Abstract
The chromatin structure of eukaryotic telomeres plays an essential role in telomere functions. However, their study might be impaired by the presence of interstitial telomeric sequences (ITSs), which have a widespread distribution in different model systems. We have developed a simple approach to study the chromatin structure of Arabidopsis telomeres independently of ITSs by analyzing ChIP-seq data. This approach could be used to study the chromatin structure of telomeres in some other eukaryotes. The analysis of ChIP-seq experiments revealed that Arabidopsis telomeres have higher density of histone H3 than centromeres, which might reflects their short nucleosomal organization. These experiments also revealed that Arabidopsis telomeres have lower levels of heterochromatic marks than centromeres (H3K9Me2 and H3K27Me), higher levels of some euchromatic marks (H3K4Me2 and H3K9Ac) and similar or lower levels of other euchromatic marks (H3K4Me3, H3K36Me2, H3K36Me3 and H3K18Ac). Interestingly, the ChIP-seq experiments also revealed that Arabidopsis telomeres exhibit high levels of H3K27Me3, a repressive mark that associates with many euchromatic genes. The epigenetic profile of Arabidopsis telomeres is closely related to the previously defined chromatin state 2. This chromatin state is found in 23% of Arabidopsis genes, many of which are repressed or lowly expressed. At least, in part, this scenario is similar in rice.
INTRODUCTION
Telomeres prevent chromosome fusions and degradation by exonucleases and are implicated in DNA repair, homologous recombination, chromosome pairing and segregation. Telomeric DNA usually contains tandem repeats of a short GC-rich motif, which can also be found at interstitial chromosomal loci (1–5). These interstitial telomeric sequences (ITSs) have a widespread distribution in different model systems, including Arabidopsis, and have been related to chromosomal aberrations, fragile sites, hot spots for recombination and diseases caused by genomic instability, although their functions remain unknown (5–8).
Two major chromatin organizations can be found inside the cell nucleus: heterochromatin and euchromatin. Heterochromatic regions are highly condensed in interphase nuclei giving rise to the so-called chromocenters and usually associate with repetitive and silent DNA, although certain level of transcription is required for their establishment and maintenance. By contrast, euchromatic regions have an open conformation and are often related to the capacity to be transcribed. Both kinds of chromatin organizations exhibit defined epigenetic modifications that influence their biochemical behavior. In Arabidopsis, chromocenters contain pericentromeric heterochromatin, which associates with the 178-bp satellite repeats (also known as 180-bp repeats) and with other repetitive DNA sequences including mobile elements and ITSs (9–15). Arabidopsis heterochromatin is characterized by high levels of cytosine methylation, which can be targeted at CpG, CpNpG or CpNpN residues (where N is any nucleotide), and by H3K9Me1,2, H3K27Me1,2 and H4K20Me1. In turn, Arabidopsis euchromatin is characterized by H3K4Me1,2,3, H3K36Me1,2,3, H4K20Me2,3 and by histones acetylation (16). In addition, many genes that localize in Arabidopsis euchromatin are labeled with H3K27Me3, a repressive mark that is thought to regulate tissue-specific gene expression (17–19).
The analysis of telomeric chromatin structure from ChIP, ChIP-on-chip or ChIP-seq experiments might be challenged by the presence of ITSs (20). This problem might also be extensive to other repetitive sequences. Here, we have developed an approach to study the epigenetic modifications of Arabidopsis telomeres independently of ITSs by analyzing genome-wide ChIP-seq data. The ChIP-seq experiments revealed that Arabidopsis telomeres have higher density of histone H3 than centromeres. These experiments also revealed that Arabidopsis telomeres have lower levels of heterochromatic marks than centromeres (H3K9Me2 and H3K27Me), higher levels of some euchromatic marks (H3K4Me2 and H3K9Ac) and similar or lower levels of other euchromatic marks (H3K4Me3, H3K36Me2, H3K36Me3 and H3K18Ac). Interestingly, the ChIP-seq data also revealed that Arabidopsis telomeres exhibit higher levels of H3K27Me3 than centromeres. At least, in part, this scenario is similar in rice.
