNA in a molar ratio of 1.0:1.1. Crystallization trials were performed using the hanging-drop vapor diffusion method at room temperature by mixing equal volumes of 100 µM protein–RNA solutions with the respective crystallization reservoir solution. Crystals of the Sa Hfq–A7 RNA complex appeared within 3 days from a solution containing 12% MPD (2-methyl-2,4-pentanediol), 0.1 M Na cacodylate pH 6.5, 0.2 M Zn(OAc)2 and 0.1 M KCl. The MPD concentration was increased to 35% for cryo-protection. The crystals were flash frozen in a nitrogen stream at 100 K. X-ray intensity data were collected to 2.20 Å resolution under cryo-conditions at the Advanced Light Source (ALS) in Berkeley, CA, at beamline 8.3.1 and processed and scaled using MOSFLM and SCALA (52,53). The crystals take the trigonal space group P3 with cell dimensions a = b = 156.4 Å, c = 34.6 Å (Table 1).
Table 1.
Selected crystallographic data and statistics
| Space group | P3 |
| Cell dimensions | |
| a, b, c (Å) | 156.4, 156.4, 34.6 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 78.10 – 2.20 (2.32–2.20)a |
| Reflections (#) | 144 061 (20 896) |
| Rmerge (%)b | 6.1 (29.3) |
| I/σ(I) | 13.4 (2.9) |
| Completeness (%) | 99.4 (99.0) |
| Refinement | |
| Rwork/Rfree (%)c | 19.6/ 25.9 |
| Atoms (#) | |
| Protein | 6934 |
| RNA | 176 (8 nt) |
| Solvent | 242 (29 Zn2+ atoms) |
| Average B factor, protein (Å2) | 47.3 |
| Average B factor, RNA (Å2) | 58.8 |
| Root-mean-square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.003 |
| Ramachandran analysis | |
| Most favored (%) | 92.6 |
| Add. favored (%) | 7.3 |
| Gen. allowed (%) | 0.1 |
| Disallowed (%) | 0 |
aValues in parentheses are for the highest resolution shell.
bRmerge = ∑∑|Ihkl(j) − Ihkl)|/∑∑Ihkl, where Ihkl(j) is the observed intensity and Ihkl is the final average intensity value.
cRwork =∑||Fobs| − |Fcalc||/∑|Fobs| and Rfree =∑||Fobs| − |Fcalc||/∑|Fobs|, where all reflections belong to a test set of 5% randomly selected reflections.
Structure determination and refinement
The structure was solved by molecular replacement using Phaser (54) and one hexamer of the Hfq apo structure from S. aureus (PDB code: 1KQ1) as the search model. The initial protein model was subjected to rigid body refinement followed by simulated annealing in CNS (55). After successive rounds of manual fitting and model building in COOT (56), the model was subjected to positional and B-factor refinement in CNS. After manual building of the RNA into the positive electron density of the resulting difference density maps, the model was refined further with PHENIX (57). A final composite omit map was generated by CNS.
The asymmetric unit of the S. aureus Hfq–A7 crystal contains 14 Hfq subunits (residues 5–65 in chains D, E, M; residues 6–65 in chains C, F, I and residues 5–66 in chains A, B, G, H, J, K, N and S) as well as two tetra adenosine oligoribonucleotides (A4), 29 Zn2+ ions and 207 water molecules. To reflect the presence of only four adenosines, we henceforth refer to the structure as the Sa Hfq–A4 complex. Residues 66–77 or 67–77 of the respective subunits are disordered. The model was refined to final Rwork and Rfree values of 19.6% and 25.9%, respectively. Selected data collection and refinement statistics are listed in Table 1. The structure was validated with PROCHECK and shows excellent stereochemistry (Table 1). The sugar puckers of the eight adenosine ribonucleotides were analyzed using CURVES+ (58). All figures were created with PyMol (59).
Coordinates and structure factors
The coordinates and structure factors for the Sa Hfq–A4 complex are deposited in the PDB under the PDB ID code 3QSU.
Fluorescence polarization
Fluorescence polarization measurements were performed with a PanVera Beacon 2000 instrument (Invitrogen, Madison, WI, USA) at 295 K. Hfq was serially titrated into 1 ml of binding buffer containing 25 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA and 0.5 nM fluorescein-labeled oligoribonucleotide. Samples were excited at 490 nm, and emission was detected at 530 nm. Fluorescence polarization of the 5′-fluorescein-labeled oligoribonucleotides increases due to the specific binding of the protein and plateaus after all high-affinity binding sites are saturated. Data were analyzed using Kaleidograph assuming a 1:1 stoichiometry between one Hfq hexamer and one molecule of labeled RNA. Using non-linear least squares analysis, the respective binding isotherms were fitted to the equation:
P = {(Pbound − Pfree) [protein] / (Kd + [protein])} + Pfree, with Pbound being the maximum polarization at saturation, P is the polarization at a given protein concentration, Pfree is the polarization of free fluorescein-labeled RNA and Kd is the equilibrium dissociation constant. The final value of each measurement is the average of at least three individual polarization measurements and each binding experiment was carried out at least three times.
RESULTS AND DISCUSSION
Global structure of the S. aureus Hfq–A4 complex
Previous studies indicate that Hfq harbors specific and independent sRNA and mRNA recognition sites, which enable simultaneous binding of two RNAs and the formation of an active regulatory ternary complex (60,61). U-rich RNA sequences, which are often found at the 3′-end of sRNAs, generally bind to the proximal side of Hfqs of both Gram-negative and Gram-positive bacteria, although with different binding modes that appear to be influenced by the presence of additional non-uridine nucleotides (31,39,40). By contrast, the binding site for A-rich RNA sequences and the poly(A) tails of mRNAs is located on the distal side of Hfq in the Gram-negative bacterium E. coli (35,60,61). As the Hfq proteins from E. coli and S. aureus exhibit a remarkable difference in their electrostatic surface potentials as well as their RNA-binding properties (13,35) it is unclear whether Sa Hfq would bind A-tract oligoribonucleotides similar to Ec Hfq. To this end, multiple attempts were made to crystallize Sa Hfq in complex with A-rich RNA.
