Abstract
Methods for generating loss-of-function mutations, such as conventional or conditional gene knockout, are widely used in deciphering gene function in vivo. By contrast, inducible and reversible regulation of endogenous gene expression has not been well established. Using a mouse model, we demonstrate that a chimeric transcriptional repressor molecule (tTS) can reversibly inhibit the expression of an endogenous gene, Nmyc. In this system, a tetracycline response element (TRE) artificially inserted near the target gene’s promoter region turns the gene on and off in a tetracycline-inducible manner. NmycTRE mice were generated by inserting a TRE into the first intron of Nmyc by the knockin technique. NmycTRE mice were crossed to tTS transgenic mice to produce NmycTRE/TRE: tTS embryos. In these embryos, tTS blocked Nmyc expression, and embryonic lethality was observed at E11.5d. When the dam was exposed to drinking water containing doxycycline (dox), normal endogenous Nmyc expression was rescued, and the embryo survived to birth. This novel genetic modification strategy based on the tTS–dox system for inducible and reversible regulation of endogenous mouse genes will be a powerful tool to investigate target genes that cause embryonic lethality or other defects where reversible regulation or temporary shutdown of the target gene is needed.
INTRODUCTION
In the post-genomic era, annotating the in vivo function of newly identified genes remains a significant challenge for scientists. One of the most powerful tools for studying gene functions in mouse models is gene targeting or random mutagenesis (chemical or insertion) (1). With these methods, it is difficult to control spatially and temporally the resulting gene loss of function. Inactivation of a gene in the zygote may lead to embryonic or neonatal lethality, which precludes any efforts to study the gene function later in life (1). Alternatively, genome adaptation to a mutation during development may compensate the gene loss so that the phenotype is not detectable in adults (2). To overcome these limitations, conditional gene inactivation was developed using site-specific recombination systems, such as the Cre/loxP system, which permitted temporal and spatial inactivation by controlling the recombinase activity (3). Similar to conventional gene targeting, the conditional gene knockout irreversibly disrupts the target gene expression, and transient inhibition of endogenous genes in a specific developmental stage is impossible (4,5). Recently, an inducible RNAi transgenic system using a reversible knockdown of endogenous genes was been established, providing an alternative to Cre-mediated conditional gene inactivation (4,6,7). However, its application is limited by potential shRNA toxicity and low efficiency in vivo (8,9).
An inducible gene regulation system using epigenetic repression to control gene expression in in vitro and in transgenic mice has been reported. In this system, genes are turned on or off by an artificial transcriptional repressor (tTS, also known as tTRKRAB) in the presence or absence of doxycycline (dox) (6). tTS is a fusion transcriptional repressor composed of the tetracycline repressor (tetR) and a Krüppel-associated box (KRAB) domain that binds to the tetracycline responding elements (TREs) and recruits the corepressors causing transcriptional repression of the local gene (6). A similar system has been used in mice to eliminate leakiness in inducible overexpressed transgenes (10). tTS has been shown to regulate inducibly and reversibly the expression of endogenous genes, such as Hoxa2, Mlc1 and Htr1a (11–13). However, a comprehensive investigation to use the system in regulation mouse endogenous gene is still absent. Furthermore, in these published reports, the TRE was inserted into the promoter region, such as Hoxa2 and Htr1a, which may alter promoter activity outside of the tTS regulation (11), or as Mlc1, the information and side effect of TRE insertion was not revealed. Thus, the regulation of endogenous genes by tTS requires further optimization and confirmation.
In this article, the TRE site was inserted in an intron near the promoter region to test whether the target gene is still regulated by tTS binding. We selected Nmyc as a target gene to show the reversible and inducible regulation of the gene expression by the tTS–dox system. Nmyc is a member of myc family of proto-oncogenes, which plays important roles in development and tumorigenesis (14). Homozygous Nmyc knockout mice die prenatally at E11.5d (15,16), precluding assessment of the function of Nmyc in late life. Conditional deletion of Nmyc identified its roles in cerebellar development and digit separation (17,18). Here, we demonstrate in multiple tissues that the transcriptional activity of the Nmyc promoter is tightly regulated by tTS in a tetracycline-inducible manner. Blocking Nmyc expression resulted in embryonic lethality in the absence of dox. The lethality was rescued by administering dox of dam for a certain duration, but the surviving offspring could exhibit cerebellar defects and syndactyly, similar to phenotypes in conditional knockout mice. The severity of the cerebellar defects depended on the initial time point and duration of the dox administration and the time window of Nmyc functioning critically in postnatal cerebellar development was measured. Our results confirmed the tTS–dox system is a reliable one to regulate endogenous gene in an inducible and reversible manner.