MATERIALS AND METHODS
Determination of the relative amounts of (CCCTAAA)4 sequences at telomeres and ITSs
To analyze the chromatin structure of Arabidopsis telomeres using genome-wide ChIP-seq experiments, we had to define a specific DNA sequence that revealed telomeres but not ITSs. For that purpose, we estimated the number of times that the sequence (CCCTAAA)4 appears at internal chromosomal loci and at telomeres in the Arabidopsis thaliana (Col-0) genome. First, we performed Blast analyses at the Map Viewer web site in National Center for Biotechnology Information (NCBI) to determine the number of times that the sequence (CCCTAAA)4 appears at internal chromosomal loci (http://www.ncbi.nlm.nih.gov/mapview). In the case that a specific ITS contained five perfect tandem telomeric repeats, the Blast analyses revealed two overlapping (CCCTAAA)4 sequences. If the ITS contained six perfect tandem telomeric repeats, the Blast analyses revealed three overlapping (CCCTAAA)4 sequences and so on. We found 118 (CCCTAAA)4 sequences at internal positions in the five chromosomes of Arabidopsis, including subtelomeric regions.
To estimate the number of times that the sequence (CCCTAAA)4 is found at Arabidopsis telomeres, we assumed that Arabidopsis thaliana (Col-0) telomeres are composed of perfect telomeric repeats that spread about 3750 bp (21,22). We estimated that the five Arabidopsis chromosomes should contain about 5350 overlapping (CCCTAAA)4 sequences at telomeres [(3750/7) × 10]. Therefore, when the frequency of reads containing the sequence (CCCTAAA)4 is determined in input samples of Arabidopsis ChIP-seq experiments, only 2% of these reads should correspond to ITSs [(118 × 100)/(118 + 5350)]. In consequence, the frequency of the (CCCTAAA)4 sequence should essentially reveal telomeres in Arabidopsis ChIP-seq experiments. In the case of rice (Oryza sativa ssp. japonica cv. Nipponbare), the frequency of the (CCCTAAA)4 sequence should also reveal telomeres. The rice genome contains a similar number of (CCCTAAA)4 sequences at internal chromosomal loci than Arabidopsis (127), has more chromosomes (twelve) and similar telomeric length (23).
Determination of enrichment values
We used the Sequence Read Archive database at NCBI to study different epigenetic modifications in Arabidopsis and in rice telomeres. In Arabidopsis, we analyzed all the experiments from study SRP002100 (Gene Expression Omnibus accession number GSE28398). This ChIP-seq study was performed using aerial tissue (24). We determined the number of reads containing the (CCCTAAA)4 sequence and the number of reads containing the sequence TTGGCTTTGTATCTTCTAACAAG, which is a conserved region of the 178-bp satellite repeats present at centromeres. This sequence served as heterochromatic reference (12,15). Although a fraction of the 178-bp sequences associates with CENH3 chromatin, surrounding 178-bp repeats associate with H3 chromatin. The conserved sequence that we have selected spans from positions 56 to 79 of the repeats and do not contain motifs specifically associated with CENH3 chromatin (15). Thus, this sequence is present at the centromeric 178-bp repeats that associate with CENH3 chromatin and also at the 178-bp repeats that associate with H3 chromatin. It allowed us to analyze the chromatin organization of Arabidopsis 178-bp satellite repeats as an average, which is known to be heterochromatic (9–11,15,25).