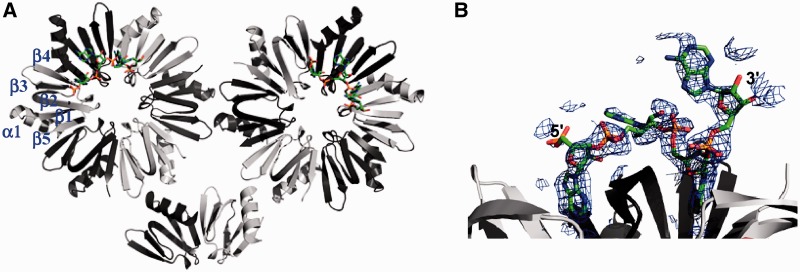
Crystals of the Sa Hfq–A7 complex were grown from solutions of 12% MPD. The structure of the complex was solved by molecular replacement using the apo Sa Hfq crystal structure (PDB ID code 1KQ1) as the search model and refined to Rwork and Rfree values of 19.6% and 25.9%, respectively, to 2.20 Å resolution (Table 1). The asymmetric unit contains 14 Hfq protomers, which form two biologically relevant hexamers and one-third of a hexamer, which is completed by the crystallographic 3-fold axis of the crystal (Figure 1A). Each subunit takes the canonical Hfq fold with an N-terminal α helix, α1 (residues 6–19) followed by five β-strands with the topology β5 (residues 60–65)-β1 (residues 21–26)-β2 (residues 29–39)-β3 (residues 43–48)-β4 (residues 51–56).
Figure 1.
The structure of S. aureus Hfq bound to A4. (A) Ribbon diagram of the asymmetric unit of the Sa Hfq–A4 complex, which contains 14 protomers, looking into the distal face. Contiguous subunits are colored light and dark grey. The secondary structure of one subunit is labeled and the RNA is shown as atom-colored sticks. (B) The simulated annealing omit electron density map of the oligoribonucleotide contoured at 1 σ.
As observed in the structures of the apo Sa Hfq and Sa Hfq–(AU5G) complex, the last 11 or 12 residues (∼66–77) of each protomer are disordered suggesting their relative unimportance in binding to small sized RNA [see also (62)]. Superimposition of RNA-bound Sa Hfq with the apo Sa Hfq structure reveals no meaningful structural changes upon RNA binding (root-mean-square deviation = 0.82 Å for all Cα atoms). This confirms that this A-tract RNA-binding pocket is preformed as had been observed previously in the Ec Hfq–A15 complex (35).
Each Hfq hexamer binds to its distal face one RNA fragment, which displays continuous electron density for only four of the seven adenosine ribonucleotides (Figure 1B). The underlying reason for the presence of only four of the nucleotides is unclear, as crystal packing would not appear to interfere with their binding. Degradation of A7 to A4 is possible but less likely because one would not expect the decay to stop at A4. Moreover, using A4 RNA in crystallization trials did not yield any crystals with this crystallization solution. Regardless, each A4 binds essentially identically to their Hfq proteins. All riboses take the C2′-endo sugar pucker, thus differing from the preferred C3′-endo pucker taken by double-stranded RNA. The conformation of the adenine bases alternates between anti and syn except for the most 3′-base, which also takes an anti conformation likely due the lack of an additional 3′-nucleotide to hold it in the energetically less favorable syn conformation (63). The adenine bases take these alternating glycosidic conformations in order to fit the A-tract-binding site of Sa Hfq optimally (described later). Interestingly, the solvent exposed amino acid side chains of residues His53 and His58, which reside on the proximal face, coordinate Zn2+ ions that are present in the crystallization buffer (Supplementary Figure S1). As pore residue His58 is involved in specific RNA binding to the proximal side of Sa Hfq (39) we quantified the RNA-binding properties of S. aureus Hfq in the presence and absence of Zn2+ ions by fluorescence polarization. Neither the addition of 100 mM zinc acetate to the RNA-binding assay buffer, which is equivalent to the initial Zn2+ ion concentration in the crystallization drop, nor a 2-day incubation of Hfq with the mother liquor (apart from the precipitant MPD) altered the affinity of Sa Hfq for either U-rich (AU5G) or A-rich (A6G or A7) RNA (data not shown). This result argues against the possibility that the coordination of Zn2+ ions interfered with or changed the binding mode of the A-tract RNA. Thus, the central pore on the proximal site of Sa Hfq does not constitute the physiologically relevant binding site for A-tract RNA. Rather, and as seen in the Ec Hfq–A15 complex structure, the binding site for A-tract RNA is located on the distal face of the Sa Hfq protein.
The (R–L) bipartite RNA-binding motif of the Hfq distal face
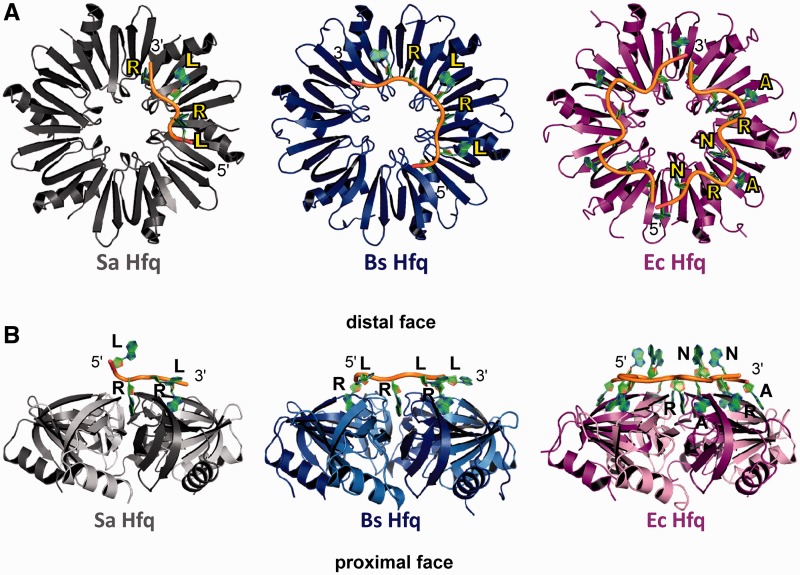
The A4-binding site of Sa Hfq closely resembles that of the (AG)3A Bs Hfq and in part that of the A15-binding site of Ec Hfq, however, the mode of RNA binding displays differences from both, especially from Ec Hfq (Figures 2 and 3). In the Sa Hfq–A4 and the Bs Hfq–(AG)3A complexes the RNA sits flat on top of the Hfq protein and does not have multiple nucleotides projecting into the solvent (Figure 2B). More important, in lieu of the (A–R–N) tripartite-binding motif of Ec Hfq, Sa Hfq and Bs Hfq employ a bipartite-binding motif, which we now name the R–L motif in which the R-site is a purine nucleotide-binding site and the L-site is the R-site linker (Figure 2). The previously identified A-site of Ec Hfq, which is a specific adenosine-binding site for A-rich RNA, is not present in either the Sa or Bs Hfq. Hence, the six R–L motifs of Sa Hfq and Bs Hfq provide the capacity to bind optimally 12 ribonucleotides to its distal-face A-tract-binding site as compared to the 18 ribonucleotides that Ec Hfq can bind. As a consequence of not having an A-site, the sugar–phosphate backbones of the A4 RNA and (AG)3A RNA follow a more circular, less weaving path as compared to the A15 path on Ec Hfq (Figure 2A).
