Abstract
Morpholino oligomers (MOs) have been widely used to knock down specific genes in zebrafish, but their constitutive activities limit their experimental applications for studying a gene with multiple functions or within a gene network. We report herein a new design and synthesis of caged circular MOs (caged cMOs) with two ends linked by a photocleavable moiety. These caged cMOs were successfully used to photomodulate β-catenin-2 and no tail expression in zebrafish embryos.
INTRODUCTION
Manipulation of a specific gene’s function without cross-effects will open exciting opportunities for the study of gene networks. Among many methods, light has stood out as an excellent non-invasive external trigger due to high spatiotemporal resolution. Light-activated (also ‘caged’) molecules have been used for controlling biological processes for a few decades (1). However, only during recent years have they been widely used in more complex biological applications, such as gene regulation. Photocaged moieties can be used to temporarily ‘cage’ bioactive molecules whose activities can be restored by light irradiation which removes the caging groups. This method has been used to control gene expression in cells or animals (2–17), and using caged biomolecules, especially, nucleic acids, is one of the most important ways of achieving this control. Photoregulation of antisense oligonucleotides, antigens, siRNAs, microRNAs, DNAzymes and RNAzymes has been developed for the specific interaction with target genes (18–53). However, it is still challenging to effectively and readily switch gene expression ‘on’ and/or ‘off’ and provide high spatiotemporal resolution with caged nucleic acids.
For specific photomodulation of gene expression in zebrafish, several methods have been reported to cage nucleic acids and study their ability to photomodulate gene functions. One method is based on statistical caging strategy with the attachment of multiple caging moieties to the nucleobases or the backbone of phosphate groups. Ando et al. caged a green fluorescent protein (GFP) mRNA and a DNA plasmid through non-specific labeling of the backbone phosphates approximately once every 35 bases with coumarin-based photolabile protecting groups. These caged mRNA and DNA constructs enabled the photoregulation of GFP expression in zebrafish and subsequent study of the role of lhx2 in zebrafish forebrain growth (49,52). More recently, Deiters et al. reported morpholino oligomers (MOs) with multiple photocaged monomeric building blocks and demonstrated the utility of these caged MOs in light-activated control of gene function in zebrafish embryos (26). For the introduction of a single caging moiety, Tang et al. (37) applied a heterobifunctional linker to link an antisense strand to a partially complementary sense strand and succeeded in photomodulating gene expression of chordin and bozozok in zebrafish embryos. Based on the same method, caged MOs were reported to silence the no tail (ntl) gene by Chen (18,30,38). The rational design of caged MOs showed that caged MOs could be modeled as a competitive thermodynamic equilibrium among caged MO, uncaged MO and uncaged MO/mRNA in in vivo environments. More recently, Mayer et al. showed their ‘Photomorph’ to silence GFP expression by using a caged sense RNA to temporarily block antisense MO until light-induced strand break of the sense RNA to release the antisense MO (27).
Caged nucleic acids with multiple caging moieties sterically inhibited the interaction with their targets. However, incomplete uncaging was unavoidable with limited intensity and duration of light irradiation. The employment of a single caging group led to more complete uncaging, but the relative thermostability of the caged and uncaged oligonucleotides, antisense strand/target duplex was difficult to balance, especially, in vivo study. The number of paired bases, the positions of paired bases (blunt or staggered), the length of photolinkers, and many other factors all had effect on the photomodulation of gene regulation in zebrafish (30). Uncaged antisense oligodeoxynucleotides (ODNs) were not quickly and fully accessible to mRNA binding, and optimized candidates in vitro might not be the best one in vivo due to complicated environments in biological systems (54). Currently there is still a need for the design of new nucleic acid tools for gene regulation in vivo.
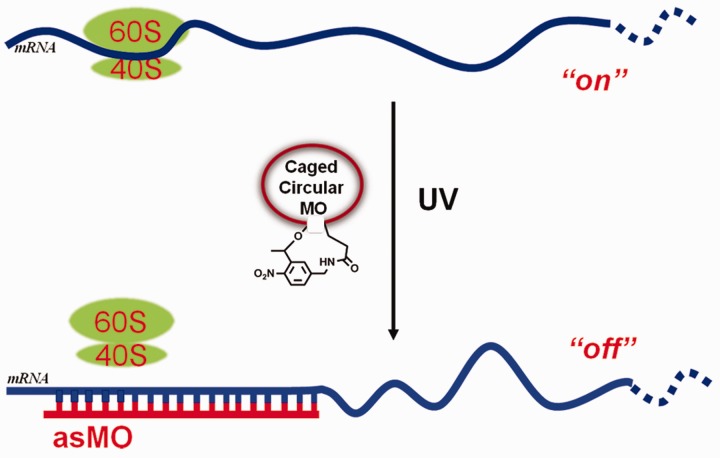
A circular ODN may be able to overcome current shortages of caged ODNs with the need of multiple caging groups or a thermodynamic equilibrium due to the application of only a single photolabile linker and the absence of a sense strand. Several photolabile circular oligonucleotides have been reported recently. Richards et al. synthesized a photolabile circular DNAzyme enzymatically using T4 ligase and realized photomodulation of RNA digestion (29). Previously, we also succeeded in photomodulating RNA digestion by RNase H through the synthesis of light-activated circular antisense ODNs (24). We sought to apply this circularization strategy to cage MOs for photomodulation of gene expression in zebrafish and expect caged circular MOs (caged cMOs) may be structurally unavailable to stably bind target mRNA [while our manuscript is required for resubmission to include photomodulation of a second gene, a related cMO was recently reported for gene photomodulation in zebrafish online 11 June 2012 (55)]. Here we report this new method to cage antisense MOs (MO-cat2, MO-ntl) by linking two ends of linear MOs with a heterobifunctional photocleavable linker (PL) and study their photomodulation of β-catenin-2 and ntl expression in zebrafish embryos. The circularization of the MOs temporarily restricts the opening of MO ring until irradiation, which provides the photocontrol of duplex formation between MO and mRNA. Once light-activation cleaves the chemical bond of PL, uncaged MOs will quickly and completely interact with their target mRNAs without the need for the competition with their sense strands, and the photomodulation of gene regulation will then be readily achieved, as shown in Figure 1.