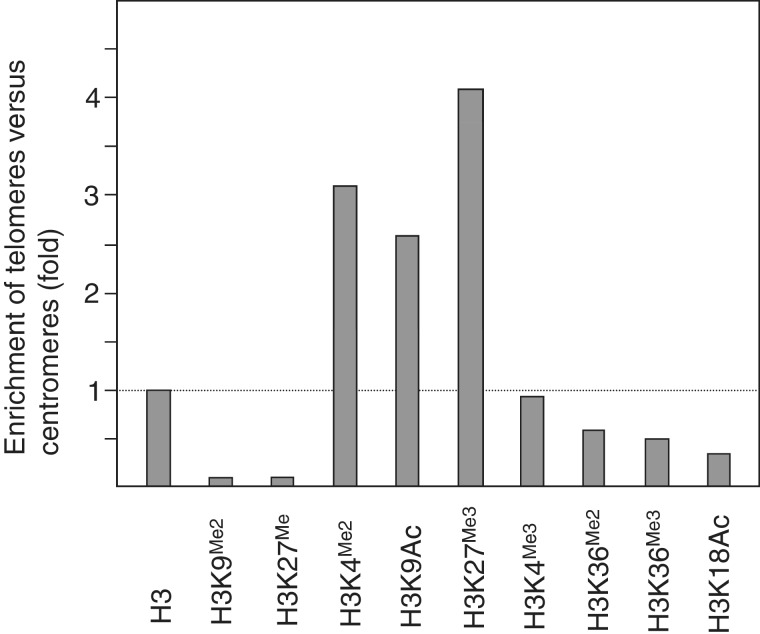
The study mentioned earlier in the text focused on nine epigenetic modifications named H3K4Me2, H3K4Me3, H3K9Me2, H3K27Me, H3K27Me3, H3K36Me2, H3K36Me3, H3K9Ac and H3K18Ac. The number of telomeric and centromeric reads corresponding to all these epigenetic marks were determined (Supplementary Table S1) and normalized against the input sample. Then, enrichment values of telomeres versus centromeres were calculated and normalized against histone H3 occupancy. The resulting values are represented in Figure 1. Similar results to those shown in Figure 1 were obtained when a different conserved sequence from the 178-bp satellite repeats was used to estimate relative enrichment values (data not shown). This sequence (CATATTTGACTCCAAAACACTAA) contains the dinucleotide TG at positions 160–161 of the 178-bp repeats and is not frequent in CENH3 chromatin (15).
Figure 1.
Epigenetic features at Arabidopsis telomeres. Different histone H3 epigenetic modifications were analyzed at telomeres and at centromeres using study SRP002100 (Gene Expression Omnibus accession number GSE28398). Relative enrichment values of telomeres versus centromeres were determined and represented as indicated in the ‘Materials and Methods’ section.
The enrichment values for H3K4Me3 and H3K9Ac were also calculated by analyzing experiments from a different ChIP-seq study (SRP002650; GSE22276), which was performed using Arabidopsis leaves (26). The enrichment values obtained for these two epigenetic marks were very similar to those shown in Figure 1: 0.7 for H3K4Me3 and 3.3 for H3K9Ac.
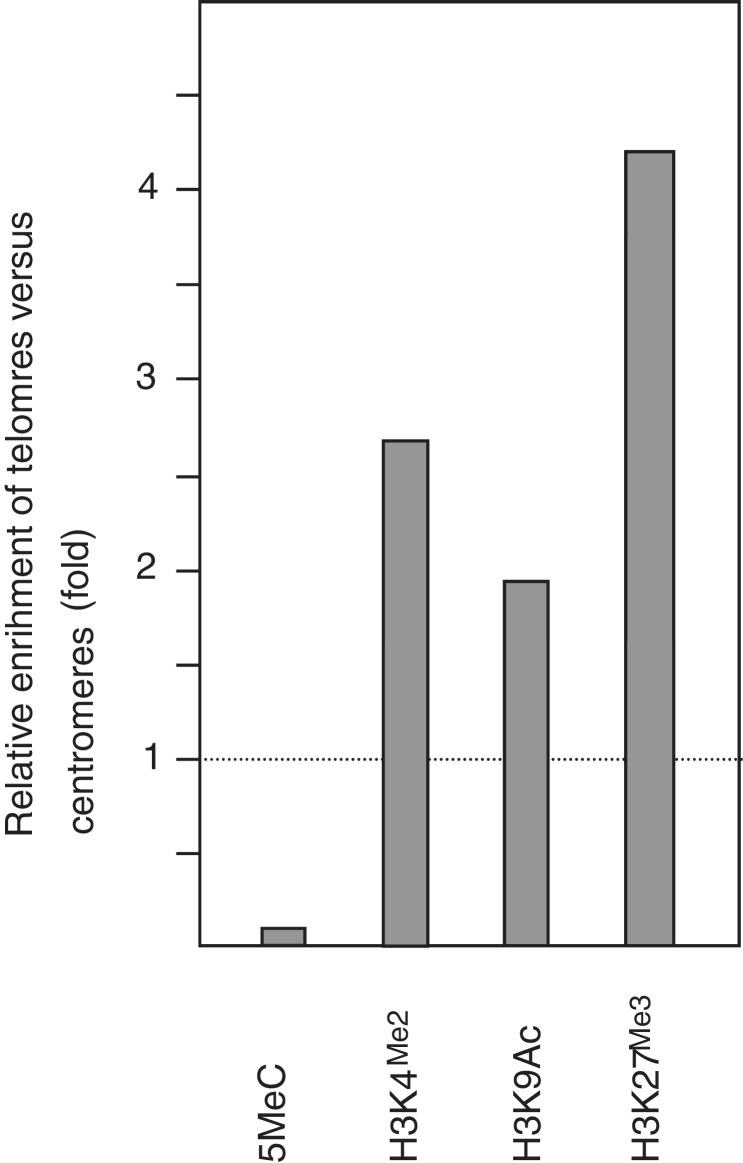
For rice (O. sativa ssp. japonica cv. Nipponbare), enrichment values were calculated by analyzing experiments SRX016118, SRX016122, SRX016126 and SRX016130 from study SRP001788 (GSE19602). These experiments were performed using four-leaf stage seedlings (27). The number of reads containing the (CCCTAAA)4 sequence and the number of reads containing the sequence CGTTCGTGGCAAAAACTCACTTCGT, which is part of the CentO-1 centromeric satellite repeat (positions 1–25) (28), were determined for four epigenetic modifications (H3K4Me3, H3K9Ac, H3K27Me3 and DNA methylation; see Supplementary Table S2). The CentO repeats, which are known to undergo DNA methylation similarly to the 178-bp repeats from Arabidopsis, served as heterochromatic reference (29–31). Since study SRP001788 did not include input samples, relative enrichment values were calculated. The number of reads corresponding to the telomeric sequence was divided by the number of CentO-1 reads for every epigenetic modification. Then, relative enrichment values were calculated by normalizing against the resulting value for DNA methylation. To obtain a graphic representation with similar values to those shown in Figure 1, all the relative enrichment values were divided by 10.
