Abstract
Combinatorial interactions of transcription modulators are critical to regulate cell-specific expression and to drive direct cell reprogramming (e.g. trans-differentiation). However, the identification of key transcription modulators from myriad of candidate genes is laborious and time consuming. To rapidly identify key regulatory factors involved in direct cell reprogramming, we established a multiplex single-cell screening system using a fibroblast-to-monocyte transition model. The system implements a single-cell ‘shotgun-transduction’ strategy followed by nested-single-cell-polymerase chain reaction (Nesc-PCR) gene expression analysis. To demonstrate this, we simultaneously transduced 18 monocyte-enriched transcription modulators in fibroblasts followed by selection of single cells expressing monocyte-specific CD14 and HLA-DR cell-surface markers from a heterogeneous population. Highly multiplex Nesc-PCR expression analysis revealed a variety of gene combinations with a significant enrichment of SPI1 (86/86) and a novel transcriptional modulator, HCLS1 (76/86), in the CD14+/HLA-DR+ single cells. We could further demonstrate the synergistic role of HCLS1 in regulating monocyte-specific gene expressions and phagocytosis in dermal fibroblasts in the presence of SPI1. This study establishes a platform for a multiplex single-cell screening of combinatorial transcription modulators to drive any direct cell reprogramming.
INTRODUCTION
A defined combination of transcription modulators is key to target-directed cell reprogramming or trans-differentiation. Retro/lentiviruses have enabled genetic approaches in cultured primary cells to ectopically express genes of interest (1–6). Viral screens using focused overexpression libraries can identify putative reprogramming factors, for example, by identifying a defined set of transcription factors that are selectively required to induce pluripotent stem cells (2–4,7) or trans-differentiate from mature somatic cell into another mature somatic cell (1,8–11). Despite the power of viral technologies, pooled screens for cell reprogramming have generally adopted a one-by-one elimination approach or testing for exhaustive combinations and interrogating phenotypes in cell culture from transgenic mice (1,3,10) and/or aberrantly cloned cell lines (12). These approaches require both considerable specialty and a substantial investment in time.
We therefore implemented a rapid and direct approach by allowing multiple viruses to infect at random and identify specific viral transcripts that give rise to target-specific expressions at a single cell level. Our focus was identifying essential interplay of transcription ‘modulating’ factors (TMs) through lentivirus that were selectively expressed at a single-cell level. This type of screen holds promise for discovery of novel combination of TMs for regenerative medicine (1,13) or for any screening assay, which requires selectable phenotype such as biomarkers and cancer stem cells (14,15).
We and others previously described that the genetic interactions is key to defining the transcription regulatory network of human monocytes (16,17). We thus hypothesized that introducing the transcriptional network of monocytes into fibroblasts would promote trans-differentiation into functional monocytes characterized by the expression of both CD14 and HLA-DR surface antigens. In this study, we transduced a pool of 18 monocyte-enriched transcription modulators into human dermal fibroblasts and isolated CD14 and HLA-DR-positive single cells after 2 weeks. A highly multiplex nested-single-cell-polymerase chain reaction (Nesc-PCR) analysis showed heterogeneous infection patterns but revealed significant enrichments of SPI1 and HCLS1 transcripts when selected for monocyte markers. On the basis of our single-cell screening method, we further demonstrate that SPI1 promoted and HCLS1 enhanced marker expressions and function specific to monocytes in dermal fibroblasts.
MATERIALS AND METHODS
Complementary DNA and virus preparation
Gateway-compatible human full-length complementary DNA (cDNA) entry clones derived from RIKEN BRC clone bank [http://www.brc.riken.jp/ (23 July 2012, date last accessed)], Invitrogen (Carlsbad, CA, USA) and OpenBiosystems (Huntsville, AL, USA) were recombined into 150 ng of pENTR lentivirus vector (CSII-EF-RfA-IRES2-VENUS; Supplementary Figure S1) overnight at room temperature using Gateway LR clonaseII enzyme mix (Invitrogen). After 2 h of Protease K treatment at 55°C, recombined plasmids were transformed into OneShot® Stbl3 competent Escherichia coli (Invitrogen) following manufacturer’s protocol. Plasmids derived from single colony were expanded and purified using PureYield Plasmid Midiprep System (Promega). For every 8.5 µg of the plasmid, 5 µg of HIV-gp and 5 µg of VSV envelop genes were co-transfected onto 4 × 106 293T cells (prepared the day before at 37°C, 10% CO2) using FuGeneHD (Roche) in OPTI-MEM (WAKO) medium containing 5% FBS at 37°C 5% CO2. Between 24 h and 72 h of incubation, supernatant-containing virus were collected and centrifuged at 19 400rpm for 2 h at 20°C. The pallet was then dissolved in 100 µl HBSS buffer (WAKO), followed by titer check and stored at −80°C freezer for later use.