MATERIALS AND METHODS
Animals
C57BL/6J and EIIA-Cre transgenic mice were housed in a pathogen-free facility and maintained in controlled conditions (21–24°C; 12-h light–dark periods). For inducible and reversible experiments, mice were exposed to 2 mg/ml dox (Sigma-Aldrich, St Louis, USA) dissolved in 5% sucrose supplied as a drinking water, which was refreshed every 2 days. For dose effect experiments, different dosages of dox were in drinking water and sucrose was added so that the ratio of dox to sucrose was constant. All animal study protocols were reviewed and approved by the Institutional Animal Care and Use Committee in Shanghai Research Center for Model Organisms.
Generation of NmycTRE−EGFP-Neo (NmycEGFP) and NmycTRE knockin mouse
The targeting vector was generated basing on homologous recombination mediated by the recE and recT proteins in EL250 bacterial cells (19). The bMQ-165E1 clone from the mouse bacterial artificial chromosome library (Source Biosciences, Nottingham, UK) was used as the source of two homologous arms. The TRE site was amplified from pLVCT-tTRKRAB (Addgene, Cambridge, MA, USA). The Nmyc intron 1 fragment and upstream of exon2 were polymerase chain reaction (PCR)-amplified from BAC bMQ-165E1, which included the sequence from chromosome 12: 12 948 341–12 947 191. The EGFP and polyA fragment were PCR amplified from pIRES2-EGFP (Clontech, Mountain View, CA, USA). All PCR products were sequenced bidirectionally. The TRE site, Loxp site, Nmyc intron1 fragment, EGFP and polyA fragment were each cloned separately into PL451 plasmid to generate the knockin fragment. The final targeting construct contained a 2.73-kb 5′-homology arm, the 4.4-kb knockin fragment and a 4.39-kb 3′-homology arm. The targeting vector was linearized by NotI digestion and electroporated into SCR012 embryonic stem (ES) cells (Chemicon, Temecula, CA, USA). The ES colonies were selected by G418/gancyclovir (Sigma-Aldrich). The survival ES cell colonies were identified by PCR amplification using primer sets I/II and III/IV (Figure 2c) and confirmed by sequencing. Positive ES cell clones were expanded and injected into C57BL/6J blastocysts to generate the chimeric offspring. The chimeric mice were mated with C57BL/6J mouse. The F1 offspring were genotyped by tail genomic DNA. The NmycEGFP allele was identified by PCR amplification using primers V and VI. The F1 offspring carrying the NmycEGFP allele was backcrossed to EIIA-Cre mouse to delete the Loxp flanked fragment and generate the offspring with the NmycTRE allele. The NmycTRE alleles were identified by PCR amplification using primers VII and VIII. These mice were then backcrossed to C57BL/6J to generate the offspring with the NmycTRE allele but without Cre allele. The absence of the Cre allele was confirmed by PCR with primers Cre-F and Cre-R. The detailed information of primers is described in the Supplementary Material.
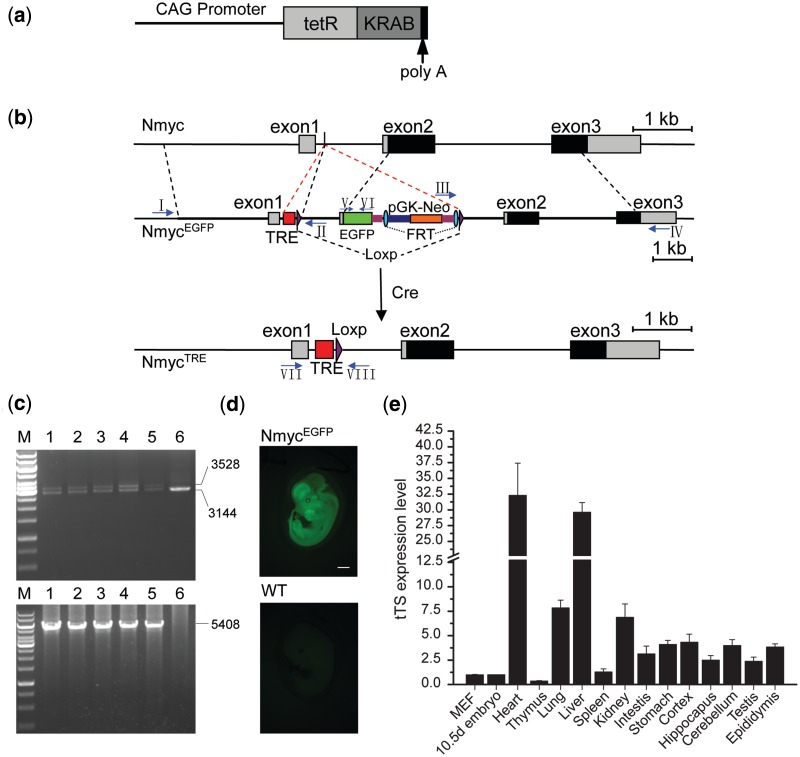
Figure 2.