RESULTS
Epigenetic marks at Arabidopsis telomeres
To study the epigenetic modifications present at Arabidopsis telomeres, we analyzed ChIP-seq data. We determined the relative enrichment of telomeres versus centromeres, which served as heterochromatic reference. Previously, we had to find a specific sequence that represents telomeres but not ITSs in ChIP-seq analyses. Since Arabidopsis ITSs are mostly composed of very short stretches of perfect telomeric repeats interspersed with degenerated repeats (7,8,32,33), a short stretch of perfect telomeric repeats might essentially represent telomeres. Blast analyses of the Arabidopsis genome revealed that 98% of the (CCCTAAA)4 sequences are found at telomeres, whereas only 2% of these sequences localize at ITSs (see ‘Materials and Methods’ section). Therefore, the (CCCTAAA)4 sequence is essentially found at Arabidopsis telomeres. We chose this sequence to analyze telomeres in ChIP-seq experiments. For centromeres, we selected a conserved sequence from the 178-bp centromeric satellite repeats. It is well known that these repeats are heterochromatic and localize to chromocenters (9–13,15,25).
We analyzed ChIP-seq data from a study that focused on different histone H3 modifications in Arabidopsis. The epigenetic modifications analyzed in this study were two characteristic heterochromatic marks (H3K9Me2 and H3K27Me) and six marks associated with euchromatin (H3K4Me2, H3K4Me3, H3K36Me2, H3K36Me3, H3K9Ac and H3K18Ac). In addition, this study also focused on unlabeled histone H3 and on H3K27Me3, a repressive mark found in many euchromatic genes (17–19). We determined the enrichment of telomeres versus centromeres and found that the levels of unlabeled histone H3 were 1.6 times higher at telomeres (Supplementary Table S1). In all eukaryotic organisms analyzed, the nucleosomal spacing of telomeres is ∼20 bp shorter than the average bulk nucleosome organization (34). This important chromatin feature is probably related to the straight nature of telomeric DNA and might condition the epigenetic behavior of telomeric nucleosomes (32,35–38). The high density of histone H3 determined here at telomeres might reflect the short spacing of telomeric nucleosomes, which has been previously revealed by micrococcal nuclease digestion experiments. Alternatively, we cannot rule out other possibilities including a nucleosomal spacing of the nucleosomes associated with the 178-bp repeats longer than the average bulk nucleosome organization.
