Abstract
Therapeutic gene transfer by replication-defective viral vectors or, for cancer treatment, by replication-competent oncolytic viruses shows high promise for treatment of major diseases. To ensure safety, timing or dosing in patients, external control of therapeutic gene expression is desirable or even required. In this study, we explored the potential of artificial aptazymes, ligand-dependent self-cleaving ribozymes, as an innovative tool for regulation of therapeutic gene expression. Importantly, aptazymes act on RNA intrinsically, independent of regulatory protein–nucleic acid interactions and stoichiometry, are non-immunogenic and of small size. These are key advantages compared with the widely used inducible promoters, which were also reported to lose regulation at high copy numbers, e.g. after replication of oncolytic viruses. We characterized aptazymes in therapeutic gene transfer utilizing adenovectors (AdVs), adeno-associated vectors (AAVs) and oncolytic adenoviruses (OAds), which are all in advanced clinical testing. Our results show similar aptazyme-mediated regulation of gene expression by plasmids, AdVs, AAVs and OAds. Insertion into the 5′-, 3′- or both untranslated regions of several transgenes resulted in ligand-responsive gene expression. Notably, aptazyme regulation was retained during OAd replication and spread. In conclusion, our study demonstrates the fidelity of aptazymes in viral vectors and oncolytic viruses and highlights the potency of riboswitches for medical applications.
INTRODUCTION
Gene therapy and virotherapy are in pre-clinical and clinical development for the treatment of various major diseases. In gene therapy, corrective genes or genes encoding antigens, therapeutic proteins or regulatory RNAs are transferred into patients. In contrast, virotherapy is a modality for cancer treatment by tumor-restricted virus infection, cell lysis and spread. These strategies strongly rely on the development of efficient gene transfer vectors and oncolytic viruses, respectively. As viruses naturally possess efficient strategies for transferring their genetic material into mammalian cells, viruses that have been rendered replication-deficient are widely utilized gene transfer vectors (1). For virotherapy applications, oncolytic viruses have been engineered to possess tumor-restricted replication competency (2). They can be additionally ‘armed’ with therapeutic genes in order to combine oncolysis with gene therapy regimens for multi-modal anti-tumor activity (3). Robust and efficient genetic tools for an external control of therapeutic gene expression are needed for timing and dosing of gene drugs in clinical applications and as a safety measure. This is especially of interest for emerging applications of ‘armed’ oncolytic viruses, which replicate in tumors and thereby amplify therapeutic gene delivery. Inducible promoters have been widely explored for the regulation of gene therapies (4,5). However, they possess several disadvantages. Most inducible promoter systems depend on the expression of heterologous transcription factors as sensors for inducing drugs. These transcription factors need to be encoded by the vector drawing on valuable genomic space and in most cases they are immunogenic and pre-determined on a specific ligand. Moreover, impaired regulation of inducible promoters (leakiness) was found at high copy numbers either after high titer virus transduction or after virus genome replication of oncolytic viruses (6–9). This might reflect a problem of stoichiometry resulting from the complex mode of action of promoters, i.e. their dependence on protein–DNA interactions for transactivation.
We reasoned that gene control systems that are encoded in cis within mRNAs are well-suited candidates for circumventing problems associated with the use of transcription factor-dependent mechanisms in gene therapy and virotherapy. Interestingly, nature has invented such simplified control devices of gene expression: riboswitches are wide-spread mRNA-encoded sensors that regulate gene expression in response to metabolites, second messengers or toxic agents in prokaryotes, fungi and plants (10,11). They are composed of an aptamer domain determining the ligand specificity and an expression platform that modulates the gene expression in response to ligand binding to the aptamer. This modular architecture has resulted in the development of artificial riboswitches for gene regulatory devices in diverse synthetic biology applications (10,12–14). In addition to such artificial RNA switches, we have previously engineered aptazymes that rely on a mRNA-encoded, hammerhead ribozyme (HHR)-mediated self-cleavage reaction as expression platform (15–17). The advantage of triggering a cleavage reaction is that momentarily irreversible effects such as RNA degradation can be implemented, also allowing regulation of other RNA classes such as tRNAs and rRNAs (18,19). Of note, such systems require little vector space, are non-immunogenic and, due to their RNA-based intramolecular mode of action, should function independently of gene copy numbers. Furthermore, aptamers and aptazymes with desired qualities can be selected in vitro and can be customized, for example for OFF and ON switch applications, or for induction by novel ligands with desired pharmacological properties (20–22). For all these reasons, synthetic aptazymes show high potentials for medical applications. Nonetheless, only limited applications of aptazymes in mammalian cells have been reported to date. One reason is that aptazymes selected in vitro or in bacteria or yeast frequently lost their activity in mammalian cells (23,24).
The aim of this work is the development of aptazymes as an alternative tool for regulation of gene expression in a viral context for applications in gene therapy and virotherapy (Figure 1). Specifically, we explored adenoviruses, which have been widely investigated in pre-clinical and clinical studies, mostly for genetic vaccination, cancer gene therapy and viral oncolysis (25–27). In fact, they are the most frequently used gene transfer vectors in clinical studies (JGM Gene Therapy Clinical Trials Database). Adeno-associated viruses (AAVs) are further important gene therapy vectors that have recently shown encouraging results both pre-clinically and in patients for the treatment of inherited diseases (28). A key advantage of adenoviruses and AAVs is that they can be genetically engineered in different ways to match specific requirements for therapeutic applications (27,29,30). However, efficient tools for external control of gene expression are still sought for. Therefore, we explored the suitability of a theophylline-responsive aptazyme previously developed in mammalian cells (17) for applications in adenoviruses and AAVs. Specifically, we investigated strategies for optimized aptazyme insertion into transgenes and for aptazyme-mediated regulation in the context of gene transfer by replication-deficient adenovectors (AdVs), AAVs and oncolytic adenoviruses (OAds).
Figure 1.