Figure 1.
Controlling gene expression using a caged cMO. Photolysis promotes the uncaging of caged cMO and its subsequent binding to mRNA.
MATERIALS AND METHODS
General methods
Commercially available MOs with a 5′-end amine on resin and a 3′-end amine (Fmoc protected) were purchased from Gene-Tools, LLC. All oligodeoxynucleotides were purchased from Sangon Biotech (Shanghai) Co. Ltd. The concentrations of all oligodeoxynucleotides and MOs were measured in water at 260 nm using a Beckman DU 800 Nucleic Acid/ Protein Analyzer. All high-performance liquid chromatography (HPLC) was performed on a Varian ProStar 210 equipped with Agilent Zorbax Oligo column or DNAPac PA-200 ion-exchange HPLC column (Dionex, 4 × 250 mm). All mass spectrometric data were obtained using Waters ESI or Shimadzu AXIMA-CFR Plus MALDI-TOF-MS. Zebrafish embryos were injected using microinjector (Harvard Apparatus, PLI-100). All photoirradiation experiments of zebrafish embryos were carried out with UV lamp (365 nm, 11 mW/cm2).
Synthesis of the PL (Supplementary Scheme S1)
An amount of 0.2 g 1-[5-(aminomethyl)-2-nitrophenyl]ethanol (56) was reacted with ethyl trifluoacetate (1.2 ml, 10 eq.) in 10 ml ethanol with 0.5 ml triethylamine (TEA) at room temperature overnight. The reaction mixture was concentrated to dryness, and the intermediate was used in the next step without further purification. The dry residue was dissolved in 10 ml acetonitrile, followed by the addition of 0.39 g (1.5 eq.) N,N′-Disuccinimidyl carbonate (DSC) and 0.42 ml (3 eq.) dry TEA. The mixture was then stirred at room temperature for 6 h. After removal of acetonitrile, the residue was dissolved in methylene chloride, washed with saturated sodium chloride aqueous solution, and dried with anhydrous Na2SO4. The product was isolated using silica gel-column chromatography with ethyl acetate and petroleum ether. The isolated yield was 65% for two steps. 1H-NMR (400 M, CDCl3), δ = 1.79 (d, 3H), 2.79 (s, 4H), 4.55–4.80 (m, 2H), 6.32 (m, 1H), 7.48 (d, 1H), 7.62 (s, 1H), 7.83 (s, 1H), 8.07 (d, 1H) 13C-NMR (100 M, CDCl3) 21.5, 25.3, 42.9, 76.45, 125.4, 125.9, 129.0, 137.4, 143.6, 146.5, 150.6, 157.4, 157.8, 169.0 Mass: calculated 433.07, measured: 456.06 (M+Na)+, 275.06 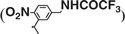 .
.
Synthesis and purification of caged cMOs
To remove Fmoc groups from the protected 3′-amine end of the commercially available MOs, MOs on resin were treated with 20% piperidine in 1,3-Dimethyl-2-Imidazolidinone (DMI) twice and vortexed for 2 min each time, followed by washing with DMI three times and then acetone twice. The obtained resin was suspended in 100 μl DMI solution with PL (1.0 mg, 1.5 μmol) and TEA (1 μL, 6.9 μmol) and vortexed for 24 h. The resin was then washed with DMI three times and acetone twice. The trifluoacetate protecting group of PL was removed by soaking the resin in cold diluted ammonium hydroxide (0.1 M) for 1 h, followed by washing the resin with water and acetonitrile. Succinic anhydride, N-Hydroxy benzotrizole (HOBt) and TEA in DMI were then added to the above dried resin and the mixture was incubated at room temperature for 4 h. The resin was then thoroughly washed with DMI and acetone.
MO was cleaved from resin and deprotected using 0.3 ml of concentrated ammonium hydroxide at 40°C for 24 h. The solution was chilled for 10 min and centrifuged at 2500 rpm for 5 min. The supernatant was collected, followed by the addition of 10 ml acetone. Precipitated white solid was collected by centrifuging 5 min and further washed with 5 ml acetonitrile. The white solid was dissolved in 1 ml water and the concentration was determined by the measurement of absorbance at 260 nm using a Beckman DU 800 Nucleic Acid/ Protein Analyzer.
Roughly 10 nmol MO after cleavage and deprotection was dissolved in 2 ml 0.1 M TEM buffer with 0.3 M NaCl and 5 mM HOBt. An amount of 1 mg 1-Ethyl-3 -(3-dimethylaminopropyl)carbodiimide hydrochloride (EDAC) was added to above solution with vortexing. The mixture was shaken at room temperature overnight. To remove the salts, small molecules and small fragments of MO, the reaction solution was passed through a NAP-10 column or was dialyzed against 10% ethanol in water using molecular porous membrane tubing (Spectra/Por Dialysis Membrane, MWCO 3500 from Spectrum, Houston, TX, USA). Concentrated phosphate buffer and acetonitrile were added in accordance with phase A of HPLC purification buffers (20 mM, pH 7.2, 10% acetonitrile). The 2-fold complementary ODN was then added to the above MO solution for further ion-exchange HPLC purification.
For purification of the caged cMO-cat2, Agilent ion-exchange column (Zorbax Oligo) was used. The running conditions for HPLC purification: Phase A, 20 mM phosphate buffer; B, A + 2 M sodium chloride. The following step-wise gradient was used with flow rate of 1 ml/min at 40°C: B, 0% in 10 min, 0–30% in 30 min. The column temperature was maintained at 40°C. The caged cMO was collected in the first 10 min. For purification of the caged cMO-ntl, Dionex Ion-exchange column (DNAPac PA-200) was used. The running conditions for HPLC purification: Phase A, 20 mM phosphate buffer with 10% of acetonitrile; B, A + 1 M sodium chloride. The following step-wise gradient was used with flow rate of 0.9 ml/min at 40°C: B, 0% in 8 min, B, 60–90% in 10 min, B, 90% for 3 min, 0% for 4 min.