Figure 2.
The RNA-binding motifs of Hfq in Gram-positive and Gram-negative bacteria. (A) View looking down onto the distal face of Sa Hfq (left, colored grey), Bs Hfq (middle, colored blue) and Ec Hfq (right, colored magenta). Note the assigned colors will be used in all subsequent figures. Bound RNA is shown as a cartoon with sugar phosphate backbone colored yellow and the purine bases colored green. Each protein is labeled and colored appropriately. The purine nucleotide sites are labeled R, the linker sites are labeled L and the A-sites, which are found only in the Ec Hfq, are labeled A. The 5′- and 3′-ends of the RNA are labeled. (B) Side view of the Sa Hfq–A4 (left), Bs Hfq–(AG)3A (middle) and Ec Hfq–A15 (right) complexes. Each Hfq is labeled and colored as in (A) and the ‘distal’ and ‘proximal’ faces are labeled. Contiguous subunits are colored light and dark grey, blue or magenta in the respective complexes.
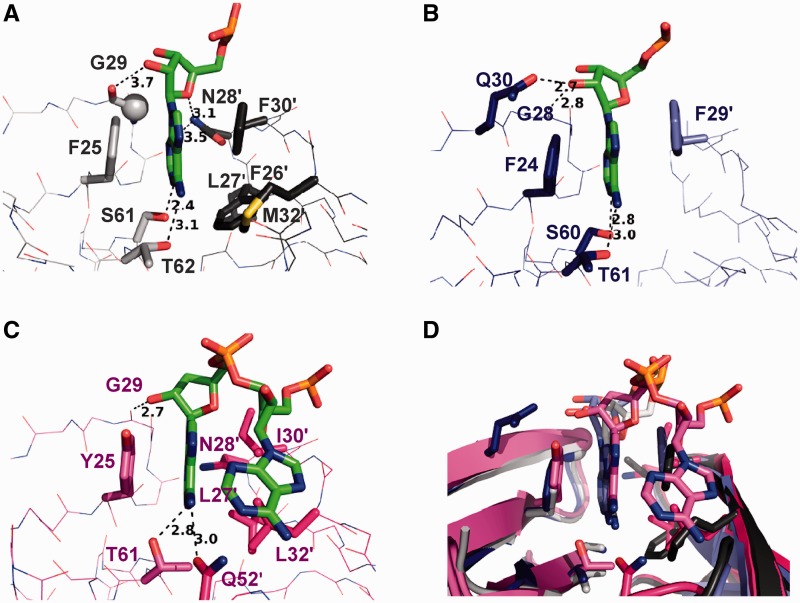
Figure 3.
Adenosine binding to the R-site of Hfq of Gram-positive and Gram-negative bacteria. (A) View into the R-site of Sa Hfq. Interacting residues are shown as grey sticks and labeled. The adenosine moiety is shown as green sticks. Interactions are indicated by dashes and distances (Å) are given for most hydrogen bonds. Residues from the neighboring subunit are labeled with primes. Residue Gly29 (G29) is represented as a grey ball. The main chain atoms of all non-interacting residues are shown as lines and not labeled. (B) View into the R-site of Bs Hfq. Interacting residues are shown as blue sticks and labeled. The adenosine moiety is shown as green sticks. Interactions are indicated by dashes and distances (Å) are given for most hydrogen bonds. Residues from the neighboring subunit are labeled with primes. The main chain atoms of all non-interacting residues are shown as lines and not labeled. (C) View into the R-site of Ec Hfq. Interacting residues are shown as magenta sticks and labeled. The adenosine moiety is shown as green sticks. Interactions are indicated by dashes and distances (Å) are given for most hydrogen bonds. The main chain atoms of all non-interacting residues are shown as lines and not labeled. The adenosine moiety that binds to the nearby A-site is also shown. Residues from the neighboring subunit are labeled with primes. (D) Overlay of an adenosine-bound R-site of Sa Hfq, Bs Hfq and Ec Hfq. Each protein is colored as in (A), (B) and (C). The RNA is shown as sticks for each but with the carbon atoms of A4 colored grey, the carbon atoms of (AG)3A colored slate and the carbon atoms of A15 colored magenta. Note the deeper pocket binding of the adenosines in the Gram-positive Hfq proteins.
The majority of the Sa Hfq–A4 contacts occur in a crevice equivalent, but not identical, to the R-sites of Ec Hfq and Bs Hfq (Figure 3). This binding pocket is found between β strands 2 and 2′ of two neighboring subunits and two of the four adenosines of each of the independent A4 are bound within (Figure 2). The adenine base inserts into this highly aromatic pocket and stacks against the side chains of residues Phe25, Phe30′ and Phe26′, where the prime indicates the residues from the neighboring subunit, and makes van der Waals contacts with Leu27′ and Met32′ (Figure 3A). The distinctive aromatic character of this crevice of the Gram-positive Hfq proteins favors or results in a downward rotation of the adenosine such that the base sticks deeper into the cleft when compared to adenosine binding to the purine nucleotide specificity R-site of Ec Hfq (Figures 2B and 3D). In addition to these stacking interactions hydrogen bonds between the side chain amide of Sa Hfq residue Asn28′ and the sugar O4′ and adenine N3 nitrogen further strengthen ribonucleotide binding (Figure 3A). The 2′-OH group of the ribose makes an additional, albeit relatively weak, hydrogen bond with the peptide backbone carbonyl oxygen of residue Gly29 (Figure 3A), thereby increasing significantly the preference of Sa Hfq for A-rich RNA over A-rich DNA (39). Additional Sa Hfq–RNA interactions include hydrogen bonds between the N1 atom and exocyclic N6 amino group of the adenine and the hydroxyl groups of Ser61 and Thr62, respectively, which are analogous to the Thr61′ and Gln52–adenine contacts seen in the Ec Hfq–A15 complex (Figure 3A and C). Thus, the R-site of the Sa Hfq makes a large number of contacts to the bound adenosine, which anchors the distal-face-bound RNA to the protein.