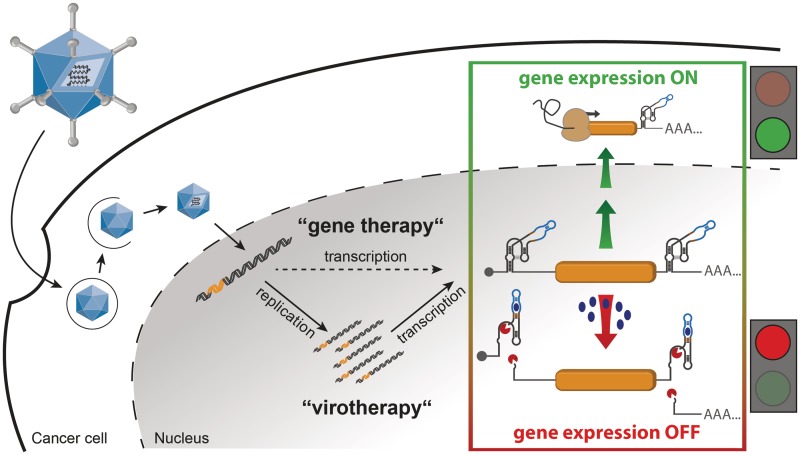
Schematic outline of our strategy for aptazyme-regulated transgene expression in mammalian cells by adenoviral vectors (AdVs) or oncolytic adenoviruses (OAds). In our strategy, control of transgene expression by replication-deficient AdVs and replication-competent OAds is achieved by insertion of an aptazyme, a synthetic ligand-dependent self-cleaving ribozyme, into the 5′- and/or 3′-UTR of the transcription unit. Upon infection, the viral genome is transmitted to the nucleus, where replication occurs for OAds in tumor cells, but not for AdV. Then, transgenes are transcribed. The applied OFF switch, given by an aptazyme, allows for transgene expression in the absence of ligand (gene expression ON). Upon addition of the ligand (e.g., the small molecule theophylline), the aptazymes can fold into an active conformation and mRNA self-cleavage occurs (gene expression OFF).
MATERIALS AND METHODS
Mammalian cell culture maintenance
Human cell lines HEK293 (embryonic kidney), SK-MEL-28 (malignant melanoma), HeLa (cervical cancer) and A549 (lung adenocarcinoma) were cultivated in Dulbecco’s modified Eagle’s medium (Invitrogen, Karlsruhe, Germany) supplemented with 10% heat-inactivated fetal bovine serum (PAA, Pasching, Austria), 100 IU/ml penicillin and 100 µg/ml streptomycin (both from Invitrogen). Cells were grown at 37°C in a humidified atmosphere of 5% CO2.
Plasmid construction
All cloning steps were conducted by applying standard molecular biology techniques. Used oligonucleotides were synthesized by Eurofins MWG Operon (Ebersberg, Germany). Cloned plasmids were purified by standard anion exchange columns (Qiagen, Hilden, Germany) and verified by DNA sequencing (GATC Biotech, Konstanz, Germany). Necessary cloning enzymes were obtained from Fermentas (St. Leon-Rot, Germany); DNA polymerase was obtained from BioCat (Heidelberg, Germany). Used nomenclature was as follows: HHR N107 (31); P1-F5, theophylline-dependent aptazyme (17). The term ‘in’ describes a mutated aptazyme or HHR sequence harboring a point mutation A14G (corresponding to the HHR nomenclature within the catalytic core of the HHR domain, which was reported to inactivate HHR self-cleavage (31). Branchpoint polypyrimidine tract and splice acceptor element (BPSA) (32).
For detailed cloning procedure, see Supplementary Methods. Briefly, for cloning of pS-Luc (firefly luciferase) variants (5′/5*/3′/5′5*/5′3′/5*3′/5′5*3′ and HHR 5′3′/inHHR 5′3′/in5′3′), pGL3 promoter (Promega, Mannheim, Germany) and pS-ctrl (pShuttle, with inserted polyA-SV40 promoter-Luc-polyA cassette from pGL3-promoter) were used as cloning vectors. For a schematic outline of the pS-Luc constructs, see Figure 2A. For cloning of transgene variants, pSCMV-CCL5 (Chemokine (C-C motif) ligand 5), psiCheck-2 (Promega) and pIND(SP1)/Hygro/β-galactosidase (lacZ; Invitrogen) were used as template vectors. For cloning of pSΔ24BPSA-Luc variants (P1-F5 5′/3′/5′3′), pGL3BPSA-Luc and pSΔ24BPSA-Luc were used as cloning vectors (33).
Figure 2.
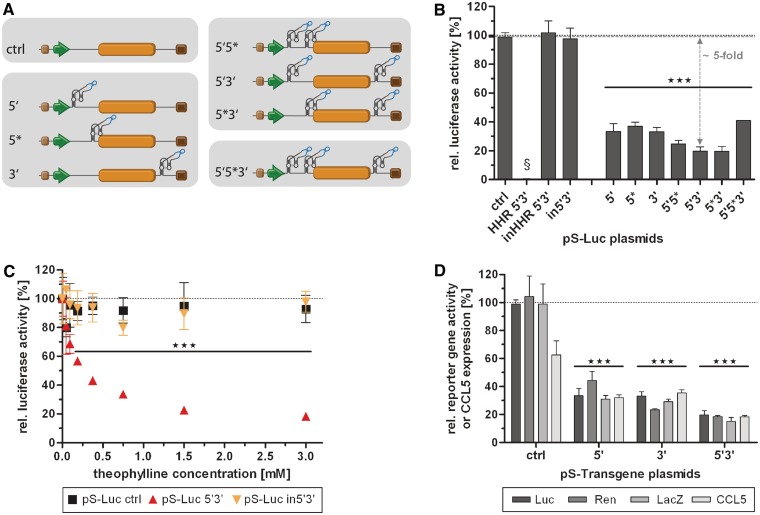
Characterization of different aptazyme insertion sites, and combinations thereof, using plasmid transfection. (A) Schematic representation of the analyzed aptazyme reporter shuttle plasmids (pS), which were also subsequently used for cloning of AdV genomes. Insertion sites for the aptazyme P1-F5 were within the 5′- and/or 3′-UTR of the transgene (orange box) driven by a SV40 promoter (green arrow). Brown box, synthetic polyadenylation signal; dark brown box, polyadenylation signal (pA). Nomenclature: 5′-UTR insertion distant (5') or proximal (5*) to the start codon, 3′-UTR insertion (3'), or in combinations thereof (5′5*, 5′3′ and 5′5*3'); ctrl, no aptazyme insertion. (B–D) HEK293 cells were transfected with the indicated reporter plasmids, harvested 48 h post transfection and gene activity/expression was measured. Shown are relative reporter gene activities or relative CCL5 expression for plasmids in the presence of theophylline (3 mM, except in C) in % of expression levels in the absence of theophylline (set as 100%, dotted line) of a representative experiment. Columns/symbols show mean values of triplicate transfections, error bars show SD. Significance for both theophylline-dependent down-regulation of gene expression for individual constructs and relative reporter gene expression in comparison to ctrl is indicated with ***(P < 0.001). (B) Regulation of gene expression for plasmids with firefly luciferase reporter gene by theophylline (3 mM). Dashed line, reference gene activity level for calculation of aptazyme performance. Controls are: in5′3′, inactive P1-F5 aptazyme in 5′- and 3′-UTR; HHR 5′3′, hammerhead ribozyme in 5′- and 3′-UTR; inHHR 5′3′; inactive HHR in 5′- and 3′-UTR. §, absolute reporter gene expression for HHR 5′3′ was <1% compared with inHHR 5′3′. (C) Dose-dependent regulation of the 5′3′ firefly luciferase construct and of the controls in5′3′ and ctrl by theophylline. (D) Regulation of gene expression for constructs 5′, 3′ and 5′3′ containing different reporter genes (Luc, firefly luciferase; Ren, renilla luciferase; LacZ, β-galactosidase) or the gene for human chemokine CCL5 by theophylline (3 mM).