Caged cMOs were collected in first 10 min. The unreacted linear MOs formed stable duplexes with their complementary ODN strands and the duplexes moved much more slowly through ion-exchange column due to negative charges of the complementary ODNs. After removing water using a SpeedVac Concentrator, the residue was desalted and concentrated using Millipore-Amicon Ultra-0.5 ml Centrifugal Filters (Cutoff = 3000). The aqueous residue was then freeze-dried to give final white power. The collected coupling yields of caged cMOs were 20–50% of cleaved MO mixtures after coupling, purification and desalting.
MOs were characterized with ESI–MS (1% TFA in CH3OH/H2O 5/95) or MALDI–TOF–MS. Samples were analyzed using a saturated solution of sinapinic acid in 50% acetonitrile/water as a matrix for MALDI measurement. Samples were analyzed according to manufacturer’s manual. MO-cat2: before cyclization, 9036.0; after cyclization, 9020.0; MO-ntl: before cyclization, 9117.7; after cyclization, 9099.5.
Fluorescence measurement of molecular beacon with caged cMO-ntl
A complementary ODN molecular beacon (MB-30: 5′-6-Fam-ATCGGAAATATGCCTGCCTCAAGTCCCGAT-Dabcyl-3′, HPLC purified) was used for the fluorescence binding assay experiment. The concentrations of stock solutions of MB-30, native MO-ntl, caged cMO-ntl in ultra-pure water were determined by the absorbance at 260 nm.). The final concentration in 1× PBS buffer (SIGMA) of molecular beacon was 100 nM, while the concentrations of linear MO-ntl and caged cMO-ntl were 200 nM. To prepare uncaged MO-ntl solution, caged ciruclar MO-ntl solution was irradiated using a UV lamp (365 nm, 11 mW/cm2) for 10 min. After a short vortex, the above solutions were incubated for 5 min at 37°C. Fluorescence spectra were measured immediately with a Cary Eclipse fluorescence spectrophotometer with the excitation wavelength at 488 nm at 37°C
Gel-shift assay for evaluation of MO/target binding
To determine the binding of a complementary oligodeoxynucleotide to caged cMO-ntl before and after light irradiation, gel mobility shift assays were performed. An amount of 100 nM ODN-38 (5′-ACATTCGATCGGAAATATGCCTGCCTCAAGTCTGTACT-3′) was added to 200 nM linear, caged circular and uncaged MOs in 20 mM phosphate buffer (pH 7.2). The ODN/MO mixtures were annealed by incubation at 37°C for 30 min and then were put into ice water. One microliter DNA loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol FF, 30% glycerol in water) was then added to above solutions. All solutions were loaded into 15% native polyacrylamide gels. The gels were then electrophoresed at 100 V for 2 h using 1× TBE buffer (pH 8.2). Gels were exposed and incubated in 1× SYBRR Gold nucleic acid gel stain solution for 20 min. The images were acquired by Bio-rad ChemiDoc XRS system.
Zebrafish embryo injection and phenotype observation
Embryos bred from wild-type zebrafish were collected at one-cell stage. Linear and caged cMOs were each injected into zebrafish embryos (1 mM, 5 nl for MO-cat2 and 25 μM, 5 nl for MO-ntl) at one- or two-cell stage. The embryos were then put into an incubator at 29°C. At 3 hpf, half of embryos injected with caged cMO were irradiated with 365-nm light (11 mW/cm2) for 10 min. The irradiated embryos were then put back to the incubator. At 24 hours post-fertilization (hpf), zebrafish phenotypes were observed. The embryos were kept in the incubator for another 7 days and the survived zebrafish was imaged again.
RESULTS AND DISCUSSION
Synthesis and purification of caged cMOs
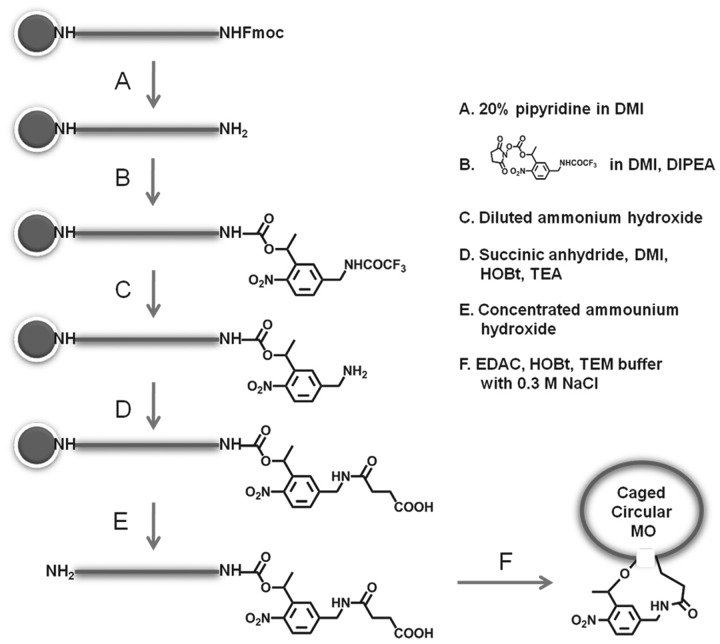
Synthesis of caged cMOs started with MOs on resin, as shown in Figure 2. In order to introduce primary amine and carboxylic acid at two ends of MOs, the custom-synthesized MOs with Fmoc-protected amine at 3′-end and 5′-amine preloaded solid resin were purchased from Gene Tools. To simplify tough purification steps of modified intermediate MOs, the addition of PL and introduction of acid moiety were carried out on solid resin consecutively. Then, the obtained MOs were deprotected and cleaved from the resins and were circularized through reaction of their 5′-amine and 3′-acid groups
Figure 2.
Synthetic route of the caged cMOs.