This binding pocket, like the previously described corresponding pockets in the Ec Hfq–A15 and Bs Hfq–(AG)3A structures, is named the R-site because binding by either adenosine or guanosine, is feasible at least in silico (Supplementary Figure S2). However, our modeling does reveal that adjustment to the guanine position or R-site pocket is needed in order to alleviate steric clash between the exocyclic N2 atom and the Cβ methylene group of residue Ser61 and that such changes are likely the origin of the previously reported weak binding of G6 to Sa Hfq (Kd > 4 µM) (39). Furthermore, the interactions between the hydrogen bond donor amide group of residues Asn28 and the hydrogen bond acceptors O4′ and N1 of the R-site-bound adenosine favor this nucleotide over guanosine, the N1 of which is a hydrogen bond donor. Rotation of the Asn28 side chain to allow the carbonyl oxygen to interact with the N1 of a bound guanine would result in the loss of the hydrogen bond to the ribose O4′ and potentially unfavorable van der Waals contacts between the two oxygen atoms. Thus, adenosine binding to the R-site of Gram-positive Hfq proteins is significantly preferred over guanosine binding. Although this site could be renamed the A-site to reflect this preference, we maintain the R-site moniker to reflect its high structural homology with the R-site of the Ec Hfq and to avoid confusion with the Ec Hfq A-site that is structurally distinct (35). The smaller pyrimidine bases, cytosine and uracil, would not bind this pocket effectively due to their poorer stacking and inability to make multiple hydrogen bonds. Of note, the nucleotide-free R-sites do not have tightly bound water molecules to any of the adenosine-binding amino acid residues, a likely reflection of the highly aromatic and non-polar nature of the pocket.
As noted, the recently published structure of Bs Hfq in complex with the RNA oligopurine aptamer, (AG)3A, reveals a very similar R-site-binding mode to that observed in the Sa Hfq–A4 complex wherein only adenosines occupy the R-site pockets (Figure 3A and B) and again the mechanism of discrimination that appears to favor adenosine-only binding to the R-sites for reasons that may be similar to those described above. Regardless, the R-site-binding mechanisms of Sa Hfq and Bs Hfq do differ in several important details (Figure 3A and B). For example, Sa Hfq makes more hydrophobic and stacking interactions to the R-site-bound adenosine by the additional involvement of residues Phe26′, Leu27′ and Met32′ and makes hydrogen bonds between residue Asn28′ and the bound adenosine. By contrast Sa Hfq lacks the equivalent strong Gln30-(3′-OH) hydrogen bond (Sa Hfq residue Gln31 is 3.5 Å from the 3′-OH). Interestingly, neither Sa Hfq nor Bs Hfq makes direct contacts with the phosphate group that connects the R- and L-site-bound nucleotides. This phosphate group is located on the rim of the distal-face pore and perhaps is positioned there to accommodate or is the result of the alternating glycosidic torsion angles of the R- and L-site-bound purines. Finally, it should be noted that the 3′-adenosine, for which the electron density is relatively poor, is located above the distal face and makes no contacts with the Sa Hfq distal surface (Figure 2B). This ‘site’ thus resembles the N-site (formerly the E-site or entrance/exit site) that is observed six times in the Hfq–A15 complex structure but only once per Sa Hfq–A4 complex structure and never in the Bs Hfq–(AG)3A complex. This adenosine does, however, make contacts with the proximal face of a nearby Hfq hexamer including stacking with Tyr56 and engaging in a weak hydrogen bond between the cyclic N7 atom and Nζ of residue Lys41, which also contacts the 5′-phosphate group of the adenosine. These latter interactions are likely of limited physiological relevance.
Loss of the A-site in Sa Hfq–A4 and creation of the L-site
In the Ec Hfq–A15 complex structure, the ribonucleotides in the R-site and the A-site are close with their bases almost within stacking distance (Figure 3C) (35). As noted, Sa Hfq as well as Bs Hfq lacks this A-site entirely and does not have an alternative A-site. The decisive factor for the missing A-site is the difference of only a few amino acid residues in the otherwise highly conserved R-sites of Sa, Bs and Ec Hfq and the altered conformations of β3 and β4. Specifically, Sa Hfq has a phenylalanine at position 30 instead of the isoleucine found in Ec Hfq (Supplementary Figure S3). Simply, the larger aromatic ring, which is also a phenylalanine in Bs Hfq, would clash with the sugar ring of any A-site-bound ribonucleotide thereby inhibiting binding to this site (Figure 4A). Whereas a phenylalanine or tyrosine at residue 30 is highly conserved amongst Hfq proteins from multiple Gram-positive bacteria including the food-borne pathogens L. monocytogenes and B. cereus, this position in Gram-negative Hfq proteins is typically an isoleucine (Supplementary Figure S3). The second component of the loss of the A-site from Hfq proteins in Gram-positive bacteria arises from the insertion of an extra residue in the loop that connects β3 to β4 (Figure 4B). This insertion is found in all Gram-positive bacteria and when compared to Hfq proteins from Gram-negative bacteria, causes a shift in the sequence alignment at position 49/50 (for Ec and Sa Hfq, respectively) (Supplementary Figure S3). However, this insertion does not result in a longer loop between β3 and β4. Indeed, the corresponding loop of Ec Hfq is three amino acid residues instead of the two residue-loops seen in Sa Hfq and Bs Hfq. Rather, the β3 and β4 strands of Sa and Bs Hfq, and likely all Gram-positive bacteria, are each one amino acid residue longer than the corresponding β strands of the Hfq proteins from Gram-negative bacteria. As a consequence the region of the protein encompassing the C-terminal end of β3 to the β3/β4 loop and the N-terminal beginning of β4 is one amino acid residue longer in Hfqs from Gram-positive bacteria than in Hfqs of Gram-negative bacteria. Additionally both β3 and β4 are more twisted towards the A-site and their inward orientation causes the connecting loop to protrude 2.4 Å further into the A-site-binding pocket (Figure 4B). This conformation would result in the nucleotide clashing with the hydroxyl side chain of residue Ser48, which now occupies part of the A-site, and thereby precludes nucleotide binding (Figure 4A). Finally, although the amino acid side chains of residues His53 and Met32 do not interfere with A-site binding per se, their proximity to a possible A-site base disallows a nucleotide to create an alternative A-site (Figure 4A).