Recombinant AdVs and OAds
For a schematic outline of the adenovirus genomes generated and used in this study, see Figures 3A and 4A.
Figure 3.
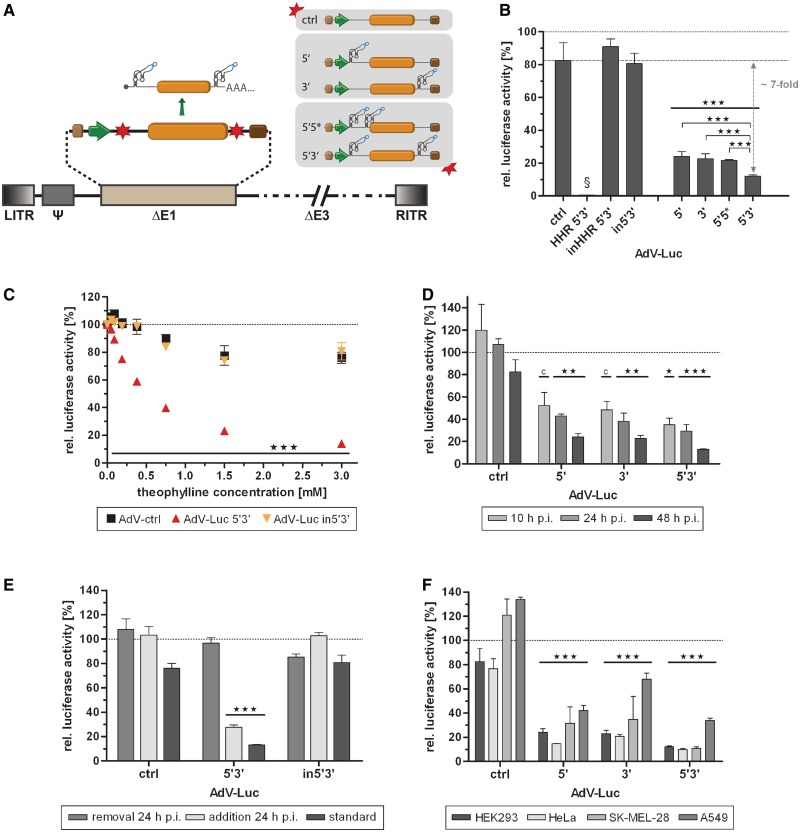
Application of the aptazyme for regulation of gene expression by an adenoviral vector (AdV) in a replication-competent and replication-deficient model. (A) Schematic outline of a replication-deficient, first generation AdV. The essential E1 genes were replaced (ΔE1) by the shown firefly luciferase transcription units. Nomenclature for aptazyme and control constructs is as in Figure 2A. LITR, left inverted terminal repeat; Ψ, packaging signal; RITR, right inverted terminal repeat; ΔE3, E3 region deleted (not required for virus replication); other viral genes and elements are not shown (dashed lines). (B–F) Cells were transduced with indicated AdVs, and gene expression was measured at 48 h (B–F) or at indicated time points (D) post transduction. Shown are relative luciferase activities for AdVs in the presence of theophylline (3 mM, except in C) in % of expression levels in the absence of theophylline (set as 100%, dotted line) of a representative experiment. Columns/symbols show mean values of triplicate transductions, error bars show SD. Significance for both theophylline-dependent down-regulation of gene expression for individual constructs and relative reporter gene expression in comparison to ctrl is indicated with *** over bars (P < 0.001). (B) Transduction of E1-complementing HEK293 cells with indicated AdVs (5 TCID50/cell). Dashed line, reference gene activity level for calculation of aptazyme performance. §, absolute reporter gene expression for HHR 5′3′ was <1% compared with inHHR 5′3′. Theophylline-dependent regulation of gene expression was significantly superior for 5′3′ compared with both 5′ and 3′ as indicated by *** (P < 0.001). (C) Dose-dependent regulation of transgene expression by theophylline after transduction of HEK293 cells with indicated AdVs (5 TCID50/cell). (D) Regulation of transgene expression at 10, 24 and 48 h after transduction of HEK293 cells with the indicated AdVs (5 TCID50/cell). *(P < 0.05) and **(P < 0.01) as calculated above; c, P < 0.05 for relative reporter gene expression in comparison with ctrl only. (E) Transduction of HEK293 cells with indicated AdVs (5 TCID50/cell). Theophylline was added either after transduction as before (standard), was added after transduction and removed at 24 h, or was added at 24 h post transduction. (F) Transduction of HEK293 (5 TCID50/cell), HeLa (50 TCID50/cell), SK-MEL-28 (100 TCID50/cell) or A549 (50 TCID50/cell) cells with indicated AdVs.
Figure 4.