We found that the levels of heterochromatic marks analyzed (H3K9Me2 and H3K27Me) were lower at telomeres than at centromeres. On the contrary, the levels of some euchromatic marks were higher at telomeres (H3K4Me2 and H3K9Ac; Figure 1). These results indicate that Arabidopsis telomeres exhibit euchromatic features. However, not all the euchromatic marks analyzed were enriched in telomeres versus centromeres. That was the case for H3K4Me3, H3K36Me2, H3K36Me3 and H3K18Ac. The levels of these marks were similar or even lower at telomeres than at centromeres. Interestingly, Arabidopsis telomeres were also enriched in H3K27Me3 (Figure 1). Since we have previously found that telomeres in Arabidopsis are marked with H4K16Ac (14), we conclude that Arabidopsis telomeres are labeled with H3K4Me2, H3K9Ac, H3K16Ac and H3K27Me3.
Epigenetic marks at rice telomeres
To study whether some of the epigenetic marks that characterize Arabidopsis telomeres are present in other plant species, we analyzed a genome-wide study performed in rice where the levels of H3K4Me2, H3K9Ac, H3K27Me3 and DNA methylation were analyzed (27). Since the (CCCTAAA)4 sequences present at rice ITSs represent ∼1% of the total, these sequences also reveal telomeres in rice ChIP-seq experiments (see ‘Materials and Methods’ section). As heterochromatic reference, we analyzed a sequence from CenO, which is a satellite repeat present in rice centromeres. We found that the relative enrichment levels of telomeres versus centromeres were more than one order of magnitude higher for H3K4Me2, H3K9Ac and H3K27Me3 than for DNA methylation (Figure 2). These results strongly suggest that rice telomeres are also labeled with H3K4Me2, H3K9Ac and H3K27Me3. However, these results are still compatible with the existence of low levels of DNA methylation at rice telomeres, which can also be present at Arabidopsis telomeres (14).
Figure 2.
Epigenetic features at rice telomeres. Different histone epigenetic modifications were analyzed at telomeres and at centromeres using study SRP001788 (GSE19602). Relative enrichment values of telomeres versus centromeres were determined and represented as indicated in the ‘Materials and Methods’ section.
DISCUSSION
ChIP-seq data confirm that Arabidopsis telomeres exhibit euchromatic features
We have previously analyzed the chromatin structures of Arabidopsis telomeres and ITSs by ChIP, digestion with the restriction endonuclease Tru9I, which digested ITSs, and hybridization with a telomeric probe. These experiments revealed that, although subtelomeric regions and ITSs associate with heterochromatic marks, Arabidopsis telomeres exhibit euchromatic features (14). More specifically, we found that telomeres have lower levels of heterochromatic marks than ITSs (H3K9Me2, H3K27Me and DNA methylation) and higher levels of euchromatic marks (H3K4Me2, H3K9Ac and H4K16Ac). Here, we have shown by analyzing ChIP-seq data that telomeres have lower levels of heterochromatic marks than centromeres (H3K9Me2 and H3K27Me) and higher levels of some euchromatic marks (H3K4Me2 and H3K9Ac). Thus, these data confirm our previous results and validate the methodological approach presented here.
The experimental approach that we have used here to study the chromatin structure of Arabidopsis telomeres independently of ITSs could be applied to other eukaryotes. It will depend on how well genomes are assembled, including subtelomeric regions, and also on the structure of ITSs, including their length and number of perfect tandem repeats.
Arabidopsis telomeres are labeled with H3K27Me3
We have shown that Arabidopsis telomeres are labeled with H3K273Me, a repressive mark that usually do not colocalize with DNA methylation. Recently, ∼4400 genes have been found to be labeled with this epigenetic modification. These genes are often expressed in a tissue-specific manner and many of them might be involved in plant development. H3K27Me3 is established by the Polycomb Repressive Complex 2. This complex is conserved along evolution and is present in Arabidopsis, where its disruption causes serious developmental problems (18,19,39–42). It will be interesting to ascertain whether some of these problems are related to telomeres biology.