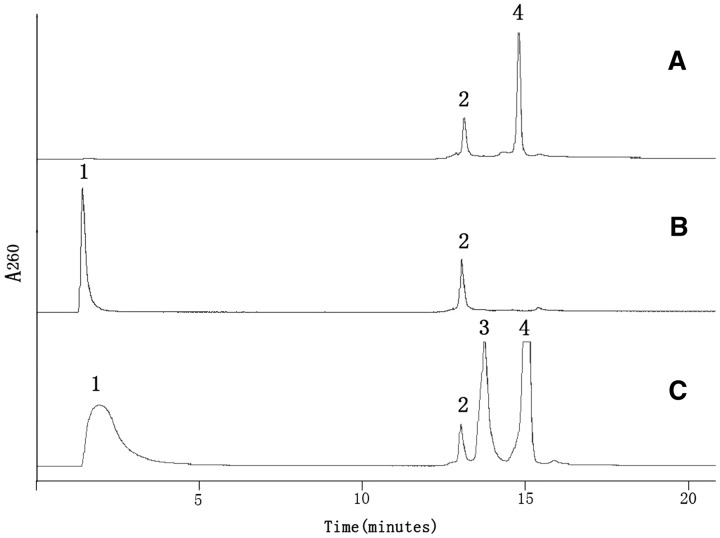
Further separation of caged cMOs from the components of the reaction mixtures, especially, the linear MO, is challenging. Usually 0.1 M sodium hydroxide is used to deprotonize nucleobases to charge MO for the purification by ion-exchange HPLC as in Chen’s MOs (38). However, these harsh conditions are not favorable for HPLC purification of the caged cMOs. We developed a new method to purify a caged cMO by mixing with a complementary ODN. When the complementary ODN binds to unreacted linear MO to form a duplex, it will move through ion-exchange column much more slowly than the uncharged caged cMO itself. As shown in Figure 3, the caged cMO-ntl had a retention time of <5 min, while the duplex of linear MO-ntl and the complementary ODN-38 had a much slower retention time of 14 min, followed by the peak of ODN-38 with a retention time of 15 min on HPLC traces.
Figure 3.
HPLC traces of caged cMO-ntl separation. (A) The complementary oligodeoxynucleotide (ODN-38) to MO-ntl, (B) The linear MO-ntl, (C) Cyclization reaction mixtures with 2-fold amount of its complementary ODN-38. Peak 1 is pure MO, peak 2 is not related to any oligo, but due to the change of high salt buffer, peak 3 is the duplex of linear MO-ntl and its complementary ODN-38, and peak 4 is the complementary ODN-38 of MO-ntl only.
Interaction of caged cMOs with their complementary oligodeoxynucleotides
Binding ability of a complementary ODN to the caged cMO is the key point to determine whether the caged cMO is able to effectively photomodulate gene expression. As shown in Supplementary Figure S1, when 2-fold of 25-mer sense ODN (ODN-25: 5′-GTTTGGAAGTCGCTCAGGCTAAAG G-3′) which was fully complementary to MO-cat2 was used, it still bound to caged cMO-cat2 and moved through ion-exchange column slowly. This result showed that 25-mer complementary ODN was not long enough for caged cMO-cat2 to be completely immune to target binding. This phenomenon was also observed in our previous study (24). However, a 30-mer ODN (ODN-30: 5′-CCTTTGTTTGGAAG TCGCTCAGGCTAAAGG-3′) was designed. ODN-30 has hairpin structure with 6-nt base pairs (2CG, 3AT and 1GT) which leads to its immunity to the binding of caged cMO-cat2. This observation enables caged cMOs to be used for photoregulating their binding with much longer and structural target mRNA. This property is also used for the purification of caged cMOs.
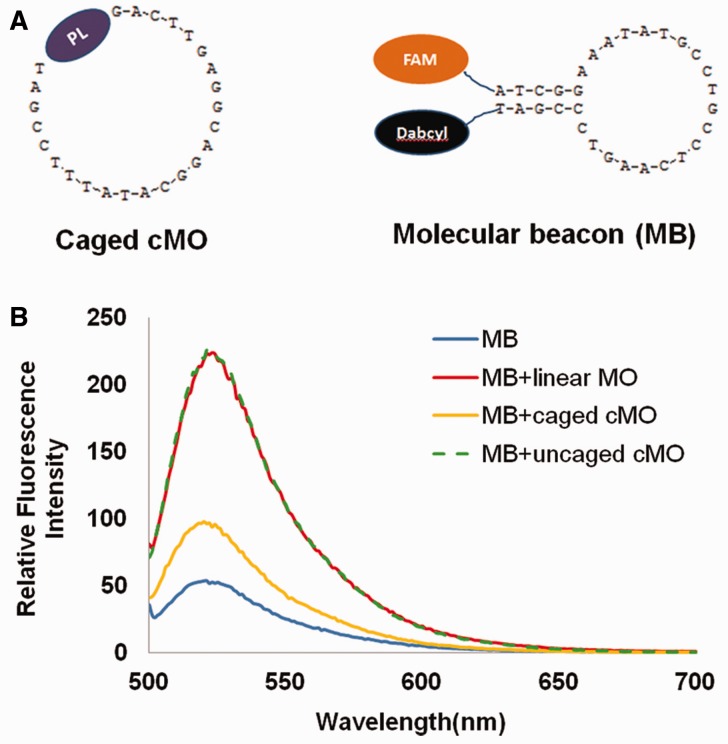
To further quantify the interaction of a caged cMO with its complementary ODN, MB-30 was designed for its binding with MO-ntl by assaying increase in fluorescence intensity when native MO-ntl, caged cMO-ntl or uncaged MO-ntl was present in solutions. As shown in Figure 4, fluorescence intensity from beacon solution slightly increased in the presence of caged cMO-ntl. However, fluorescence intensity reached up to 4.1-fold upon the addition of uncaged MO-ntl, which is almost the same fluorescence intensity with the addition of the same concentration of native MO-ntl. These results were also confirmed with gel-shift assay. Supplementary Figure S2 also shows that caged cMO cannot stably bind its target sequence while photoactivation can fully recover the binding of MO with its target sequence.
Figure 4.
(A) Structures of caged cMO-ntl and its complementary molecular beacon (MB) with FAM and Dabcyl; (B) Relative fluorescence intensity of MB, MB + linear MO-ntl, MB + caged cMO-ntl and MB + caged cMO-ntl + 10 min irradiation at a concentration of 100 nM MB in PBS buffer at 37°C, λex = 488 nm.