Figure 4.
Sa Hfq does not utilize an A-site to bind A-rich RNA. (A) Overlay of Sa Hfq–A4 and Ec Hfq–A15 complex structures. Key side chains are shown as sticks and labeled for Sa Hfq. The carbon atoms of the Sa Hfq side chains are colored grey whereas the carbon atoms of the Ec Hfq side chains are colored magenta. The Ec Hfq A-site and the Sa Hfq L-site adenosines are shown in thick magenta and grey sticks, respectively, whereas the rest of the respective RNAs is shown in thin magenta and grey lines. Black double-headed lines indicate steric clash between the A-site adenosine and side chains of Sa Hfq and the distances in Angstrom are shown. The locations of the respective R-site, A-site and L-site are labeled accordingly. The black swirl denotes the shift of the ‘A-site’ adenosine to its new L-site position in Sa Hfq and the approximate rotation that is necessary for this movement is given in degrees. The same steric hindrances between S48 and F30 and the A-site adenosine are found in Bs Hfq thereby precluding A-site binding to that protein. (B) Ribbon diagram of the overlay of the crystal structures of Sa Hfq–A4 (grey), Bs Hfq–(AG)3A (blue), Ec Hfq–A15 (magenta), and St Hfq–U6 (orange) complexes. β strands 3 and 4 are labeled. An A-site-bound adenosine from the Ec Hfq–A15 complex is shown as magenta sticks and labeled Ec A. Note the different twists of β strands 3 and 4 in Gram-positive and Gram-negative bacteria whereby the inward orientation of the β3 and β4 strands in Gram-positive bacteria obstructs an A-site adenosine from binding at this position. Sa Hfq residue S48 and corresponding Ec Hfq residue N48 are shown as blue and magenta sticks, respectively, to highlight their altered locations.
As a consequence of these protein sequence-derived structural constraints and in order to avoid steric clashes with multiple residues, the adenosine nucleotide located adjacent to the R-site-bound nucleotide rotates ∼40° to occupy a new location on the distal face (Figure 4A). Here the adenine base stacks in parallel with the side chain of residue Gln31, whilst the peptide amide of Gln31 participates in a hydrogen bond with the sugar O4′ (Figure 5A). Furthermore, the base has now switched from the anti to syn conformation. The same conformational changes are taken by the guanosine nucleotides in the Bs Hfq–(AG)3A complex and identical stacking and van der Waals interactions are seen between Bs Hfq residue Gln30 and the L-site guanine base that connects adjacent R-sites (Figure 5A, B and D) (46). There is no equivalent site in Ec Hfq (Figure 5C).
Figure 5.
The L-site of Sa Hfq and Bs Hfq. (A) View of the L-site of Sa Hfq. The L-site-bound adenosine is show as green sticks and selected residues as labeled dark grey sticks. Two linked R-sites are labeled as is the L-site. The 5′- and 3′-ends of the RNA are labeled. Proximal secondary structures are shown as grey cartoons. Interactions between Hfq and the L-site adenosine are depicted by dashes. (B) View of the L-site of Bs Hfq. The L-site-bound adenosine is show as green sticks and interacting residues as labeled blue sticks. Two linked R-sites are labeled as is the L-site. The 5′- and 3′-ends of the RNA are labeled. Proximal secondary structures are shown as blue cartoons. Interactions between Hfq and the L-site adenosine are depicted by dashes. (C) View of two contiguous R-sites of Ec Hfq. The R-site, A-site and N-site adenosines are show as green sticks and labeled. Residues that correspond to L-site interacting residues in Sa or Bs Hfq are shown as labeled magenta sticks. The interaction between the A-site 5′-phosphate group and K31 is shown as a dashed line. The 5′- and 3′-ends of the RNA are labeled. Proximal secondary structures are shown as magenta cartoons. (D) The R–L RNA-binding mode is conserved amongst Hfq proteins from Gram-positive bacteria. View of the R–L motif of Sa Hfq and Bs Hfq after the superimposition of the structures of the Sa Hfq–A4 and Bs Hfq–(AG)3A complexes. Each protein is colored as in (A) and (B). The RNA is shown as sticks but with the carbon atoms of A4 colored white and the (AG)3A carbon atoms colored slate. Note that whereas Bs Hfq residue R32 interacts with the L-site-bound guanine, the corresponding Sa Hfq residue, K33, does not interact with the L-site-bound adenine.