Virus titer check
One microliter of concentrated virus from the previous step was serially diluted (1:1000, 1:3000, 1:9000, 1:27 000 and 1:36 000) in MEM-α containing 10% FBS/L-glutamine/antibiotics (WAKO) and transduced onto 3000 of 293 T cells seeded in a black clear-bottom 96-well plate (BD) for 3 days. The nucleus was stained using Hoechst (Invitrogen) for 30 min at 37°C 5% CO2, and the plate was subjected to Cellomics ArrayScan XTi Reader (ThermoScientific) for image analysis. FITC and Hoechst filters were used to detect Yellow Fluorescent Protein derivative (Venus) and nuclear staining, respectively, at 10× magnification, and 10 images were taken per each well. Cellomics bio-application protocol ‘SpotDetector’ was used to segment Hoechst+ nucleus and to count Venus overlapping cells. The titer was calculated by
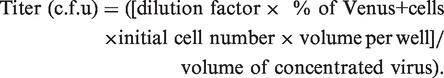 |
Cell culture and virus transduction
Human CD14+ primary monocytes were purchased from Lonza, and THP1 cells (16) were cultured in monocyte medium containing RPMI1400 (WAKO) containing 10% FBS/5M HEPES/2.5M sodium pyruvate/3.4 µg β-mercaptoethanol/l-glutamine/antibiotics (GIBCO). Human dermal fibroblasts (Passages 5–8) were purchased from RIKEN BRC and prepared at 5 × 104 cells in culture media: MEM-α containing 10% FBS/l-glutamine/antibiotic (WAKO) at 37°C 5% CO2 1 day before virus transduction. A total of 10 multiplicity of infection (MOI, 0.55 MOI per TM) were pooled into 3 ml of culture media containing 80 mg/ml of Polybrene (SIGMA) to increase transduction efficiency. The virus was transduced and incubated for 2 weeks in 37°C 5% CO2 with MEM-α containing 10% FBS/l-glutamine/antibiotic in the period of first 5 days and in the monocyte medium for the remaining 9 days.
Immunostaining and cell sorting
Two weeks post-virus transduction, cells were detached using Accutase and stained with human anti-CD14/AlexaFluora700 (BioLegend) and human anti-HLA-DR/APC (BD) antibodies for 30 min in ice. Fluorescence-activated cell sorting (FACS) analysis was performed on BD FACS Aria II (BD) for sorting Venus+/CD14+/HLA-DR+ single cells. To ensure a successful capture of single cells, we verified the cell-sorting accuracy before every experiment by observing the cells under a fluorescent microscope to confirm the alignment (center of the well) and to detect a single cell body. We detected 72 ± 10% positive single cells. Thereafter, single cells were sorted directly into 96-well plate containing the pre-amplification mixture. Pre-amplified samples positive for ACTB expression were used for the subsequent analysis.
Candidate gene extraction
Illumina Human WG-6 version 3.0 microarrays were used to analyze differentially expressed transcripts between human dermal fibroblasts and CD14+ monocytes (18) (GSE27304). After quantile normalization, top 20 most differentially expressed transcripts annotated as ‘regulation of transcription’ [based on Gene Ontology (19)] in monocytes when compared with fibroblasts were selected for lentivirus production (Student’s t-test: P value < 0.001). The viral transcripts, GAS7 and EGR2, could not be detected using Nesc-PCR; hence, they were removed from the virus pool.
Nested-single-cell PCR
The primer mix for pre-amplification consisted of 20 µM of sense primers targeting ORF region of each gene (total 36) plus 100 µM of Attb2 anti-sense primer or 20 µM of anti-sense primer for endogenous transcripts for primary fibroblasts and monocytes. The primer mixture is further diluted to 200 nM and mixed with OneStep qRT-PCR 2 × Reaction Mix, SuperScriptIII RT/Platinum TaqMix, RNAse OUT (Invitrogen) and DNAse/RNase-free distilled water (GIBCO) to total volume of 9 µl per reaction. After single-cell sorting, the samples were incubated at 55°C for 25 min, 95°C for 2 min and 18 cycles of 95°C for 15 s and 60°C for 4 min. The final samples were diluted 1:5 with DNase/RNase-free water and stored in −20°C for later use. Primer mixture for PCR amplification consisted of 20 µM of both sense and antisense primer pairs targeting ORF region of each gene plus 0.3 µM of UPL probe for specificity (ROCHE). Additional 2× assay loading reagent (Fluidigm) and water were added to adjust final volume of 5 µl per reaction. Diluted pre-amplified cDNA (1.25 µl) is mixed with 2× FastStart Universal Probe Master (ROX; ROCHE) and 20× GE Sample loading reagent (Fluidigm) and water to final volume of 5 µl per reaction. The primer mix and the sample mix were loaded onto 48.48 Dynamic Array (Fluidigm) and primed and mixed using the MX IFC controller (Fludigm). The dynamic array was further subjected to BioMark Array System (Fluidigm) at 95°C for 10 min and 40 cycles of 95°C for 15 min and 60°C for 1 min.