Photoregulation of β-catenin-2 expression in zebrafish embryos using caged cMO-cat2
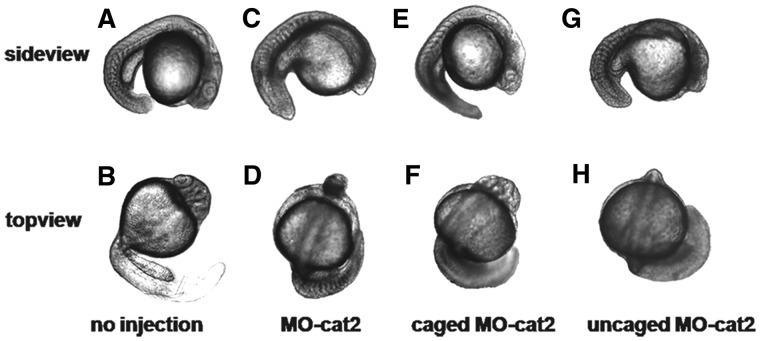
The morpholino sequence (MO-cat2, 5′-CCTTTAGCCTGAGCGACTTCCAAAC-3′) for targeting β-catenin-2 was first used as a model to show photomodulation of β-catenin-2 expression in zebrafish embryos. β-catenin-2 is essential for the development of dorsal axial structures, and gene knockdown causes the pin head and trunk patterning defects. This MO sequence has been proven effective in silencing β-catenin-2 expression and resulted in the ventralized phenotypes including the loss of notochord in zebrafish (57). Embryos bred from wild-type zebrafish were used for this study. Light irradiation (365 nm, 11 mW/cm2, 10 min) did not cause adverse effects on early embryo development. Embryos with 10 min irradiation and no injection developed normally as shown in Figure 5A and B for the sideview and topview. As a positive control, we injected zebrafish embryos with 1 mM linear MO-cat2 (total embryos, n = 80) and these embryos developed clear morphogenesis of missing β-catenin-2 at 24 hpf as shown in Figure 5C and D. For embryos injected with 1 mM caged cMO-cat2 (total embryos, n = 204) targeting β-catenin-2 at one-cell or two-cell stage, half of the embryos were subject to global irradiation after 3 h post-fertilization (hpf), and the other half were still covered with aluminum foil to avoid light irradiation. As we expected, 91% embryos with caged cMO-cat2 injection had the same normal development as the uninjected ones if the embryos were kept in dark (Figure 5E and F). However, light activation triggered uncaging of the caged cMO-cat2 and promoted its binding to target β-catenin-2 mRNA. Of irradiated embryos, 93% showed block-shaped somites and/or pin head (Figure 5G and H). These phenotypes were identical to the embryos injected with the linear MO-cat2. Light irradiation by itself under our conditions did not show any toxic effects on the development of zebrafish embryos, so the phenotypes can be attributed to the uncaged MO-cat2.
Figure 5.
Transmitted light images of representative 24-hpf zebrafish embryos with side view and top view. (A, B) Uninjected embryos irradiated for 10 min developed normally. (C, D) Embryos with 1 mM linear MO-cat2 injection exhibited clear β-catenin-2 defective phenotype. (E, F) Injection with 1 mM caged cMO-cat2 had no effect in the dark. (G, H) Injection with 1 mM caged cMO-cat2 and 10-min UV-irradiation at 3 hpf gave β-catenin-2 defective phenotype.
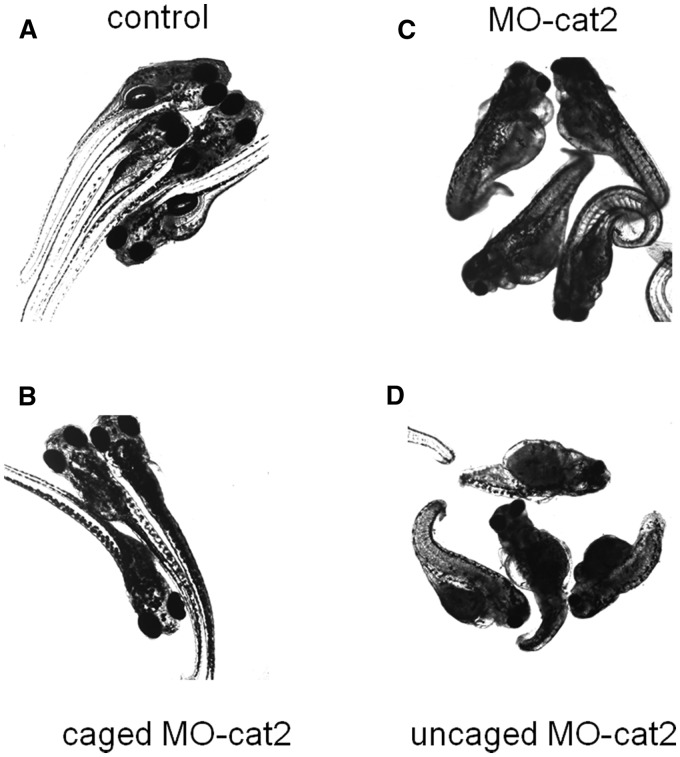
To further verify long-term effect of caged cMO and uncaged MO on zebrafish development, embryos were kept in an incubator for another 7 days and were imaged again. Without injection, the fish showed exactly normal development. Whereas injection with the linear MO-cat2 caused zebrafish to show narrow head and/or much shorter tail (loss of notochord) which were the typical phenotypes for β-catenin-2 defective mutants. Zebrafish embryos injected with the caged cMO-cat2 at one-cell or two-cell stage showed the same normal development as the uninjected embryos if no light was applied. However, embryos injected with the caged cMO-cat2 and irradiated with 365-nm light showed the clear phenotypes which were similar to the phenotypes of embryos injected with the linear MO-cat2, as shown in Figure 6. These results indicate embryos injected with caged cMO and uncaged MO maintained their phenotypes for quite long time.
Figure 6.
Transmitted light images of representative 8-day zebrafish embryos. (A) Uninjected embryos UV-irradiated at 3 hpf for 10 min developed normally. (B) Injection with 1 mM caged cMO-cat2 had no effect on embryos in the dark. (C) Embryos injected with 1 mM the linear MO-cat2 exhibited a clear β-catenin-2 defective phenotype. (D) Injection with 1 mM caged cMO-cat2 and 10-min UV-irradiation at 3 hpf gave no-β-catenin-2 phenotype of embryos.