Biochemical consequences of the R–L motif
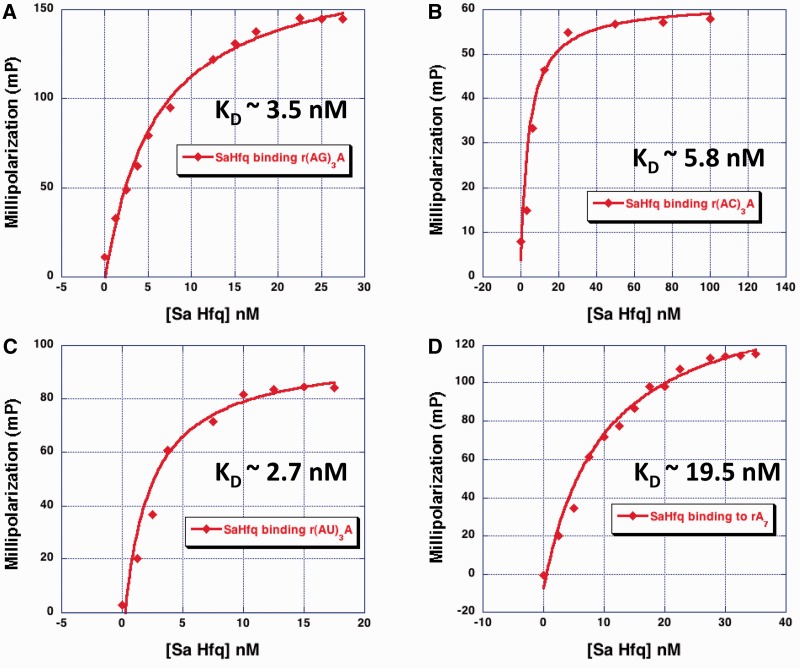
The structures of the Sa Hfq–A4 and Bs Hfq–(AG)3A complexes suggest that the R–L motif of Hfq proteins from Gram-positive bacteria might be able to bind sequences of the type (A–N)n, where N is any base. To test this hypothesis, the binding affinities of Sa Hfq for a series of heptamers were determined using a fluorescence polarization-based binding assay. These oligoribonucleotides included (AG)3A, (AA)3A, (AC)3A and (AU)3A. Our initial assumption was that each oligoribonucleotide binds the R–L sites on the distal face, where the adenosine binds the R-site and the following nucleotide of the dinucleotide repeat (G or A or C or U) binds the L-site. The results of these binding studies show that Sa Hfq-bound (AG)3A with a Kd = 3.5 nM; (AU)3A with a Kd = 2.7 nM; (AC)3A with a Kd = 5.8 nM; and (AA)3A with a Kd = 19.5 nM (Figure 6 and Table 2). Hence, Sa Hfq binds these sequences with high affinity with only a slight preference for which nucleotide occupies the L-site. The high-affinity binding of Sa Hfq to (AG)3A can be rationalized by the facile modeling of a hydrogen bond between residue Lys33 and either the exocyclic O6 or N7 atom or both of the guanine base (Supplementary Figure S4). This interaction would resemble that of the Bs Hfq Arg32–guanine interaction (36). Modeling of a uracil at the L-site suggests that residue Lys33 could make a good hydrogen bond with the O4 oxygen atom of the pyrimidine ring and a good contact between Lys33 and the N3 nitrogen atom of the cytosine ring is also possible (Supplementary Figure S4). Of course, stacking and van der Waals interactions between residue Gln31 and all L-site-bound nucleotides also contribute significantly to the binding strength.
Figure 6.
Sa Hfq binds RNA (A–N)3A sequences with high affinity. (A) Sa Hfq–(AG)3A-binding isotherm. (B) Sa Hfq–(AC)3A-binding isotherm. (C) Sa Hfq–(AU)3A-binding isotherm. (D) Sa Hfq–(AA)3A (or A7)-binding isotherm. The abscissa is the concentration of protein and the ordinate is millipolarization.
Table 2.
Dissociation constants (Kd) for selected Hfq–RNA complexesa
| Oligoribonucleotide | S. aureus Hfq Kd (nM) | S. aureus Hfq (Lys33Ala) Kd (nM) | E. coli Hfq Kd (nM) |
|---|---|---|---|
| (AA)3A = (A7) | 19.5 ± 6.4 | 406.3 ± 111.9 | 390 ± 20b |
| (AG)3A | 3.5 ± 0.3 | 11.9 ± 3.0 | NDc |
| (AC)3A | 5.8 ± 7.3 | >1000 | ND |
| (AU)3A | 2.7 ± 2.3 | 308.7 ± 40.2 | ND |
| U6 | 69.8 ± 7.0 | 72.7 ± 21.9 | ND |
| A16 | 4.2 ± 0.5 | ND | 0.4 ± 0.5 |
| A27 | 109 ± 13 | ND | 0.85 ± 0.08 |
| (GGA)2 | 450 | ND | 88 ± 28b |
| (GGA)9 | 350 | ND | 16 ± 1b |
aEach value is the average of three individual experiments and the standard deviations.
bTaken from (35).
cND, not determined.
Intriguingly a positively charged amino acid is found at the position corresponding to residue Lys33 of Sa Hfq (Arg32 in Bs Hfq) in most Gram-positive Hfq proteins (Supplementary Figure S3). As noted previously, the guanidinium side chain of Bs Hfq residue Arg32 can interact with either the O6 or N7 or both atoms of the L-site-bound guanine base, although this interaction is not seen for every guanine, thereby favoring this purine at this site (46). Replacement of Bs Hfq residue Arg32 by an alanine obliterates the ability of Bs Hfq to bind shorter (AG)nA sequences (46). The corresponding lysine in Sa Hfq is not observed to interact with the L-site adenine despite its ability to form a hydrogen bond to the N7 hydrogen bond acceptor atom of this purine (see modeled hydrogen bond Supplementary Figure S4).