Single-cell data analysis and statistics
Expression data from BioMark (Fluidigm) were processed in R Bioconductor (20). To illustrate gene expression and for statistical analyzes, we first set all Ct values >35 to 35 and inversed the expression by subtracting the values by 35. To test the statistical significance of marker enrichment analysis, we performed chi-square test to compare observed data (detected number of exogenous TM) with data we would expect to obtain (marker+ cells and marker − cells). The data are presented in terms of percentages.
qRT-PCR
Reverse transcription of total ribonucleic acid (RNA) was achieved with PrimeScript™ Reverse Transcriptase (Takara) and random hexamers in accordance with the manufacturer’s protocol. Beta actin (ACTB) messenger RNA was used as a control for data normalization. The PCR primers used for each gene in this analysis are provided in Supplementary Table S2. PCR amplification was performed on an ABI PRISM® 7500 Sequence Detection System (Applied Biosystems). For amplification, SYBR Premix Ex Taq™ II (Takara) was used as instructed in the manual. Changes of gene expression were determined using the 2−ΔΔCt method.
Phagocytosis assay
Combinations of vector control, SPI1 and HCLS1, were transduced onto human dermal fibroblasts seeded at 5000 cells in a black clear bottom 96-well plate for a period of 20 days in RPMI1400 (WAKO) containing 10% FBS/5 M HEPES/2.5 M sodium pyruvate/3.4 µg β-mercaptoethanol/l-glutamine/antibiotics (GIBCO) at 37°C 5% CO2. Each plate contained six replicate wells of the same condition. The experiment was performed two independent times. One vial (2 mg) of the pHrodo BioParticles fluorescent particles (Invitrogen) were diluted in 4 ml of uptake buffer consisting of HBSS (WAKO), 20 mM HEPES, pH 7.4 and replaced the culture medium with 100 µl of the prepared pHrodo BioParticles suspension. The plate was incubated in 37°C 5% CO2 for 7 h, and cells were stained with Hoechst (Invitrogen) for 1 h at 37°C 5% CO2. The images were taken using the Leica XT microscope (Leica) and quantified using Cellomics ArrayScan XTi Reader (ThermoScientific).
siRNA transfection and RNA extraction
Reverse transfection of 1 × 106 THP1 cells in each 60-mm cell culture dish was performed with 20 nM of each stealth negative control siRNA, SPI1 siRNA (Stealth RNAi: UGGUGCCCUAUGACACGGAUCUAUA) or HCLS1 siRNA (Silencer® Select, Invitrogen; siRNA ID s6484) in Opti-MEM and Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Total RNA was extracted 48 h after transfection using miRNAeasy kit (Qiagen) according to manufacturer’s instructions.
RESULTS
Single-cell screening system
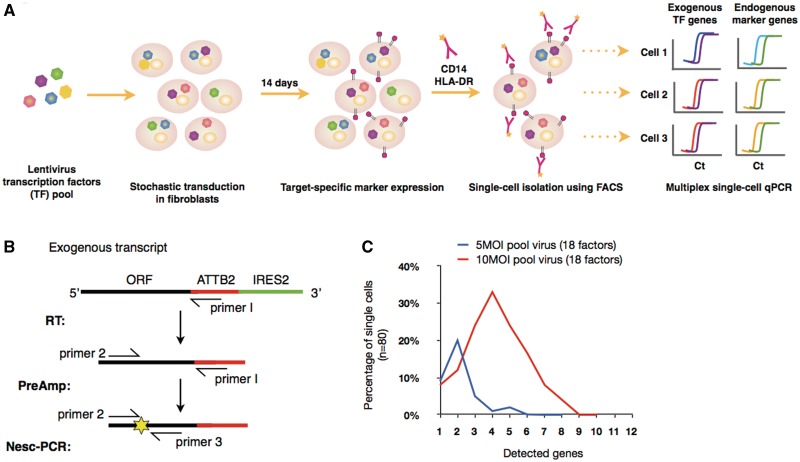
To rapidly screen for TMs involved in the direct monocyte specification, we designed an experimental workflow that integrates lentivirus technology (5,6), FACS and single-cell profiling analysis (21–23) (Figure 1). Human primary dermal fibroblasts were simultaneously transduced with a lentivirus pool of 18 monocyte-enriched TMs, selected based on gene-specific expression in primary CD14+ monocytes when compared with dermal fibroblasts (Supplementary Table S1, see Section ‘Materials and Methods’). The stochastic nature of the virus infection allows for a mixed combination of TMs to be expressed per infected single cell, giving rise to target-specific expressions from a sub-population that accompanies the defined combination of monocyte-specifying TMs. A multiplex expression profiling of both exogenous and endogenous transcripts from single cells allows for the immediate identification of TMs essential for direct cell reprogramming.
Figure 1.