The histone code hypothesis involves a complex interplay of writing, reading and erasing activities that regulate the levels and the output of different epigenetic modifications (43). We have previously found that Arabidopsis telomeres are labeled with H3K4Me2, H3K9Ac and H4K16Ac. Here, we have confirmed these results and have also shown that Arabidopsis telomeres are labeled with H3K27Me3. The analysis of telomeres in mutants affected in the establishment, recognition and/or erasing of all these epigenetic marks open new avenues for future telomeric studies.
The chromatin organization of Arabidopsis telomeres shares similarities with the previously reported chromatin state 2
Four main chromatin states (CS1–CS4) have been recently defined in Arabidopsis based on the genome-wide distribution of 12 epigenetic marks analyzed by ChIP-on-chip (44–46). Since six of these marks were also analyzed here (H3K4Me2, H3K4Me3, H3K9Me2, H3K27Me, H3K27Me3 and H3K36Me3), we compared both studies and found that the chromatin organization of Arabidopsis telomeres is most similar to chromatin state 2 (CS2). This chromatin state is associated with 23% of Arabidopsis genes, being many of them repressed or lowly expressed. CS2 is characterized by the presence of H3K4Me2, H3K27Me2 and, above all, H3K27Me3. In addition, CS2 is also characterized by the absence of H3K4Me3, H3K9Me2, H3K9Me3, H3K27Me, H3K36Me3, H3K56Ac, H4K20Me, H2Bub and 5MeC (45).
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2.
FUNDING
The Spanish Ministry of Education and Science [grant BFU2008-02497/BMC] and by FEDER funds. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We want to thank the laboratories of Eric Lam, Jeffrey Chen and Xing-Wang Deng for making available their ChIP-seq data at the public databases. We are especially grateful to Eric Lam for allowing us to use their data prior to publication. We also thank NCBI for providing the tools that we have used to analyze the ChIP-seq data.
REFERENCES
- 1.Blackburn E. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 2.Cech T. Beginning to understand the end of the chromosome. Cell. 2004;43:405–413. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 3.Greider C. Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harb. Symp. Quant. Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- 4.Meyne J, Baker R, Hobart H, Hsu T, Ryder O, Ward O, Wiley J, Wurster-Hill D, Yates T, Moyzis R. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma. 1990;99:3–10. doi: 10.1007/BF01737283. [DOI] [PubMed] [Google Scholar]
- 5.Richards E, Goodman H, Ausubel F. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin K, Yan J. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutation Res. 2008;658:95–110. doi: 10.1016/j.mrrev.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Regad F, Lebas M, Lescure B. ITSs within the Arabidopsis thaliana genome. J. Mol. Biol. 1994;239:163–169. doi: 10.1006/jmbi.1994.1360. [DOI] [PubMed] [Google Scholar]
- 8.Uchida W, Matsunaga S, Sugiyama R, Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002;77:63–67. doi: 10.1266/ggs.77.63. [DOI] [PubMed] [Google Scholar]
- 9.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 10.Lindroth A, Cao X, Jackson J, Zilberman D, McCallum C, Henikoff S, Jacobsen S. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 11.Lindroth A, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Zapater J, Estelle M, Somerville C. A high repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 1986;204:417–423. [Google Scholar]
- 13.Nagaki K, Talbert P, Zhong C, Dawe R, Henikoff S, Jiang J. Chromatin immunoprecipitation reveals that the 180-bp satelite repeat is the key functional element of Arabidopsis thaliana centromeres. Genetics. 2003;163:1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaquero-Sedas M, Gámez-Arjona F, Vega-Palas M. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2010;39:2007–2017. doi: 10.1093/nar/gkq1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, Lee H, Koo D, Jiang J. Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns-from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Fischer A, Hofmann I, Naumann K, Reuter G. Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis. J. Plant Physiol. 2006;163:358–368. doi: 10.1016/j.jplph.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Johnson L, Mollah S, Garcia B, Muratore T, Shabanowitz J, Hunt D, Jacobsen S. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Clarenz O, Cokus S, Bernatavichute Y, Pellegrini M, Goodrich J, Jacobsen S. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaquero-Sedas M, Vega-Palas M. On the chromatin structure of eukaryotic telomeres. Epigenetics. 2011;6:1055–1058. doi: 10.4161/epi.6.9.16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards E, Ausubel F. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 22.Shakirov E, Shippen D. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohmido N, Kijima K, Ashikawa I, de Jong J, Fukui K. Visualization of the terminal structure of rice chromosomes 6 and 12 with multicolor FISH to chromosomes and extended DNA fibers. Plant Mol. Biol. 2001;47:413–421. doi: 10.1023/a:1011632111845. [DOI] [PubMed] [Google Scholar]