Photoregulation of ntl gene expression in zebrafish embryos using caged cMO-ntl
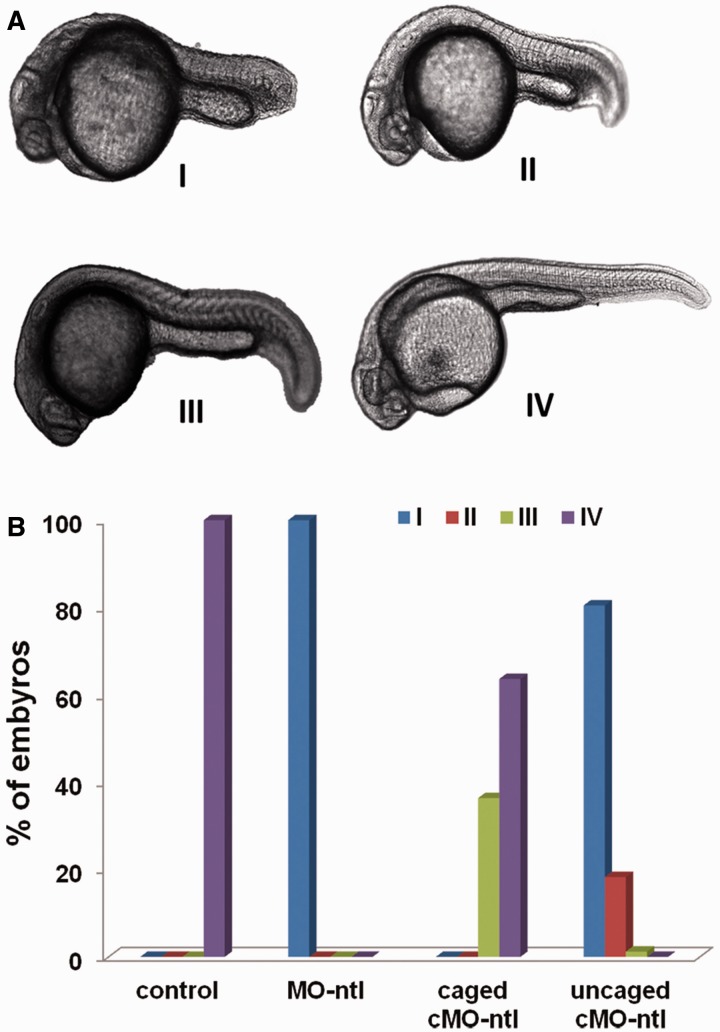
To test the specificity of a caged cMO, we targeted a second gene, ntl, which plays an important role in nodal-independent tail somite formation. The sequence (MO-ntl, 5′-GACTGAGCAGCAATTCCGT-3′) has been proved effective in knockdown of ntl gene expression and resulted in the obvious ntl phenotypes with missing notochord cells. Zebrafish embryos are sensitive to the dose of MO-ntl which causes different phenotypes that can be categorized into four classes according to their severity (Figure 7A). Light irradiation by itself did not cause the ntl phenotype under our experimental conditions. However, when embryos were injected with linear MO-ntl (125 fmol/embryo), all the embryos (total embryos, n = 72) developed into class I phenotype as shown in Supplementary Figure S4 and Figure 7B. Embryos injected with the same amount of caged cMO-ntl (n= 170) was divided into two dishes. One dish was subject to global light irradiation at 3 hpf, while the other one was still covered with aluminum foil to avoid light irradiation. As seen in Figure 7B, all the embryos with caged cMO-ntl injection and 10-min irradiation show obvious ntl phenotypes predominantly in class I and class II. While the majority of these injected most embryos covered with aluminum foil developed into the same normal phenotypes class IV as the uninjected embryos (Figure 7B). Some class III phenotype embryos were also observed. This may be due to light exposure of caged cMO-ntl during time period of MO injection. Since zebrafish embryos are sensitive to this MO-ntl sequence, even a small amount of uncaged MO-ntl could lead to the observation of a weak ntl phenotype as indicated in Figure 7B.
Figure 7.
(A) Transmitted light images of representative zebrafish embryos with different class of ntl phenotypes. (B) Phenotypic distributions for embryos injected with the indicated linear MO-ntl or caged cMO-ntl and incubated in dark or irradiated with UV light at 3 hpf.
CONCLUSION
We designed and synthesized a new kind of caged MOs with the circular structure. The synthesized caged cMOs were purified with ion-exchange HPLC through a complementary ODN-assisted method. The binding ability of caged cMOs and their target sequences was further characterized with HPLC, fluorescence emission and native PAGE gel, which indicated caged cMOs are immune to target binding. However, they are immediately and fully accessible to target binding upon light activation, which overcomes the previous shortages due to multiple caging groups or relative thermostability of a three-component system. By injecting caged cMO-cat2 and MO-ntl into zebrafish embryos, gene expression was efficiently photomodulated, and clear phenotypes of missing β-catenin-2 and ntl gene expression were observed upon light activation during development of zebrafish embryos in both 24 h and several days after fertilization. This new kind of caged cMOs will be promising for the practical applications to spatiotemporal photoregulation of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Scheme S1.
FUNDING
The National Science Foundation of China (NSFC) [21072015]; Program for New Century Excellent Talents in University [NCET-10-0203] and the National Basic Research Program of China [‘973’ Program; 2012CB720600]. Funding for open access charge: NSFC 21072015 and ‘973’ Program 2012CB720600.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Bo Zhang (Center of Developmental Biology and Genetics, Peking University) for kindly providing zebrafish embryos and instruments for microinjection and imaging, etc.), and Prof. Vasudevan Ramesh (University of Manchester, UK) and Dr Julia L. Richards for proofreading this article.