Workflow of single-cell screening and Nested-single-cell-PCR (Nesc-PCR). (A) Lentviruses encoding multiple transcription ‘modulating’ factors (TMs) are pooled and transduced into human dermal fibroblasts for a period of 2 weeks. Because of the stochastic nature of virus infection, a variety of TM combinations are expressed at random per single cell. Cells expressing a defined set of TMs give rise to the expression of target-cell-specific markers (e.g. CD14 and HLA-DR) allowing them to be individually sorted using FACS. Subsequently, single cells are profiled using a Nesc-PCR gene expression analysis by reverse transcribing (RT) the exogenous transcripts with a virus-specific primer (primer 1) followed by 18 cycles of pre-amplification with gene-specific forward primers (primer 2) and primer 1. The cDNA products are then subjected to a microfluidic qPCR for the identification of exogenous transcripts using gene-specific sense (primer 2) and anti-sense (primer 3) primer pairs and fluorescence probes (yellow star) (B). (C) The single cells transduced with pooled lentivirus (18 factors) at 5 MOI and 10 MOI were sorted and profiled using Nesc-PCR. At 10 MOI, >30% of single cells expressed 4 out of 18 TMs, while 20% of single cells expressed 2 out 18 TMs at 5 MOI.
First, to ascertain accurate detection of exogenous genes, single cells expressing Venus fluorescent protein (i.e. 18-virus pool infected fibroblasts) were individually sorted using FACS and subjected to a multiplex Nesc-PCR for the detection of 18 virus-specific transcripts (Figure 1B). Based on our statistical analysis, the single-cell transduction followed the Poisson distribution and the number of detected genes correlated with the half of estimated number of virus particles per cell (Figure 1C). This suggests that a minimum of two copies of viral transcripts can be detected, and 10 MOI of the pooled virus, or 0.55 MOI per gene, was optimal to detect multiple target genes from single cells since >50% of the population contained average number of two to six genes.
Single-cell gene expression analysis of virus-infected fibroblasts
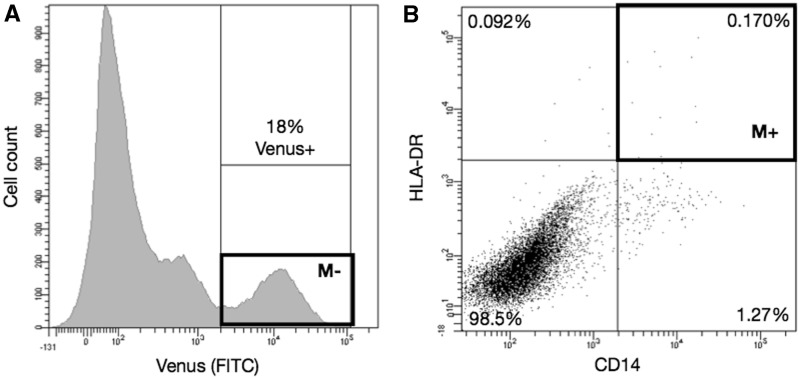
CD14 and HLA-DR proteins have been well characterized to play a role in LPS response and antigen presentation in monocytes, respectively (24,25). To determine which of the 18 factors were critical for activating monocyte gene expression, we labeled the transduced cells using CD14 and HLA-DR antibodies. Two weeks post-infection at 10 MOI, 18 ± 2% of the fibroblasts showed Venus fluorescence (Figure 2A). Predictably from a pooled virus, a minute amount of Venus+ cells were positive for both CD14 and HLA-DR (0.17 ± 0.1%; Figure 2B) but was sufficient to obtain single cells for transcript profiling using Nesc-PCR (M+, n = 86). To analyze the enrichment of TMs in marker-positive single cells, we additionally sorted an equal number of single cells from Venus+ population as a baseline control (Figure 2A, M−). Additionally, fibroblasts transduced with vector control (10 MOI) failed to express CD14 and HLA-DR surface markers (data not shown).
Figure 2.
Flow cytometry sorting of CD14 and HLA-DR-positive single cells. Two weeks post transduction of 18 monocyte-TMs, 18% of fibroblasts expressed Venus fluorescent protein (A, ‘M−’), out of which 0.17% was positive for both CD14 and HLA-DR markers (B, ‘M+’). Eighty-six single cells from M+ and M−boxes were sorted for further analysis.
Individually sorted single cells were profiled specifically targeting 18 viral-TM genes, 16 endogenous monocyte markers and 1 internal control (Supplementary Table S2). The final 35 monocyte–gene panel was used to measure by means of Nesc-PCR. To further address the effect of exogenous factors on fibroblasts, we additionally isolated and analyzed single cells from ‘unperturbed’ fibroblasts and human CD14+ primary monocytes (n = 40 each, Supplementary Figure S2A). These individual cells were profiled using the same 35-gene panel but targeting 18 endogenous TMs.