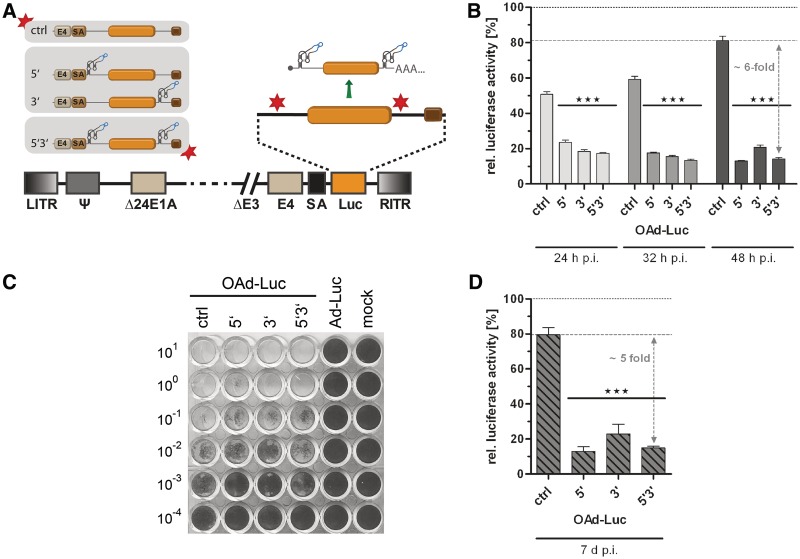
Application of the aptazyme for regulation of late transgene expression by oncolytic adenoviruses (OAds). (A) Schematic outline of replication-competent OAds. The firefly luciferase gene (orange box) with SV40 polyadenylation signal (brown box) was inserted with an upstream splice acceptor site (SA) into the OAdV genome, facilitating transgene co-expression at the late phase of virus replication. E1AΔ24, E1A gene with 24 bp deletion for tumor-selectivity. E4, E4 region. Other abbreviations as in Figure 3. Nomenclature for aptazyme and control constructs is as in Figure 2. (B) A549 cells were infected with OAds at 5 TCID50/cell and cultured in the presence or absence of 3 mM theophylline. Cells were harvested at 24 h, 32 h and 48 h post infection. Shown are relative luciferase activities for OAds in the presence of theophylline in % of expression levels in the absence of theophylline (set as 100% for each time point, dotted line) of a representative experiment. Dashed line, reference gene activity level for calculation of aptazyme performance. Columns/symbols show mean values of triplicate infections, error bars SD. Significance for both theophylline-dependent down-regulation of gene expression for individual constructs and relative reporter gene expression in comparison with ctrl is indicated with ***(P < 0.001). (C) Spread-dependent cytotoxicity of OAds in A549 cells as determined by crystal violet staining of surviving cells 20 d post infection. Numbers are viral titers in TCID50/cell used for infection. Ad-Luc, replication-deficient virus (AdV ctrl, see Figure 3). Mock, mock-infected cells. (D) A549 cells were infected using 0.05 TCID50/cell. Five days post infection, media was removed, and cells were cultured for 2 more days either in the absence or presence of 3 mM theophylline. Data presentation is as in panel B.
AdV-Luc ctrl, HHR 5′3′, inHHR 5′3′, 5′, 3′, 5′3′, 5′5* and in5′3′ are replication-deficient AdVs based on pAdEasy-1 with a pGL3-derived luciferase gene under control of the SV40 promoter. Insertion sites for HHR or P1-F5 are the same as for pShuttle plasmids described above. AdV genomes were generated by homologous recombination in BJ5183 bacteria as described (34) using the pS-Luc variants described above and pAdEasy-1 (Promega). Virus particles were produced by transfection of HEK293 cells with PacI-digested genome plasmids using Lipofectamine (Invitrogen). The recombinant viruses were amplified in HEK293 cells and purified by two rounds of CsCl equilibrium density gradient ultracentrifugation. Verification of viral genomes and exclusion of wild-type contamination were performed by polymerase chain reaction. Physical particle concentration (viral particles/ml) was determined by reading the optical density at 260 nm (virus particle yield was in the same range for all AdV variants). Infectious viral particle titers were determined by 50% tissue culture infective dose (TCID50) assay on HEK293 cells. The ratios of virus particles to infectious virus particles for virus preparations were as follows: ctrl 24; HHR 5′3′, 32; inHHR 5′3′, 10; 5′, 26; 3′, 24; 5′3′, 17 and in5′3′, 32.
OAd-Luc ctrl, 5′3′, 5′, 3′ and 5′3′ are replication-competent adenoviruses with a pGL3-derived luciferase gene inserted via a BPSA sequence into the Ad5 late transcription unit (33). Nomenclature was as described above. OAd genomes were then generated by homologous recombination of pSΔ24BPSA-Luc plasmids described above with pAdEasy-1 as described for AdVs. Virus production was carried out in A549 cells. Virus characterization was performed as described above. The ratios of virus particles to infectious virus particles for virus preparations were as follows: ctrl, 20; 5′, 6; 3′, 7 and 5′3′, 17; virus particle yield was in the same range for all OAd variants.
Transient transfections
All transient transfections of HEK293 cells were performed using Lipofectamine and Plus reagent according to the manufacturer’s protocol (Invitrogen). For transfection details and measurement of reporter activity and CCL5 expression, see Supplementary Methods. For presentation of data, background values were subtracted and values were then set in relation to the reporter activity/expression of the corresponding construct at 0 mM theophylline, which was set at 100%. Fold induction rates of aptazyme-mediated regulation were determined by relating relative reporter activity of aptazyme constructs to relative reporter activity of the control construct, both in the presence of 3 mM theophylline. Thus, these rates consider aptazyme performance only. Noteworthy, we used a maximum concentration of 3 mM theophylline, as at higher dosage, microscopically visible toxicity was observed. Therefore, absolute theophylline-mediated down-regulation is in most cases higher than as aptazyme-mediated regulation, because it considers also the residual minimal theophylline toxicity (observed as reduced activity of ctrl constructs).
Transient infections
For analysis of reporter activity, various cell lines were infected using indicated virus titers and standard procedures. For details of infection protocols and determination of luciferase activity, see Supplementary Methods. Presentation of data is as described for transient transfections.
For analysis of virus-mediated spread and cytotoxicity, A549 cells were seeded in 48-well plates to 70-90% confluence. The next day, infections were performed in serial dilutions (1:10) starting from 10 TCID50/cell. When cell lysis was observed at the lowest virus titers, cytotoxicity assay was performed.
Cytotoxicity assay
Virus-mediated spread and cytotoxicity were determined when lysis was visible at the lowest virus dilution. Then, cells were fixed and stained with 1% crystal violet in 70% ethanol, followed by washing with tap water to remove excess color. Plates were dried, and images were captured with a Perfection V500 Photo scanner (Epson Deutschland, Meerbusch, Germany).
Statistical analysis
Differences between indicated groups were analyzed using two-way analysis of variance with Bonferroni post test using GraphPad Prism version 5 for Windows (www.graphpad.com). Values of P < 0.05 were considered statistically significant.