REFERENCES
- 1.Kaplan JH, Forbush B, Hoffman JF. Rapid photolytic release of adenosine 5'-triphosphate from a protected analogue: utilization by the Na:K pump of human red blood cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 2.Su M, Yang F, Yu L, Lv C, Gu X, Wang J, Li Z, Tang X. Photoreponsive nucleic acids for gene regulation. J. Chin. Pharma. Sci. 2010;19:5–14. [Google Scholar]
- 3.Deiters A. Light activation as a method of regulating and studying gene expression. Curr. Opin. Chem. Biol. 2009;15:678–686. doi: 10.1016/j.cbpa.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey JP, Blidner RA, Monroe WT. Caged siRNAs for spatiotemporal control of gene silencin. Mol. Pharmaceutics. 2009;6:669–685. doi: 10.1021/mp900082q. [DOI] [PubMed] [Google Scholar]
- 5.Cambrige SB, Geissler D, Calegari F, Anastassiadis k, Hasan MT, Stewart AF, Huttner WB, Hagen V, Bonhoeffer T. Doxycycline-dependent Photoactivated gene expression in eukaryotic systems. Nat. Methods. 2009;6:527–531. doi: 10.1038/nmeth.1340. [DOI] [PubMed] [Google Scholar]
- 6.Lusic H, Lively MO, Deiters A. Light-activated deoxyguanosine: photochemical regulation of peroxidase activity. Mol. BioSyst. 2008;4:508–511. doi: 10.1039/b800166a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young DD, Deiters A. photochemical control of biological processes. Org. Biomol. Chem. 2007;5:999–1005. doi: 10.1039/b616410m. [DOI] [PubMed] [Google Scholar]
- 8.Tang X, Dmochowski IJ. Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst. 2007;3:100–110. doi: 10.1039/b614349k. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto H. Yin-Yang ways of controlling gene expression are now in our hands. ACS Chem. Biol. 2007;2:646–648. doi: 10.1021/cb700213e. [DOI] [PubMed] [Google Scholar]
- 10.Mayer G, Heckel A. Biologically active molecules with a ‘light switch’. Angew. Chem., Int. Ed. Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 11.Furuta T, Noguchi K. Controlling cellular systems with Bhc-caged compounds. Trends Anal. Chem. 2004;23:511–519. [Google Scholar]
- 12.Cruz FG, Koh JT, Link KH. Light-activated gene expression. J. Am. Chem. Soc. 2000;122:8777–8778. [Google Scholar]
- 13.Riggsbee CW, Deiters A. Recent advances in the photochemical control of protein function. Trends Biotechnol. 2010;28:468–475. doi: 10.1016/j.tibtech.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dmochowski IJ, Tang X. Taking control of gene expression with light-activated oligonucleotides. Biotechniques. 2007;43:161–171. doi: 10.2144/000112519. [DOI] [PubMed] [Google Scholar]
- 15.Rothman DM, Petersson EJ, Vazquez ME, Brandt GS, Dougherty DA, Imperiali B. Caged phosphoproteins. J. Am. Chem. Soc. 2005;127:846–847. doi: 10.1021/ja043875c. [DOI] [PubMed] [Google Scholar]
- 16.Link KH, Shi Y, Koh JT. Light activated recombination. J. Am. Chem. Soc. 2005;127:13088–13089. doi: 10.1021/ja0531226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin W, Albanese C, Pestell RG, Lawrence DS. Spatially discrete, light-driven protein expression. Chem. Biol. 2002;9:1347–1353. doi: 10.1016/s1074-5521(02)00288-0. [DOI] [PubMed] [Google Scholar]
- 18.Shestopalov IA, Pitt CLW, Chen JK. Spatiotemporal resolution of the Ntla transcriptome in axial mesoderm development. Nat. Chem. Biol. 2012;8:270–276. doi: 10.1038/nchembio.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morckel AR, Lusic H, Farzana L, Yoder JA, Deiters A, Nascone-Yoder NM. A photoactivatable small-molecule inhibitor for light-controlled spatiotemporal regulation of Rho kinase in live embryos. Development. 2012;139:437–442. doi: 10.1242/dev.072165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi KB, Vlachos A, Mikat V, Deller T, Heckel A. Light-activatable molecular beacons with a caged loop sequence. Chem. Commun. 2012;48:2746–2748. doi: 10.1039/c2cc16654b. [DOI] [PubMed] [Google Scholar]
- 21.Govan JM, Uprety R, Hemphill J, Lively MO, Deiters A. Regulation of transcription through light-activation and light-deactivation of triplex-forming oligonucleotides in mammalian cells. ACS Chem. Biol. 2012;7:1247–1256. doi: 10.1021/cb300161r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govan JM, Lively MO, Deiters A. Photochemical control of DNA decoy function enables precise regulation of nuclear factor kappaB activity. J. Am. Chem. Soc. 2011;133:13176–13182. doi: 10.1021/ja204980v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young DD, Lively MO, Deiters A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 2010;132:6183–6193. doi: 10.1021/ja100710j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X, Su M, Yu L, Lv C, Wang J, Li Z. Photomodulating RNA cleavage using photolabile circular antisense oligodeoxynucleotides. Nucleic Acids Res. 2010;38:3848–3855. doi: 10.1093/nar/gkq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain PK, Shah S, Friedman SH. Patterning of gene expression using new photolabile groups applied to light activated RNAi. J. Am. Chem. Soc. 2010;133:440–446. doi: 10.1021/ja107226e. [DOI] [PubMed] [Google Scholar]
- 26.Deiters A, Garne RA, Lusic H, Govan JM, Dush M, Nascone-Yoder NM, Yoder JA. Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc. 2010;132:15644–15650. doi: 10.1021/ja1053863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasini AJ, Schuler AD, Zebala JA, Mayer AN. PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis. 2009;47:736–743. doi: 10.1002/dvg.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah S, Jain PK, Kala A, Karunakaran D, Friedman SH. Light-activated RNA interference using double-stranded siRNA precursors modified using a remarkable regiospecificity of diazo-based photolabile groups. Nucleic Acids Res. 2009;37:4508–4517. doi: 10.1093/nar/gkp415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards JL, Seward GK, Wang Y-H, Dmochowski IJ. Turning the 10-23 DNAzyme on and off with light. ChemBioChem. 2009;11:320–324. doi: 10.1002/cbic.200900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CLW, Li W-H, Olson AJ, Chen JK. Versatile synthesis and rational design of caged morpholinos. J. Am. Chem. Soc. 2009;131:13255–13269. doi: 10.1021/ja809933h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou C, Young DD, Deiters A. A light-activated DNA Polymerase Angew. Chem., Int. Ed. Engl. 2009;48:5950–5953. doi: 10.1002/anie.200901115. [DOI] [PubMed] [Google Scholar]