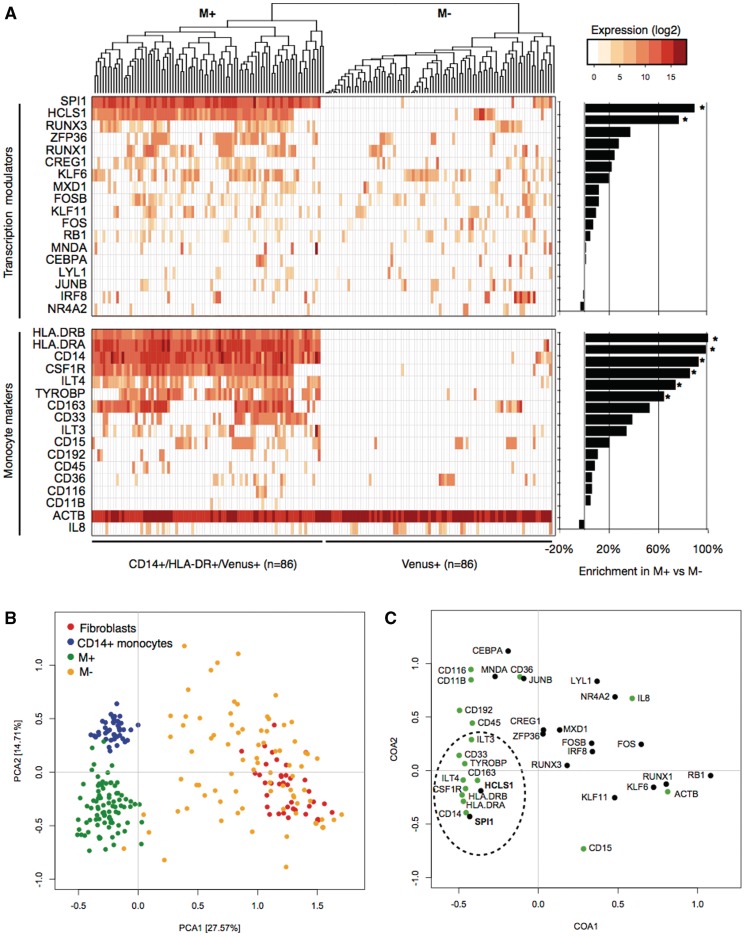
We then analyzed the ability of Nesc-PCR gene expression analysis to distinguish different cell populations based on exogenous TMs and endogenous marker expressions. Figure 3A reveals a cluster of 86 CD14+/HLA-DR+ cells (M+) and 86 Venus+ cells (M−). Most notably, single cells positive for CD14 and HLA-DR markers detected SPI1 (86/86, 100%) and 76 out of 86 expressed HCLS1 (88%), which were statistically significant when compared with the M− population where only 8/86 and 10/86 single cells were positive for SPI1 and HCLS1, respectively (chi-square test, P value < 0.001 for both). Interestingly, though eight out of nine single cells expressing exogenous SPI1 in the M− population expressed CD14 transcripts, and two out of nine expressed HLA-DRA transcripts, but no detection of HLA-DRB transcript was observed. Although the sample size was small, the analysis suggests that SPI1 alone could induce the expression of CD14. In fact, when SPI1 alone was transduced into fibroblasts in an independent experiment, 12 ± 2% of SPI1+ cells expressed the CD14 surface marker suggesting that SPI1 could elicit the expression of this marker but at a low efficiency (Supplementary Figure S3).
Figure 3.
Single-cell clustering analyses. Hierarchical clustering of single cells reveals distinct cell populations between M+ and M− (A). The gene enrichment comparisons between M+ and M− population demonstrates differential detection rate of TMs as well as monocyte-marker transcripts. (B) The principal component analysis (PCA) demonstrates variations of single cells from four populations: CD14+ primary monocytes, dermal fibroblasts, CD14+/HLA-DR+/Venus+ (M+), CD14-/HLA-DR-/Venus+ (M−). The M− population (orange) clusters near the fibroblasts (red) and shows the largest variation, mainly due to random detection of exogenous TMs. However, the M+ population (green) reveals reduced variation and clusters near the primary monocytes (blue). (C) The correspondence analysis reveals SPI1 and HCLS1 TMs (bolded; bottom-left) as key determinants of M+ population, causing the separation of M+ cells from fibroblasts and shifting the M+ cells towards monocytes. Gene enrichment analysis, chi-square test P value < 0.005.
In a drastic contrast to the M− population, the analysis of M+ population noticeably revealed that this subset, although transcriptionally heterogeneous, was almost exclusively composed of cells expressing high levels of genes characteristic of mature monocytes (Figure 3A). Single cells selected for CD14 and HLA-DR surface proteins revealed 100% detection of CD14, HLA-DRA and HLA-DRB marker transcripts, as expected. But additional markers such as CSF1R (85%), ILT4 (73%), TYROBP (63%) and CD163 (52%) transcripts were highly enriched in the M+ population when compared with the M− population (chi-square test, P value < 0.05) and significantly induced when compared with the basal expressions in dermal fibroblasts (Supplementary Figure S4).