RESULTS
Aptazyme positioning and multimerization for control of transgene expression in mammalian cells
To date, it has not been established whether aptazymes enable superior gene regulation when inserted into the 5′-untranslated region (UTR), the 3′-UTR or both UTRs of eukaryotic genes. In fact, previous studies exploring ribozymes or aptazymes in eukaryotes showed controversial results with respect to their activity or regulation in the 5′- or 3′-UTR (17,31,35). Therefore, we first tested different aptazyme insertion sites for regulation of transgene expression in mammalian cells and investigated whether multiple aptazyme insertions increase the switching performance. Luc reporter plasmids were generated with insertions of the previously described theophylline-induced aptazyme P1-F5 optimized for controlling gene expression in mammalian cells (17) in the 5′-UTR, distant (5′) or proximal (5*) to the start codon, 3′-UTR (3') or in combinations thereof (Figure 2A). Quantification of reporter activity after plasmid transfection of HEK293 cells showed theophylline dependence for all aptazyme constructs, but not for the control construct lacking the aptazyme (Figure 2B). Regulation was similar for the three insertion sites and was further increased by double insertions (although not significantly) but not in case of the triple insertion. A maximum of 5-fold aptazyme-mediated regulation was achieved for constructs 5*3′ and 5′3′ (Figure 2B; see ‘Materials and Methods’ for calculation of fold induction rates of aptazyme-mediated regulation). Furthermore, dose-dependent regulation was observed for the 5′3′ construct reaching saturation at 3 mM theophylline but not for the control lacking the aptazyme (ctrl) (Figure 2C). Aptazyme insertion into the mRNA per se (i.e., in the absence of theophylline) affected transgene expression only modestly (Supplementary Figure S1A). For example, the 5′3′ construct showed 80% of transgene expression relative to the parental construct lacking the aptazyme. A control construct composed of the constitutively active ribozyme lacking theophylline dependence inserted into the 5′3′ positions (HHR 5′3′) showed complete loss of transgene expression (Figure 2B and Supplementary Figure S1A). The latter finding demonstrates that the inserted HHRs cleave very efficiently and hence are able to control the whole population of expressed mRNAs. In further controls, an inactivating point mutation (17,31) was introduced into the catalytic core of the HHR. The inactivation of ribozymes in both the 5′3′ aptazyme construct (in5′3′) and the constitutive ribozyme control (inHHR 5′3′) resulted in high levels of gene expression, indicating that insertion of ribozyme or aptazyme sequences per se are not significantly disturbing gene expression (Figure 2B and C and Supplementary Figure S1A). Importantly, when incubated with the ligand theophylline, similar gene expression levels were observed. Taken together, the cleavage reaction triggered by theophylline in the cleavage-competent constructs described above is indeed causative for the regulatory properties of the genetic switch and not simply the binding event of the ligand to the aptamer domain of the artificial switch.
In order to demonstrate that the established ligand-dependent control is independent of the utilized mRNA, the aptazyme P1-F5 was also inserted into several other genes. Significant aptazyme-dependent regulation of transgene expression was observed for all investigated cases: the reporter genes Renilla luciferase and LacZ as well as a therapeutic gene encoding the secreted chemokine CCL5 (Figure 2D). In each case, regulation was superior for the 5′3′ construct compared with the corresponding single insertions, which is in accord with the findings for firefly luciferase. However, for CCL5 constructs, aptazyme-mediated regulation was lower compared with the reporter gene constructs, but still showed significant reduction of gene expression. Here, we also observed an inhibition of transgene expression by theophylline for the control construct. This observation might be attributed to a possible direct or indirect inhibition of the protein secretion pathway by theophylline.
Aptazyme regulation of gene expression after transduction with AdVs in absence or presence of viral genome replication and by AAV
We next generated replication-deficient adenoviral vectors (AdVs, Figure 3A) derived from the Luc constructs 5′, 3′, 5*3′, 5′3′, in5′3′, HHR 5′3′, inHHR 5′3′ and ctrl using the pShuttle plasmids analyzed in Figure 2. AdV transduction of HEK293 cells (Figure 3B and C, Supplementary Figure S1B) yielded results very similar to plasmid transfections (see Figure 2B and C, Supplementary Figure S1A): significant and dose-dependent regulation of transgene expression was observed with the utilized aptazymes with the best regulation (7-fold) again found for the 5′3′ construct. Furthermore, inhibition of transgene expression by the constitutively active control HHR was nearly complete. We observed a minor inhibition of transgene expression of control constructs (ctrl, in5′3′, inHHR 5′3′) by theophylline (Figure 3B and C). Moreover, gene expression was only modestly affected by aptazyme insertion per se (Supplementary Figure S1B). For example, the 5′3′ AdV showed 63% of transgene expression relative to the control without aptazyme. Of note, AdVs replicate in HEK293 cells, which complement the essential viral genes deleted in the AdV genomes. In a time-course experiment, we observed similar theophylline-dependent regulation of transgene expression at 10 h (before viral genome replication) as well as at 24 and 48 h (after viral genome replication) post infection (p.i.; Figure 3D). Importantly, we observed aptazyme-mediated regulation of transgene expression at 48 h irrespective of the time point of theophylline addition, either 1 h (6-fold) or 24 h (4-fold) post transduction (Figure 3E). Furthermore, transgene expression was restored at 48 h when theophylline was added 1 h post transduction and removed at 24 h (Figure 3E), demonstrating reversibility of the aptazyme-mediated control of gene expression.
Finally, we tested aptazyme-dependent regulation of AdV transduction in three further mammalian cell lines, SK-MEL-28, HeLa and A549, in which AdVs cannot replicate (Figure 3F). We observed significant regulation of transgene expression by aptazymes for 5′, 3′ and 5′3′ viruses with best regulation for the 5′3′ virus (8 -/11-/4-fold in HeLa, SK-MEL-28 and A549, respectively). Interestingly, transgene expression was differentially affected in the control construct (ctrl) upon theophylline addition, showing that the aptazyme-independent effect of theophylline is dependent on the cell line: we observed a slight reduction to approximately 80% activity in HEK293 and HeLa cells, but an increase to approximately 125% gene expression in SK-MEL-28 and A549 cells. Using HeLa cells, we also characterized dose dependency and fidelity of aptazyme regulation and found similar results compared with the HEK293 cells utilized before (Supplementary Figure S2A and B). We conclude that aptazymes enable regulation of AdV gene transfer in mammalian cells, as demonstrated for four cell types. In addition, we were able to show that regulation of gene transfer by the 5′ aptazyme strategy is not restricted to AdV and also functional in the context of another widely used gene transfer vector, AAV (Supplementary Figure S3). This result is of high relevance for the development of AAV-based gene therapies, because AAV vectors possess a small genome and thus limited capacity for heterologous DNA. Consequently, short regulatory sequences, such as aptazymes, are highly desirable to facilitate inducible gene therapy by AAV vectors.