The genetic modification system for reversible and inducible regulation of Nmyc. (a) Schematic representation of tTS transgene vector that includes a tTS expression cassette under the control of a CAG promoter. (b) Structures of wild-type Nmyc, NmycEGFP and NmycTRE. In NmycEGFP, a TRE site and one loxp site were inserted into the first intron between site 100 and 101. The 3′-region of TRE insert site in the first intron and 5′-region of ATG start codon in the exon 2 were duplicated, and the EGFP cassette was fused into the Nmyc exon 2 following the ATG start codon of Nmyc. Neo selection markers were flanked by FRT sites and another loxp site followed the 3′-FRT site. The NmycTRE allele was generated by crossing NmycEGFP mice with EIIA-Cre transgene mice. (c) The representation of PCR identifying of NmycEGFP from targeted NmycEGFP ES cell clones using primers I–IV (arrows in Panel c). Primer sets I and II, which identified the 5′-homology arm, yielded two fragments (3528 and 3144 bp) in positive clones and a single fragment (3144 bp) from negative clones (top). Primer sets III and IV, which identified the 3′-homology arm, yielded a single fragment (5408 bp) in positive clones and no product in the negative clones (bottom) (positive clones:1–5; negative clones:6; M: 1-kb ladder). (d) Fluorescence of EGFP at E12.5d embryos of NmycEGFP (top) and wild-type (bottom, as a negative control). (e) Real-time PCR of tTS expression profile in different cell and tissues of the transgenic mice (MEF: mouse embryonic fibroblasts; the tissues were all from 8-week-old adult male mice and the 10.5d embryo.) Scale bar is 1000 µm.
Generation of CAG-tTS transgenic mouse
The cytomegalovirus (CMV) promoter sequence in pcDNA3.1/Hygro vector (Invitrogen, Carlsbad, CA, USA) was removed by BglII/NheI digestion and replaced by a CAG promoter fragment excised from pLVCT-tTRKRAB (Addgene) digested by SpeI and BamHI to generate pcDNA3.1/Hygro-CAG. A PCR fragment containing the tTS cDNA with a 5′ BamHI and a 3′ KpnI site was amplified from pLVCT-tTRKRAB and ligated into pcDNA3.1/Hygro-CAG to generate pcDNA3.1/Hygro-CAG-tTS. The CAG promoter-tTS-polyA fragment was excised by digestion with BciVI and EcoRI, extracted by Qiaquick Gel Extraction kit (Qiagen, Hilden, Germany). The tTS transgenic mice were produced by microinjection of the 3.378-kb CAG-tTS-polyA fragment into the pronucleus of eggs as described (20). Transgenic founders were identified by PCR using the primer set tTS-F/tTS-R. The founders were mated with C57BL/6J and four independent lines were established. The detailed information of primers is described in the Supplementary Material. Unless otherwise specified, heterozygous transgenic mice were used for all described cross experiments.
Collection of embryos
The presence of the vaginal plug detection indicated E0.5d. Embryos and tissues were collected at the indicated days. Bright field microscopy was performed with a Nikon SMZ 1500 (Nikon, Melville, NY, USA) stereoscope. The fluorescence of embryos and tissues were directly visualized using an EGFP filter. Photographs were taken with a Nikon Digital Sight DS-U2 (Nikon) microscope camera.
Quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) and cDNA was synthesized with PrimeScript RT reagent Kit (Takara, Dalian, China). Quantitative real-time PCR was performed using a Mastercycler Realplex2 detection system (Eppendorf, Hamburg, Germany) and SYBR Premix Ex Taq mixture (Takara). Primer sequences were as follows: EGFP Forward: 5′-CCA GGA GCG CAC CAT CTT CTT CA-3′, Reverse: 5′-TGC CGT TCT TCT GCT TGT CG-3′; Nmyc Forward: 5′-AGC AGC ACA ACT ATG CTG-3′, Reverse: 5′-TTA GCA AGT CCG AGC GTG TTC-3′; Actin Forward: 5′-TAC CCA GGC ATT GCT GAC AGG-3′, Reverse: 5′-ACT TGC GGT GCA CGA TGG A-3′. For expression analysis, all samples were normalized to the Actin signal.
Western blotting
Whole embryos were homogenized with glass homogenizers in RIPA buffer containing protease inhibitor cocktail (Sigma-Aldrich). Lysates were incubated for 10 min on ice and centrifuged at 13 000g for 10 min at 4°C. The supernatant was collected, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences, Piscataway, USA). The membranes were blocked with TBST buffer containing 5% fat-free milk powder for 1 h at room temperature, and then incubated overnight at 4°C with the primary antibodies against GFP (1:1000; Roche, Mannheim, Germany), TetR (1:1000; Mobitec, Göttingen, Germany), and Gapdh (1:10 000; KangChen Biotech, Shanghai, China). After washing, membranes were incubated with fluorescent conjugated secondary antibody for 1 h (1:10 000; LI-COR Biosciences, Lincoln, USA). The protein bands of interest were analyzed with Odyssey Infrared Imaging System (LI-COR).