Role of residue Lys33 in binding (A–N)n tracts
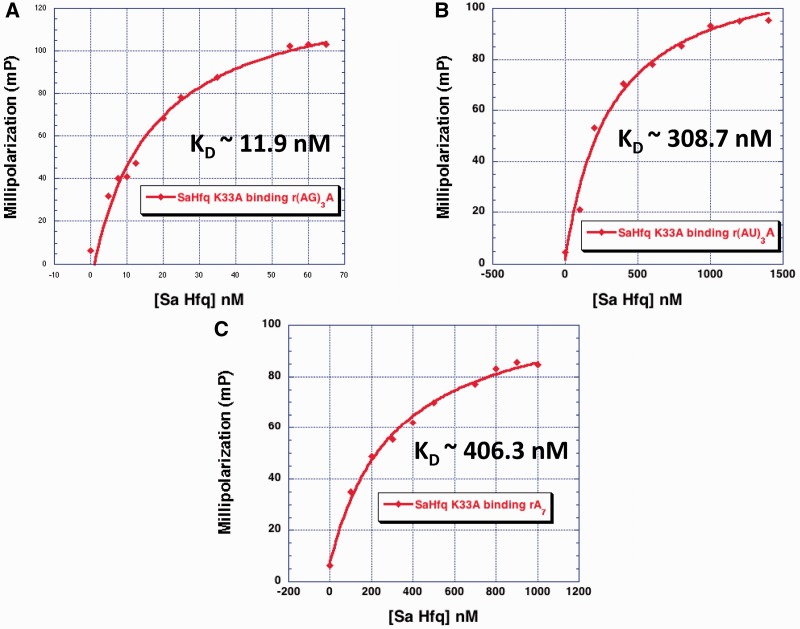
In order to assess the importance of residue Lys33 to the binding affinity of (AA)3A, (AG)3A, (AU)3A, (AC)3A and as a control, U6, which binds the proximal face of Sa Hfq (39), we replaced this residue with an alanine to generate Sa Hfq(K33A). As anticipated from our modeling, the affinity of Sa Hfq(K33A) for (AA)3A, (AU)3A and (AC)3A drops dramatically with changes ranging from 21 to >100-fold higher Kd values but does not change for U6 (Figure 7 and Table 2). Thus, the loss of this charged side chain is highly detrimental to binding. That we do not see a contact between the Lys33 side chain and N7 atom of adenine in our structure can be attributed to multiple factors, including our crystallization condition, which contains chloride ions at a relatively high concentration and hence are competitors for the amino group. Furthermore, the MPD precipitant, a dihydroxyl alcohol, lowers the dielectric constant of the solution thereby making the electrostatic between Nζ and Cl− ion yet more favorable. Notably, the interaction between the side chain guanidinium of residue Arg32 of Bs Hfq and the guanine observed in the Bs Hfq–(AG)3A complex is variable and not observed at each L-site, yet its substitution to alanine results in a dramatic loss of affinity (46). Similarly, Ec Hfq residue Lys33 also contributes significantly to the high-affinity binding of A-tract RNA to the distal face, yet this side chain is not seen to interact with the bound RNA in the Ec Hfq–A15 complex (35).
Figure 7.
Sa Hfq residue K33 is important for binding (A–N)3A sequences. (A) Representative Sa Hfq (K33A)–(AG)3A-binding isotherm. (B) Representative Sa Hfq (K33A)–(AU)3A-binding isotherm. (C) Representative Sa Hfq (K33A)–(AA)3A (or A7)-binding isotherm. The abscissa ordinate is the concentration of protein and the ordinate is millipolarization.
Unexpectedly, the binding affinity of Hfq(K33A) for (AG)3A changed only 3.4-fold suggesting that any interaction between Lys33 and an L-site guanine is not a significant contributor. Although a seemingly paradoxical result, inspection of our model of the Sa Hfq–(AN)3A complexes, provides a rationale for these data (Supplementary Figure S4). A guanine with a glycosidic angle in the syn conformation at the L-site is able to make a strong hydrogen bond from its exocyclic N2 to the preceding 3′-phosphate group and hence locks the base into this conformer whilst maintaining its optimal van der Waals/stacking interactions with residue Gln31. When any of the other bases occupies the L-site, there is either no contact to this phosphate group or in the case of the Hfq–(AU)3A complex the interaction is weak (the O2–PO4 distance is 3.4 Å with poor geometry). Hence the loss of any Lys33 hydrogen bond would allow the base greater conformational freedom and necessitate a greater loss of entropy in order to select the syn conformer for optimal binding to the L-site. In accord, anti glycosidic angles are energetically preferred by pyrimidine and adenosine nucleotides with a C2' endo sugar pucker whilst the syn conformation is favored by guanosine nucleotides with C2' endo sugar puckers (63).
Sa Hfq binding to polypurine sequences shows dependence on length and purine identity
To explore further the biochemical consequences of the different distal-face RNA-binding modes of Gram-positive and Gram-negative Hfq proteins, we quantified the affinities of Hfq from E. coli and S. aureus for other RNA sequences (Table 2). The binding affinity of Sa Hfq for A-rich RNA increases as a function of length whereby a string of 16 consecutive adenosines (A16) binds with a Kd of 4.2 nM, which is ∼5-fold lower than that of A7. However the binding affinity plunges with longer poly(A) RNAs as the Kd for A27 binds Sa Hfq 26-fold less tightly when compared to A16 (Table 2). In marked contrast, Ec Hfq binds the small A-rich tract (AA)3A with a Kd of 390 nM but A16 and A27 with essentially equal picomolar affinities (Table 2 and Supplementary Figure S5). This different behavior towards longer poly(A) RNAs likely stems in part from the reduced capacity of the Sa Hfq for ribonucleotide binding as a consequence of its bipartite distal face binding mode; only 12 nt are accommodated most efficiently, although the A16 would fill all sites and possible interact favorably with Hfq residues located either on the distal face or the lateral rim such as Phe15 or Lys16, whereas Ec Hfq can bind 18 nt. Another contributing factor is the different electrostatic surface potentials of the distal faces of the Sa and Ec Hfq, which appear to allow the positively charged Ec Hfq to steer longer ribonucleotides more productively onto the distal face (13). Once beyond a critical length perhaps the loss of entropy is a significant negative factor in Sa Hfq–distal face binding.
Notably, polynucleotide phosphorylase, which is capable of adding polynucleotide tracts and more commonly purine-rich tracts to the 3′-end of RNAs that are undergoing destruction, is common to all bacteria. Hence, additional fluorescence polarization studies were undertaken to assess the ability of Sa Hfq to bind other polypurine tracts containing an (A–R–N)N motif with guanosines occupying the R and N sites. Such sequences were shown previously to bind tightly to Ec Hfq (35). Sa Hfq can bind these oligonucleotides but with lower affinity than Gram-negative Ec Hfq, whereby the binding constants for (GGA)2 and (GGA)9 are Kd = 350 and 450 nM, respectively, versus Kd values of 88 and 16 nM, respectively, for Ec Hfq (35). This suggests that binding of Sa Hfq to such (A–R–N)-like polypurine RNA tracts is likely not physiologically important possibly due to the apparently energetically expensive looping out and solvent exposure of the intervening guanosines between AG dinucleotide repeats in the (GGA)2 and (GGA)9 oligoribonucleotides.