Aptazyme regulation of transgene expression by an OAd
Next, we aimed at superimposing aptazyme-mediated regulation on transgene expression by OAds (see Figure 1). We and others previously reported a mechanism that enables replication-dependent, thus tumor-specific transgene expression by OAds. Here, the applied transgene is inserted into the late adenoviral transcription unit via an alternative splice site. By this means, gene expression is initiated from the viral major late promoter, and a heterologous promoter is not required (29,33). We generated OAds with aptazyme P1-F5 in the 5'-UTR and/or 3'-UTR of the Luc gene inserted downstream of the splice acceptor site (Figure 4A). We observed significant regulation of transgene expression at 24, 32 and 48 h p.i. of A549 (Figure 4B) and HeLa (Supplementary Figure S4A) cells with increasing aptazyme-mediated regulation up to 6-fold at 48 h p.i. Interestingly, the OAd single aptazyme insertion construct 5′ enabled as effective regulation as the double insertion construct 5′3′ at 48 h p.i. For the ctrl OAd construct lacking aptazymes, we detected a slight unspecific inhibition of transgene expression by theophylline alone to approximately 80% (A549) or 70% (HeLa) at 48 h p.i. However, gene expression was more affected by aptazyme insertion per se (to approximately 20%) than it was the case for plasmid transfections and AdV transduction (Supplementary Figure S1C). When analyzing the time course of OAd replication, we determined a delay in genome replication (Supplementary Figure S4B) and late gene expression (mRNA and protein, Supplementary Figure S4C and D) of up to 4 h caused per se by aptazyme insertion. Possibly, this delay is, at least in part, responsible for the reduced transgene expression of the aptazyme-encoding OAds at 48 h p.i. as mentioned above. However, the influence of aptazyme insertion on spread-dependent cytotoxicity of OAds, as observed after low titer infection and 20 days incubation, was negligible (Figure 4C). In addition, the time course of replication was minimally and transiently affected by addition of theophylline (Supplementary Figure S4B and C, 20 h p.i.). In contrast, transgene expression was stably regulated over time for the 5′3′, but not for the ctrl OAd construct (Supplementary Figure S4E). Notably, aptazyme-dependent regulation of gene expression was effective even when triggered after several cycles of OAd replication and spread (Figure 4D and Supplementary Figure S1D). This observation is important since the situation in therapeutic applications is more closely resembled by successful regulation of expression in viral infections that are no longer synchronous. At that time aptazyme-mediated regulation was 5-fold and thus similar to earlier time points after infection.
DISCUSSION
This study establishes aptazymes as genetic regulatory elements for external control of transgene expression after gene transfer by AdVs, by AAV vectors and by replication-competent oncolytic viruses and hence is the first to demonstrate aptazyme activity in an extra-chromosomal viral context. Specifically, our results show that aptazyme insertion enables a dose- and ribozyme-dependent regulation of gene transfer in mammalian cells, independent of the transgene. Furthermore, regulation is improved by simultaneous insertion of aptazymes in both the 5′-UTR and 3′-UTR. Of major importance is our observation that regulation for the different aptazyme insertion constructs for AdVs and OAds closely matched the results obtained by transient transfection of the corresponding plasmids (compare Figure 2B, 3B and 4B). The resulting maximum regulation utilizing the aptazyme P1-F5 was 5-fold for plasmids and 11-fold for viruses. However, the described control constructs utilizing a constitutively active HHR shows the maximal possible regulation for aptazymes derived from HHR N107 (used here) (31), which was > 100-fold. This indicates that the aptazyme P1-F5 still allows for further improvements and demonstrates the high potential for regulation of gene therapy by synthetic riboswitches. Gene expression by AdV transduction could be switched from ‘ON’ to ‘OFF’ (addition of theophylline at 24 h post transduction) and also from ‘OFF’ to ‘ON’ by removal of theophylline, demonstrating the fidelity and reversibility of the aptazyme system in the viral context.
Oncolytic viruses are emerging cancer therapeutics and in advanced clinical development (36,37). This includes OAds and transgene-encoding viruses, such as the GM-CSF-encoding vaccinia or herpes viruses. To fully exploit the potential of such ‘armed’ oncolytic viruses, external control of therapeutic gene expression is desirable or even required, for example as a safety measure for expression of cytokines, toxins or cell death-inducing ligands or to orchestrate therapeutic gene expression with viral replication and combined treatment regimens. However, control of gene expression from amplified oncolytic virus genomes is challenging as transcription factor-dependent inducible promoters have shown reduced regulation at high copy numbers (6–9). In contrast, aptazyme regulation was functional after adenoviral gene transfer and was not affected by virus replication and thus amplification of the virus genome, as shown for AdVs in complementing HEK293 cells and for OAds. Our data for gene expression and viral genome replication of OAds indicate a modest delay in virus replication by aptazyme insertion, indicating that the chosen insertion sites are not optimal yet and require further improvement. However, even with the present insertion sites, virus spread and oncolysis were affected only marginally.