SPI1 and HCLS1 as key determinants of monocyte expression
To further investigate the key regulatory determinants of monocyte specification, we performed eigenvector-based analyses between M+, M−, fibroblasts and monocytes to visualize the heterogeneity in single cells, as reflected by the 2-axis of the principal component analysis (PCA; Figure 3B). The analysis found that the first principal component (explaining 27.57% of the variation) grouped the primary monocytes and the M+ cells but separated fibroblasts and M− cells. These distinct clusters indicate that M+ cells deviated from the original fibroblast state and transitioned into a state comparable to that of CD14+ monocytes. However, a closer examination of the second component narrowly distinguished the monocytes from the M+ cells, indicative of partial (or incomplete) induction of monocyte network in fibroblasts. The PCA also showed a wide scatter of M− single cells largely due to random virus transductions. However, a small number of single cells clustered closely to the M+ population largely contributed by positive detection of SPI1 and CD14 expression but failed to be selected by FACS due to lack of HLA-DR protein expression.
To identify the causal genes that contribute to the separation of each cell population, we performed the correspondence analysis (CoA; Figure 4C). Complementary to PCA, the CoA identified SPI1 and HCLS1 as major determinants to discriminate the monocytes from the original fibroblasts. Although the primary monocytes expressed SPI1 and HCLS1, the exogenous expression in the M+ population was 6-fold higher (in the case of SPI1), contributing to the separation from the primary monocytes on the second axis. Interestingly, the single-cell expression profile of primary monocytes revealed that 27% and 22% monocytes expressed CEBPA and IRF8, respectively, which is in contrast to the ubiquitous expression of SPI1 and HCLS1 in all 40 monocyte single cells profiled (Supplementary Figure S2B). The heterogeneous expressions of CEBPA and IRF8 transcripts may suggest a dynamic balance of expression to maintain monocyte phenotype but are not necessary required to promote key monocyte-specific markers.
Figure 4.
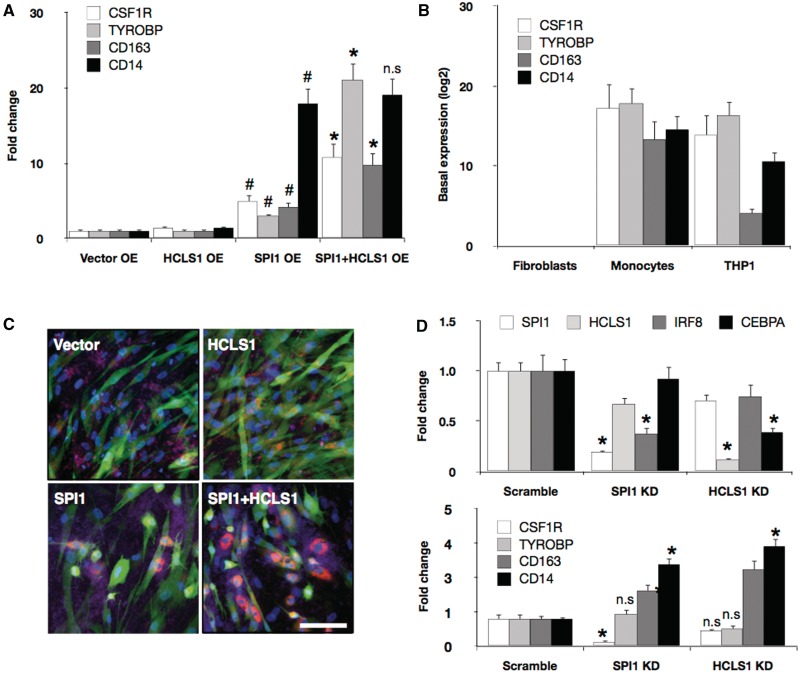
SPI1 and HCLS1 enhance monocyte marker expression and function. Vector control, HCLS1, SPI1 and SPI1/HCLS1 combination were overexpressed (OE) in dermal fibroblasts, and the marker expressions for CSF1R, TYROBP, CD163 and CD14 were quantified by means of conventional qRT-PCR expression analysis (A). (Student’s t-test: #P value < 0.05 when compared with vector control; *P value < 0.05 when compared with SPI1 or HCLS1 alone). However, the induced marker expressions were partial when compared with the basal expression levels (log-2 transformed) of dermal fibroblasts, primary monocytes and THP1 cell line (B). (C) pHrodo™ Red E. coli BioParticles® conjugate beads which is pH-sensitive Rhodamine dye specifically expressed in phagosomes were added onto fibroblasts transduced with either vector control, SPI1, HCLS1 or SPI1/HCLS1. Cells positive for Rhodamine (red) were clearly visible in both SPI1 and SPI1/HCLS1 combination, but no detection was found in cells transduced only with vector control or HCLS1. Scale bar = 100 µm. (D) Gene-specific knockdown (KD) of SPI1 and HCLS1 in THP1 cells revealed significant suppression of IRF8 and CEBPA transcripts, respectively. Knockdown of SPI1 also suppressed CSF1R, but induced CD163 and CD14 expression, which was also observed when HCLS1 transcript was ablated. P value (Student’s t-test) * < 0.001, when compared with vector control.