- 24.Luo C, Sidote D, Zhang Y, Kerstetter R, Michael T, Lam E. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for Natural Antisense Transcript production. (under review) 2012 doi: 10.1111/tpj.12017. [DOI] [PubMed] [Google Scholar]
- 25.Fransz P, Armstrong S, Alonso-Blanco C, Fischer T, Torres-Ruiz R, Jones G. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 1998;13:867–876. doi: 10.1046/j.1365-313x.1998.00086.x. [DOI] [PubMed] [Google Scholar]
- 26.Ha M, Ng D, Li W, Chen Z. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 2011;21:590–598. doi: 10.1101/gr.116467.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He G, Zhu X, Elling A, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Z, Dong F, Langdon T, Ouyang S, Buell C, Gu M, Blattner F, Jiang J. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell. 2002;14:1691–1704. doi: 10.1105/tpc.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H, Neumann P, Macas J, Jiang J. Transcription and evolutionary dynamics of the centromeric satellite repeat CentO in rice. Mol. Biol. Evol. 2006;23:2505–2520. doi: 10.1093/molbev/msl127. [DOI] [PubMed] [Google Scholar]
- 30.Yan H, Kikuchi S, Neumann P, Zhang W, Wu Y, Chen F, Jiang J. Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 2010;63:353–365. doi: 10.1111/j.1365-313X.2010.04246.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Yi C, Bao W, Liu B, Cui J, Yu H, Cao X, Gu M, Liu M, Cheng Z. The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 2005;139:306–315. doi: 10.1104/pp.105.064147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gámez-Arjona F, López-López C, Vaquero-Sedas M, Vega-Palas M. On the organization of the nucleosomes associated with telomeric sequences. Biochim. Biophys. Acta. 2010;1803:1058–1061. doi: 10.1016/j.bbamcr.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 33.Vannier J, Depeiges A, White C, Gallego M. ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000380. doi: 10.1371/journal.pgen.1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pisano S, Galati A, Cacchione S. Telomeric nucleosomes: Forgotten players at chromosome ends. Cell. Mol. Life Sci. 2008;65:3553–3563. doi: 10.1007/s00018-008-8307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fajkus J, KovariK A, Kralovics R, Bezdek M. Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 1995;247:633–638. doi: 10.1007/BF00290355. [DOI] [PubMed] [Google Scholar]
- 36.Mechelli R, Anselmi C, Cacchione S, De Santis P, Savino M. Organization of telomeric nucleosomes: atomic force microscopy imaging and theoretical modeling. FEBS Lett. 2004;566:131–135. doi: 10.1016/j.febslet.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 37.Pisano S, Pascucci E, Cacchione S, De Santis P, Savino M. AFM imaging and theoretical modeling studies of sequence-dependent nucleosome positioning. Biophys. Chem. 2006;124:81–89. doi: 10.1016/j.bpc.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Pisano S, Marchioni E, Galati A, Mechelli R, Savino M, Cacchione S. Telomeric nucleosomes are intrinsically mobile. J. Mol. Biol. 2007;369:1153–1162. doi: 10.1016/j.jmb.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 39.Feng S, Jacobsen S. Epigenetic modifications in plants: an evolutionary perspective. Curr. Opin. Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Zhang K, Sridhar V, Zhu J, Kapoor A, Zhu J. Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS One. 2007;2:e1210. doi: 10.1371/journal.pone.0001210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardner K, Allis C, Strahl B. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turck F, Roudier F, Farrona S, Martin-Magniette M, Guillaume E, Buisine N, Gagnot S, Martienssen R, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughn M, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz P, Dedhia N, McCombie W, Agier N, Bulski A. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]