Studies using artificial riboswitches in eukaryotic systems, especially in mammalian cells, have been limited. In a first example, aptamers have been introduced into mRNAs in order to achieve ligand-dependent gene expression via modification of secondary mRNA structure and thus translational control (38). Later, Mulligan and co-workers identified a nucleoside analogue as an inhibitor of ribozyme function enabling dose-dependent expression of ribozyme-containing genes (31,39). However, the underlying mechanism is not based on specific ligand-sensing as riboswitches are characterized for, but on unspecific ligand incorporation into all mRNAs which likely is connected with side effects. Alternatively, aptamers were fused with expression platforms in order to realize a specific RNA–ligand interaction. Here, we have utilized ribozyme-mediated self-cleavage as expression platforms (15–17). This approach has the advantage of triggering drastic and momentarily irreversible changes of the controlled RNA. Great prospects for therapeutic applications justify a detailed examination of the performance of aptazyme-based control mechanisms. Previous studies showed a benefit of inserting multiple ribozymes or aptazymes (31,35), but diverging results were obtained with respect to the optimum insertion site. Mulligan and co-workers found the most pronounced effects if their ribozymes were introduced into the 5′-UTR of mammalian reporter constructs (31). In Saccharomyces cerevisiae, Smolke and co-workers found the 3′-tail of mRNAs the most efficient position (35). The presented work is the first report comparing 5′ and 3′ insertion sites and combinations thereof. Our finding that the theophylline-dependent aptazyme P1-F5 is functional in both 5′-UTRs and 3′-UTRs and that regulation is improved when both are combined (for plasmid transfection, AdV transduction and OAd infection) is of importance for the development of aptazyme-based RNA switches in gene therapy and further synthetic biology applications. It is noteworthy that for the 5′3′ constructs, we observed, although with low frequency, excision of the aptazyme-flanked transgene, presumably by homologous recombination during virus replication. However, gene expression remained efficient and aptazyme regulation effective for these constructs. Thus, future applications need to consider whether such recombination effects are tolerable or whether they can be overcome by aptazyme engineering.
Of high relevance for gene regulation in therapeutic applications are the dynamics of aptazyme-mediated regulation that we report here. Ligand-induced suppression of gene expression was observed when the ligand was added immediately after plasmid transfection, AdV transduction or OAd infection, i.e. before onset of transgene expression, and also when the ligand was added 24 h after AdV transduction or 5 days after OAd infection, i.e. at a time when transgene expression and/or virus replication were fully active. Finally, our observation that transgene expression was restored 24 h after removal of the aptazyme-inducing ligand demonstrates the feasibility of sequential OFF and ON regulation of gene expression. Moreover, as gene expression remained silent in parallel cultures in which the ligand was not removed, the aptazyme proved clear functionality in the presence of active transcription. With our best constructs, we observed 7- to 11-fold aptazyme-mediated regulation of transgene expression after gene transfer by AdVs or OAds, which is superior to previously reported induction rates of aptazymes in mammalian cells (12,17). In one study, control of cytokine gene expression by triple aptazyme insertions enabled regulation of lymphocyte survival (12). Although this study showed that aptazymes with moderate gene regulatory activity can trigger biological effects, we suggest that even better switching performances are warranted to realize the full potential of RNA switches in gene therapy and virotherapy. In this regard, our observation of nearly complete shutdown of gene expression by the parental HHR is promising as it establishes the potential boundary of gene regulation achievable with ribozyme devices triggering self-cleavage of the message.
While our study especially provides proof-of-principle for aptazyme-mediated gene regulation in AdVs, AAV and oncolytic viruses, we believe that the OFF switch aptazyme, reported here, already provides a tool with different application possibilities. In the context of therapeutic gene transfer, e.g. during gene therapy or virotherapy, adjustment of gene expression level applying an OFF switch would allow for expression of the therapeutic protein within the optimal dose range (40). Moreover, an OFF switch could also serve as a safety measure in case of unwanted side effects evoked by the encoded therapeutic proteins, such as cytokines, antiangiogenic factors or apoptosis-inducing ligands. In addition, for analysis of protein function or during pharmaceutical production of potentially toxic proteins or vectors encoding this protein, the OFF switch aptazyme represents a tool for adjusting or transiently blocking expression of the protein. In contrast to transcription factor-based systems, no other potentially disturbing co-expressed proteins are required. Finally, OFF switch aptazymes could attenuate the expression of therapeutic transgenes by oncolytic viruses during initial virus replication and spread to facilitate optimal oncolysis before induction of protein synthesis from the amplified virus genomes.
Compared with the available inducible promoter systems, which were developed over decades (40), it is clear that RNA switches still have to be further improved with respect to fold induction rates in order to become an alternative for more widespread applications. In this regard, it is noteworthy that RNA-based switches can be improved with respect to pharmaceutically more suited ligands, and enhanced switching performances will likely be accessible in the near future since a variety of advanced screening and selection methods are becoming available. Notably, such customization is a further advantage of aptazymes, whereas inducible promoters are not flexible with respect to the inducing ligand. In general, successful and safe gene therapy approaches rely on regulatory tools ensuring both specific and controlled expression of the therapeutic gene. Here, regulation at different levels of therapeutic gene expression could be advantageous. For example, transcription control via promoters can be combined with RNAi by miRNA technology to achieve tissue specificity. Both approaches could be easily combined with aptazymes to ensure external regulation of gene expression. These considerations, together with our results, clearly underline that aptazyme-based switches possess multiple advantageous features for regulation of gene expression and show high potentials for biomedical applications as exemplified for gene therapy and virotherapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4, Supplementary Methods and Supplementary References [17,31–33,41–45].
FUNDING
The Helmholtz Association of National Research Centers (Helmholtz-University Group Grant VH-NG-212 to D.M.N.), the Peter und Traudl Engelhorn Stiftung (PhD fellowship to P.K.) and the VolkswagenStiftung (Lichtenberg-Professorship to J.S.H.). Funding for open access charge: Institutional (DKFZ).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Jürgen Kleinschmidt and Matthias Naumer (both DKFZ, Heidelberg, Germany) for their support with AAV technology and Kai Zanzinger (DKFZ and University Hospital, Heidelberg, Germany) for providing pSCMV-CCL5.