The (R–L) bipartite-binding motif is characteristic for Hfq proteins from Gram-positive bacteria
The structure of Bs Hfq in complex with the RNA aptamer (AG)3A (36) reveals a nearly identical binding mode to what we observe for the Sa Hfq–A4 complex wherein the adenosines are found only in the R-site, the A-site is absent, and two consecutive R-sites are linked by the guanosines that use the newly named L-site (Figures 2 and 5). Furthermore, the glycosidic torsion angles of the Bs Hfq–(AG)3A complex follow the anti (R-site)-syn (L-site) pattern as observed in the Sa Hfq–A4 complex. Thus, accumulating structural data and a highly conserved Phe/Tyr at the key Hfq residue 30 in Gram-positive bacteria (Supplementary Figure S3) indicate that Hfq homologues from a diverse array of Gram-positive bacteria will utilize the bipartite (R–L)-binding motif described herein and observed in the Bs Hfq–(AG)3A structure, to bind stretches of alternating (AG)n, (AC)n, (AA)n and (AU)n sequences with high affinity and biological significance. This stands in distinct contrast to the tripartite (A–R–N)-binding motif likely to be employed by all Gram-negative bacterial Hfq proteins, which have a different structure about the β3 and β4 loop and isoleucine or other aliphatic residue at position 30 (Figures 3C and Supplementary Figure S3). The functional consequence of the fundamentally different alternating (A–N) and polyA-binding mechanisms on the distal face of Hfq homologues from Gram-negative and Gram-positive bacteria has yet to be elucidated but is likely to continue to be an area of active investigation. Interestingly, a simple search of the S. aureus strain N315 (http://genolist.pasteur.fr/AureoList/index.html) reveals the presence of 101 A7 and 23 (AG)3A stretches in the regions queried from 30-bp upstream to 24-bp downstream of a translation initiation start site. Using the same search criteria there are seven (AC)3A and 63 (AU)3A motifs. Currently there are no reported Hfq–mRNA targets in S. aureus, however, whether or not Sa Hfq utilizes these alternating (A–N) tracts functionally in either mRNA decay or the translation control is an area under investigation.
ACCESSION NUMBERS
3QSU
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
The American Heart Association [09 GRNT2280109 to S.A.S.]; National Institutes of Health [K08 Career Development Award AI-064564 to S.A.S.]; the Danish Natural Science Research Council (to P.V.-H.); the Robert A. Welch Foundation [G-0040 to R.G.B.]; the authors also acknowledge the Advanced Light Source supported by the Director, Office of science, Office of Basic Energy Sciences, Material Sciences Division, of the US Department of Energy [contract No. DE-AC03-76SF00098], Lawrence Berkeley National Laboratory. Funding for open access charge: American Heart Association [09 GRNT2280109 to S.A.S.]; National Institutes of Health [K08 Career Development Award AI-064564 to S.A.S.]; the Danish Natural Science Research Council (to P.V.-H.) and Robert A. Welch Foundation [G-0040 to R.G.B.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the beamline scientists at ALS BL 8.3.1 for their help with data collection.
REFERENCES
- 1.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Y, Xie J. The roles of pathogen small RNAs. J. Cell. Physiol. 2011;226:986–973. doi: 10.1002/jcp.22483. [DOI] [PubMed] [Google Scholar]
- 3.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Vogel J, Wasserman KM. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorkski Pl, Borezée-Durant E, Barbet R, Jacquet E, Jacq A. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felden B, Vandenesch F, Bouloc P, Rombly P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Path. 2011;7:e1002006. doi: 10.1371/journal.ppat.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 9.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altuvia S, Wagner EGH. Switching on and off with RNA. Proc. Natl Acad. Sci. USA. 2000;97:9824–9826. doi: 10.1073/pnas.97.18.9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 17.Franze FMT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972;247: 824–831. [PubMed] [Google Scholar]
- 18.Tsui HCT, Leung HCE, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K 12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 19.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Pannekoek Y. Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol. Lett. 2009;294:216–224. doi: 10.1111/j.1574-6968.2009.01568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geng J, Song Y, Yang L, Feng Y, Qiu Y, Li G, Guo J, Bi Y, Qu Y, Wang W. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS One. 2009;4:e6213. doi: 10.1371/journal.pone.0006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–919. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 27.Møller T, Franch T, Hojrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 28.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl Acad. Sci. USA. 2011;108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS One. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentin-Hansen P, Ericksen M, Udesen C. MicroReview: The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 37.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Wang L, Zou Y, Zhang J, Gong Q, Wu J, Shi Y. Cooperation of Escherichia coli Hfq hexamers in DsrA binding. Genes Dev. 2011;25:2106–2117. doi: 10.1101/gad.16746011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olejniczak MJ. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 42.Vincent HA, Henderson CA, Stone CM, Cary PD, Gowers DM, Sobott F, Taylor JE, Callaghan AJ. The low-resolution structure of Vibrio cholera Hfq in complex with Qrr1 sRNA. Nucleic Acids Res. 2012;40:8698–8710. doi: 10.1093/nar/gks582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Almeida Ribeiro E, Jr, Beich-Frandsen M, Konarev PV, Shang W, Vecerek C, Kontaxis G, Hämmerlie H, Peterlik H, Svergun DI, Bläsi U, et al. Structural flexibility of RNA as molecular basis for Hfq chaperone function. Nucleic Acids Res. 2012;40:8072–8084. doi: 10.1093/nar/gks510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surfaces of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl Acad. Sci. USA. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with oligoriboadenylates. Biochemistry. 1980;19:6138–6146. doi: 10.1021/bi00567a029. [DOI] [PubMed] [Google Scholar]
- 46.Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–1867. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Bohn C, Rigoulay C, Bouloc P. No detectable effect on RNA-binding protein Hfq absence in Staphyolococcus aureus. BCM Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Wu N, Dong J, Gao Y, Zhang X, Mu C, Shao N, Yang G. Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS One. 2010;5:e13069. doi: 10.1371/journal.pone.0013069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søggaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leslie AGW. The integration of macromolecular diffraction data. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 53.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66: 213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeLano WL. The PyMOL Molecular Graphics System, Version 1.2r3pre. San Carlos, CA, USA: Schrödinger, LLC; 2002. [Google Scholar]
- 60.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly (A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 62.Olsen AS, Møller-Jensen J, Brennan RG, Valentin-Hansen P. C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 2010;404:173–182. doi: 10.1016/j.jmb.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 63.Neidle S. Principles of Nucleic Acid Structure. London: Elsevier; 2008. pp. 32–33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.