SPI1 is required and HCLS1 enhances expression and function specific to monocytes
To further explore the effects of combinatorial TMs, we ectopically expressed vector control, SPI1, SPI1+HCLS1 in fibroblast and measured the differential expression using the conventional qRT-PCR method. CSF1R is a well-studied monocyte membrane protein acting as a receptor for colony stimulating factor 1, and deregulation of this gene is a hallmark of many tumors (26). Although SPI1 alone could induce the expression of this monocyte marker, the signal intensity was greatly enhanced when fibroblasts were co-infected with HCLS1 (Figure 4A). Similar expression patterns were also observed for TYROBP and CD163. However, HCLS1 could not enhance the transcript level of CD14. When the induced expressions were compared with the basal expression of monocytes, the cell transformation was only partial (Figure 4B). This incomplete observation was also seen in the single-cell PCA analysis (Figure 3B), indicating that the conversion is heterogeneous and partial, although some single cells clustered closer to the primary monocytes than others. Concurrently, it is also plausible that only a subpopulation of the cells undergoes cell transformation, diluting the expression levels and thus masking the induced expression.
Next to test whether SPI1 in conjunction with HCLS1 could also enhance the functional role of monocytes, we added pHrodo™ Red E. coli BioParticles® conjugate beads, which is pH-sensitive Rhodamine dye to fibroblasts transduced with lentivirus vector control, SPI1, HCLS1 or both (Figure 4C; Supplementary Figure S5). Since the fluorescent dye of the beads specifically expresses in phagosomes, we could reliably detect the ingested beads using the fluorescent microscopy without trypsinization of the cells. We observed a clear detection of Rhodamine-positive cells after transducing SPI1 and the SPI1/HCLS1 combination. The image analysis further revealed that cells transduced in combination of SPI1 and HCLS1 ingested 33% more beads when compared with SPI1 alone (Supplementary Figure S5B).
To better understand the causal effects of SPI1 and HCSL1 involved in monocyte specification, we targeted the transcripts using siRNA oligos in THP1 (human acute monocytic leukemia cell line). THP1 has been reported to be a suitable model to study monocyte network (16) and also has been demonstrated to yield high efficiencies in siRNA transfections (16,27). Since IRF8 and CEBPA have been implicated as key regulators of monocyte network, we specifically measured the expression changes after the knockdown (16,18,28,29). Interestingly, ablating SPI1 and HCLS1 transcripts in THP1 cells could significantly repress IRF8 and CEBPA expression by 2.71-fold and 2.56-fold, respectively (Figure 4D), as well as CSF1R by 6.41-fold—in the case of SPI1 (Figure 4E). Surprisingly, however, THP1 deficient in SPI1 augmented CD163 and CD14 expression, and a similar expression pattern was also observed for the HCLS1 knockdown. Although the exact mechanisms is unclear, ablation of SPI1 and/or HCLS1 in the established monocyte network may not be sufficient to suppress marker genes such as TYROBP (‘gene redundancy’) or may induce gene expressions to maintain network homeostasis (‘network robustness’). Taken together, our results revealed that SPI1 is minimally required to promote, in part, monocytic expression and phagocytic function in human dermal fibroblasts but can be greatly enhanced through the co-expression of HCLS1.
DISCUSSION
Here we described a rapid and direct method that facilitates the use of lentivirus and Nesc-PCR for identifying key genes involved in trans-differentiation. By relying on the random nature of virus infection, we implemented a single-cell ‘shotgun-transduction’ system that enabled the prompt detection, isolation and identification of combinatorial genes that are required to transform fibroblasts into a desired target cell type.
By taking advantage of viral transcripts and the single-cell detection method, we noted SPI1 and HCLS1 as a defined set of transcription modulators responsible to switch from human dermal fibroblasts to functional monocyte-like cells. SPI1, an important regulator of myeloid and B-lymphoid cell development, also acts as a key modulator of cell transformation toward monocytes together with CEBPA (9). The detection of SPI1 by the single-cell screening system decisively validates our method in identifying the strongest modulator in the group. However, we could additionally demonstrate that CEBPA was dispensable in that SPI1 alone could induce the expression of CD14 and HLA-DRA and promote phagocytosis in human fibroblasts. Although HCLS1 alone could not induce monocyte trans-differentiation, the protein acted as a strong modulator of monocyte/macrophage phenotype. HCLS1 has been implicated in response to cortisol and LPS, activating numerous functions of macrophages (30,31). Although the exact mechanism is unknown, our data and the published reports strongly suggest that HCLS1 expression correlates positively with enhanced function of monocytes/macrophages possibly via regulating CEBPA expression.