- 32.Young DD, Lusic H, Lively MO, Yoder JA, Deiters A. Gene silencing in mammalian cells with light-activated antisense agents. ChemBioChem. 2008;9:2937–2940. doi: 10.1002/cbic.200800627. [DOI] [PubMed] [Google Scholar]
- 33.Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res. 2008;36:559–569. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard JL, Tang X, Turetsky A, Dmochowski IJ. RNA bandages for photoregulating in vitro protein translation. Bioorg. Med. Chem. Lett. 2008;18:6255–6258. doi: 10.1016/j.bmcl.2008.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blidner RA, Svoboda KR, Hammer RP, Monroe WT. Photoinduced RNA interference using DMNPE-caged 2-deoxy-2-fluoro substituted nucleic acids in vitro and in vivo. Mol. BioSyst. 2008;4:431–440. doi: 10.1039/b801532e. [DOI] [PubMed] [Google Scholar]
- 36.Young DD, Deiters A. Photochemical activation of protein expression in bacterial cells Angew. Chem. Int. Ed. Engl. 2007;46:4290–4292. doi: 10.1002/anie.200700057. [DOI] [PubMed] [Google Scholar]
- 37.Tang X, Maegawa S, Weinberg ES, Dmochowski IJ. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am Chem. Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 38.Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol. 2007;3:650–651. doi: 10.1038/nchembio.2007.30. [DOI] [PubMed] [Google Scholar]
- 39.Young DD, Deiters A. Photochemical hammerhead ribozyme activation. Bioorg. Med. Chem. Lett. 2006;16:2658–2661. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 40.Tang X, Dmochowski IJ. Controlling RNA digestion by RNase H with a light-activated DNA hairpin. Angew. Chem. Int. Ed. Engl. 2006;45:3523–3526. doi: 10.1002/anie.200600954. [DOI] [PubMed] [Google Scholar]
- 41.Liu M, Asanuma H, Komiyama M. Azobenzene-Tethered T7 promoter for efficient photoregulation of transcription. J. Am Chem. Soc. 2006;128:1009–1015. doi: 10.1021/ja055983k. [DOI] [PubMed] [Google Scholar]
- 42.Heckel A, Buff MCR, Raddatz M-SL, Mueller J, Poetzsch B, Mayer G. An anticoagulant with light-triggered antidote activity. Angew. Chem., Int. Ed. Engl. 2006;45:6748–6750. doi: 10.1002/anie.200602346. [DOI] [PubMed] [Google Scholar]
- 43.Tang X, Richards JL, Peritz AE, Dmochowski IJ. Photoregulation of DNA polymerase I (Klenow) with caged fluorescent oligodeoxynucleotides. Bioorg. Med. Chem. Lett. 2005;15:5303–5306. doi: 10.1016/j.bmcl.2005.08.058. [DOI] [PubMed] [Google Scholar]
- 44.Shah S, Rangarajan S, Friedman SH. Light-Activated RNA Interference. Angewandte Chemie. 2005;117:1352–1356. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 45.Kroeck L, Heckel A. Photoinduced transcription by using temporarily mismatched caged oligonucleotides. Angew. Chem. Int. Ed. Engl. 2005;44:471–473. doi: 10.1002/anie.200461779. [DOI] [PubMed] [Google Scholar]
- 46.Hoebartner C, Silverman SK. Modulation of RNA tertiary folding by incorporation of caged nucleotides. Angew. Chem. Int. Ed. Engl. 2005;44:7305–7309. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]
- 47.Matsunaga D, Asanuma H, Komiyama M. Photoregulation of RNA digestion by RNase H with azobenzene-tethered DNA. J. Am. Chem. Soc. 2004;126:11452–11453. doi: 10.1021/ja0471976. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J. Mol. Biol. 2004;341:887–892. doi: 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 49.Ando H, Furuta T, Okamoto H. Methods in Cell Biology. Vol. 77. Academic Press (Amsterdam): Elsevier; 2004. pp. 159–171. [DOI] [PubMed] [Google Scholar]
- 50.Dussy A, Meyer C, Quennet E, Bickle TA, Giese B, Marx A. New light-sensitive nucleosides for caged DNA strand breaks. ChemBioChem. 2002;3:54–60. doi: 10.1002/1439-7633(20020104)3:1<54::AID-CBIC54>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Asanuma H, Tamaru D, Yamazawa A, Liu M, Komiyama M. Photoregulation of the transcription reaction of T7 RNA polymerase by tethering an azobenzene to the promoter. ChemBioChem. 2002;8:786–789. doi: 10.1002/1439-7633(20020802)3:8<786::AID-CBIC786>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 52.Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 53.Zhang K, Taylor J-S. A caged ligatable DNA strand break. J. Am. Chem. Soc. 1999;121:11579–11580. [Google Scholar]
- 54.Tang X, Swaminathan J, Gewirtz AM, Dmochowski IJ. Regulating gene expression in human leukemia cells using light-activated oligodeoxynucleotides. Nucleic Acids Res. 2008;36:559–569. doi: 10.1093/nar/gkm1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamazoe S, Shestopalov IA, Provost E, Leach S, Chen JK. Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew Chem Int Ed Engl. 2012;51:6908–6911. doi: 10.1002/anie.201201690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Z, Bai Z, Ruparel H, Kim S, Turro NJ, Ju J. A photocleavable fluorescent nucleotide for DNA sequencing and analysis. Proc. Natl Acad. Sci. USA. 2003;100:414–419. doi: 10.1073/pnas.242729199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellipanni G, Varga M, Maegawa S, Imai Y, Kelly C, Myers AP, Chu F, Talbot WS, Weinberg ES. Essential and opposing roles of zebrafish b-catenins in the formation of dorsal axial structures and neurectoderm. Development. 2006;133:1299–1309. doi: 10.1242/dev.02295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.