REFERENCES
- 1.Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat. Rev. Genet. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 2.Cattaneo R, Miest T, Shashkova EV, Barry MA. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat. Rev. Microbiol. 2008;6:529–540. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cody JJ, Douglas JT. Armed replicating adenoviruses for cancer virotherapy. Cancer Gene Ther. 2009;16:473–488. doi: 10.1038/cgt.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goverdhana S, Puntel M, Xiong W, Zirger JM, Barcia C, Curtin JF, Soffer EB, Mondkar S, King GD, Hu J, et al. Regulatable gene expression systems for gene therapy applications: progress and future challenges. Mol. Ther. 2005;12:189–211. doi: 10.1016/j.ymthe.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toniatti C, Bujard H, Cortese R, Ciliberto G. Gene therapy progress and prospects: transcription regulatory systems. Gene Ther. 2004;11:649–657. doi: 10.1038/sj.gt.3302251. [DOI] [PubMed] [Google Scholar]
- 6.Fechner H, Wang X, Srour M, Siemetzki U, Seltmann H, Sutter AP, Scherubl H, Zouboulis CC, Schwaab R, Hillen W, et al. A novel tetracycline-controlled transactivator-transrepressor system enables external control of oncolytic adenovirus replication. Gene Ther. 2003;10:1680–1690. doi: 10.1038/sj.gt.3302051. [DOI] [PubMed] [Google Scholar]
- 7.Fecker LF, Schmude M, Jost S, Hossini AM, Pico AH, Wang X, Schwarz C, Fechner H, Eberle J. Efficient and selective tumor cell lysis and induction of apoptosis in melanoma cells by a conditional replication-competent CD95L adenovirus. Exp. Dermatol. 2010;19:e56–e66. doi: 10.1111/j.1600-0625.2009.00977.x. [DOI] [PubMed] [Google Scholar]
- 8.Rohmer S, Mainka A, Knippertz I, Hesse A, Nettelbeck DM. Insulated hsp70B' promoter: stringent heat-inducible activity in replication-deficient, but not replication-competent adenoviruses. J. Gene Med. 2008;10:340–354. doi: 10.1002/jgm.1157. [DOI] [PubMed] [Google Scholar]
- 9.Shashkova EV, Kuppuswamy MN, Wold WS, Doronin K. Anticancer activity of oncolytic adenovirus vector armed with IFN-alpha and ADP is enhanced by pharmacologically controlled expression of TRAIL. Cancer Gene Ther. 2008;15:61–72. doi: 10.1038/sj.cgt.7701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 12.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Hunsicker A, Steber M, Mayer G, Meitert J, Klotzsche M, Blind M, Hillen W, Berens C, Suess B. An RNA aptamer that induces transcription. Chem. Biol. 2009;16:173–180. doi: 10.1016/j.chembiol.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 16.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 17.Auslander S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 2010;6:807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 18.Wieland M, Berschneider B, Erlacher MD, Hartig JS. Aptazyme-mediated regulation of 16S ribosomal RNA. Chem. Biol. 2010;17:236–242. doi: 10.1016/j.chembiol.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Berschneider B, Wieland M, Rubini M, Hartig JS. Small-molecule-dependent regulation of transfer RNA in bacteria. Angew. Chem. Int. Ed. Engl. 2009;48:7564–7567. doi: 10.1002/anie.200900851. [DOI] [PubMed] [Google Scholar]
- 20.Vinkenborg JL, Karnowski N, Famulok M. Aptamers for allosteric regulation. Nat. Chem. Biol. 2011;7:519–527. doi: 10.1038/nchembio.609. [DOI] [PubMed] [Google Scholar]
- 21.Link KH, Breaker RR. Engineering ligand-responsive gene-control elements: lessons learned from natural riboswitches. Gene Ther. 2009;16:1189–1201. doi: 10.1038/gt.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 23.Link KH, Guo L, Ames TD, Yen L, Mulligan RC, Breaker RR. Engineering high-speed allosteric hammerhead ribozymes. Biol. Chem. 2007;388:779–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- 24.Wittmann A, Suess B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol. Biosyst. 2011;7:2419–2427. doi: 10.1039/c1mb05070b. [DOI] [PubMed] [Google Scholar]
- 25.Lasaro MO, Ertl HC. New insights on adenovirus as vaccine vectors. Mol. Ther. 2009;17:1333–1339. doi: 10.1038/mt.2009.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConnell MJ, Imperiale MJ. Biology of adenovirus and its use as a vector for gene therapy. Hum. Gene Ther. 2004;15:1022–1033. doi: 10.1089/hum.2004.15.1022. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto M, Curiel DT. Current issues and future directions of oncolytic adenoviruses. Mol. Ther. 2010;18:243–250. doi: 10.1038/mt.2009.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mingozzi F, High KA. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 29.Nettelbeck DM. Cellular genetic tools to control oncolytic adenoviruses for virotherapy of cancer. J. Mol. Med. (Berl) 2008;86:363–377. doi: 10.1007/s00109-007-0291-1. [DOI] [PubMed] [Google Scholar]
- 30.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D'Amato RJ, Mulligan RC. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 32.Jin F, Kretschmer PJ, Hermiston TW. Identification of novel insertion sites in the Ad5 genome that utilize the Ad splicing machinery for therapeutic gene expression. Mol. Ther. 2005;12:1052–1063. doi: 10.1016/j.ymthe.2005.07.696. [DOI] [PubMed] [Google Scholar]
- 33.Quirin C, Rohmer S, Fernandez-Ulibarri I, Behr M, Hesse A, Engelhardt S, Erbs P, Enk AH, Nettelbeck DM. Selectivity and efficiency of late transgene expression by transcriptionally targeted oncolytic adenoviruses are dependent on the transgene insertion strategy. Hum. Gene Ther. 2011;22:389–404. doi: 10.1089/hum.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu TC, Galanis E, Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat. Clin. Pract. Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- 37.Rowan K. Oncolytic viruses move forward in clinical trials. J. Natl. Cancer Inst. 2010;102:590–595. doi: 10.1093/jnci/djq165. [DOI] [PubMed] [Google Scholar]
- 38.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 39.Yen L, Magnier M, Weissleder R, Stockwell BR, Mulligan RC. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high-throughput screening of chemical libraries. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtin JF, Candolfi M, Xiong W, Lowenstein PR, Castro MG. Turning the gene tap off; implications of regulating gene expression for cancer therapeutics. Mol. Cancer Ther. 2008;7:439–448. doi: 10.1158/1535-7163.MCT-07-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 42.Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 43.Hauswirth WW, Lewin AS, Zolotukhin S, Muzyczka N. Production and purification of recombinant adeno-associated virus. Methods Enzymol. 2000;316:743–761. doi: 10.1016/s0076-6879(00)16760-6. [DOI] [PubMed] [Google Scholar]
- 44.Veldwijk MR, Topaly J, Laufs S, Hengge UR, Wenz F, Zeller WJ, Fruehauf S. Development and optimization of a real-time quantitative PCR-based method for the titration of AAV-2 vector stocks. Mol. Ther. 2002;6:272–278. doi: 10.1006/mthe.2002.0659. [DOI] [PubMed] [Google Scholar]
- 45.Chadeuf G, Favre D, Tessier J, Provost N, Nony P, Kleinschmidt J, Moullier P, Salvetti A. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J. Gene Med. 2000;2:260–268. doi: 10.1002/1521-2254(200007/08)2:4<260::AID-JGM111>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.