In this study, we have validated a highly scalable approach for screening lentiviral libraries for direct cell reprogramming. We used cell-specific markers to isolate single cells; however, this method can virtually be applied to any characteristics that allow separation of phenotypically distinct cells, such as cell proliferation dyes or functional biomarkers for the identification of transcription modulators. Overall, this study demonstrates a rapid single-cell screening workflow to readily identify and characterize new combinatorial factors, applicable to induce various cell-reprogramming pathways.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
Research fund for Foreign Postdoctoral Researcher Program from RIKEN (to J.W.S.); Research Grant for RIKEN Omics Science Center from MEXT (to Y. Hayashizaki); Grant of the Innovative Cell Biology by Innovative Technology (Cell Innovation Program) from the MEXT (to Y. Hayashizaki) and Strategic programs for R&D (Presidents discretionary fund) (super immune cell) (to Y. Hayashizaki). Funding for open access charge: Research Grant for RIKEN Omics Science Center from MEXT (to Y. Hayashizaki).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of Omics Science Center (RIKEN Yokohama) for virus production (Chiduru Suzuki, Ayumi Ogawa, Yumi Yoshimura and Akane Murakami), cloning/sequence verification (GeNas) and statistical analysis (Michiel De Hoon). We also extend our gratitude to Dr. Hiroyuki Miyoshi (RIKEN BRC) for providing the lentivirus vectors and Drs. Osamu Ohara and Yoshitaka Shirasaki (RIKEN RCAI) for the use of the Fluidigm Biomark system.
REFERENCES
- 1.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Soda Y, Izawa K, Tanabe T, Kang X, Tojo A, Hoshino H, Miyoshi H, Asano S, Tani K. Effective transduction and stable transgene expression in human blood cells by a third-generation lentiviral vector. Gene Ther. 2003;10:1446–1457. doi: 10.1038/sj.gt.3302026. [DOI] [PubMed] [Google Scholar]
- 6.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J. Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl Acad. Sci. USA. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 12.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A, et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 15.Vuk-Pavlovic S, Bulur PA, Lin Y, Qin R, Szumlanski CL, Zhao X, Dietz AB. Immunosuppressive CD14+HLA-DRlow/- monocytes in prostate cancer. Prostate. 2010;70:443–455. doi: 10.1002/pros.21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T, Nakano-Ikegaya M, Yabukami-Okuda H, de Hoon M, Severin J, Saga-Hatano S, Shin JW, Kubosaki A, Simon C, Hasegawa Y, et al. Reconstruction of monocyte transcriptional regulatory network accompanies monocytic functions in human fibroblasts. PloS One. 2012;7:e33474. doi: 10.1371/journal.pone.0033474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schutze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nat. Biotechnol. 1998;16:737–742. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- 22.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcus JS, Anderson WF, Quake SR. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 24.Hershman MJ, Cheadle WG, Wellhausen SR, Davidson PF, Polk HC., Jr Monocyte HLA-DR antigen expression characterizes clinical outcome in the trauma patient. Brit. J. Surg. 1990;77:204–207. doi: 10.1002/bjs.1800770225. [DOI] [PubMed] [Google Scholar]
- 25.Kirschning CJ, Wesche H, Merrill Ayres T, Rothe M. Human toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 1998;188:2091–2097. doi: 10.1084/jem.188.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonifer C, Hume DA. The transcriptional regulation of the Colony-Stimulating Factor 1 Receptor (csf1r) gene during hematopoiesis. Front Biosci. 2008;13:549–560. doi: 10.2741/2700. [DOI] [PubMed] [Google Scholar]
- 27.Tomaru Y, Simon C, Forrest AR, Miura H, Kubosaki A, Hayashizaki Y, Suzuki M. Regulatory interdependence of myeloid transcription factors revealed by Matrix RNAi analysis. Genome Biol. 2009;10:R121. doi: 10.1186/gb-2009-10-11-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubosaki A, Lindgren G, Tagami M, Simon C, Tomaru Y, Miura H, Suzuki T, Arner E, Forrest AR, Irvine KM, et al. The combination of gene perturbation assay and ChIP-chip reveals functional direct target genes for IRF8 in THP-1 cells. Mol. Immunol. 2010;47:2295–2302. doi: 10.1016/j.molimm.2010.05.289. [DOI] [PubMed] [Google Scholar]
- 29.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez N, Mages J, Dietrich H, Wantia N, Wagner H, Lang R, Miethke T. MyD88-dependent changes in the pulmonary transcriptome after infection with Chlamydia pneumoniae. Physiol. Genomics. 2007;30:134–145. doi: 10.1152/physiolgenomics.00011.2007. [DOI] [PubMed] [Google Scholar]
- 31.Billing AM, Fack F, Turner JD, Muller CP. Cortisol is a potent modulator of lipopolysaccharide-induced interferon signaling in macrophages. Innate Immun. 2011;17: 302–320. doi: 10.1177/1753425910369269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.