Histological analysis
Whole brains were fixed in 4% paraformaldehyde for 24 h and embedded in paraffin. The samples were sagittally sectioned (3 µm), stained with hematoxylin and eosin (H&E) stain, and examined with Nikon Eclipse 90i microscope.
Micro-CT imaging of mouse limb microarchitecture
After anesthetization, the hindlimb of mice were examined with an eXplore Locus Micro CT Scanner (GE Healthcare, London, Canada). Imaging was obtained at 80 kV and 450 µA with a focal spot of 45 µm. CT images were reconstructed with eXplore Lucus Microview Software (version 2.2) (GE Healthcare).
Statistical analysis
The data from same litters were analyzed. Data are expressed as mean ± SEM. The statistical difference between groups was analyzed using a one-way analysis of variance (ANOVA) and a P < 0 05 was considered significant for all statistical analyses.
RESULTS
tTS regulating endogenous gene expression
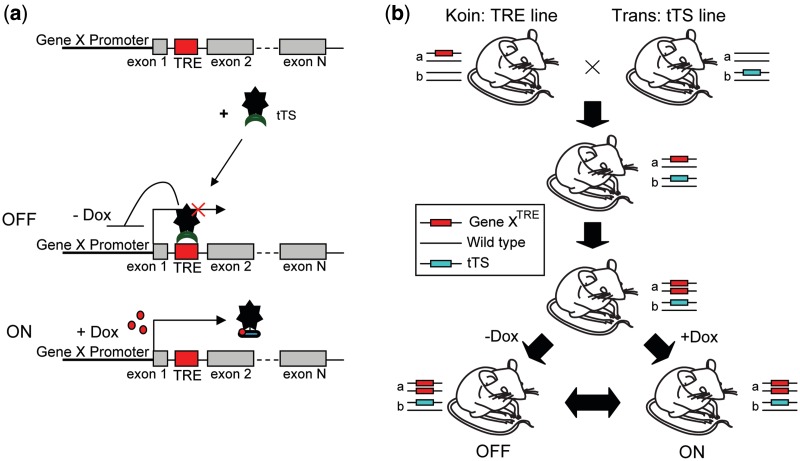
Figure 1 elucidates how the tTS–dox system regulates endogenous gene expression. There are three elements in the system (Figure 1a): dox, tTS and the endogenous target gene X with a TRE sequence. To generate a TRE knockin mouse line, the TRE site was inserted into the intron of target gene X by homologous recombination. In the tTS transgenic mouse line, the tTS is regulated by a ubiquitous or tissue-specific promoter. After these two lines are crossbreed, offspring that are homozygous for Gene XTRE and heterozygous for the tTS transgene have gene X expression regulated by tTS using dox as an inducer (Figure 1b).
Figure 1.
Schematic diagram of the genetic modification system for the regulation of endogenous gene. (a) Schematic representation of the principle of regulation model. (b) Generation of the inducible mouse by crossing the TRE line and tTS line. The inducible mouse is homozygous for the Gene XTRE locus and carries one copy of tTS allele. The expression of Gene X is driven by endogenous promoter of Gene X in the presence of dox and is repressed when tTS binds to TRE site in the absence of dox.
Construction of NmycTRE−EGFP-Neo mouse
To evaluate how well tTS can function as a regulator of endogenous mouse genes, Nmyc was chosen as target gene X. The tTS expressing in transgenic mice was driven by a CMV early enhancer/chicken β-actin (CAG) promoter (Figure 2a). One line with ubiquitous tTS expression was named ‘tTS’ (Figure 2e) and was selected for further study. The transgenic mice had no obvious abnormalities and were fertile.
To reduce the risk of interfering with the normal transcription of Nmyc by the inserted TRE site, TRE site was inserted into the first intron with less potential transcription factor-binding sites that were identified by JASPAR database analysis. To further confirm the safety of TRE insertion, the transcriptional activity of Nmyc promoter including the first intron was assayed by a luciferase reporter system, as described in Supplementary Figure S1a. No significant difference was observed after TRE site was inserted into the first intron (Supplementary Figure S1b). To generate Nmyc TRE knockin mice (NmycTRE−EGFP-Neo, abbreviated as NmycEGFP), a TRE–loxP–Nmyc first intron–enhanced green fluorescent protein (EGFP)–Flippase Recognition Target (FRT)–neomycin resistance gene (Neo)–FRT–loxP cassette was inserted into the first intron of the murine Nmyc gene (Figure 2b). To monitor the transcriptional activity of the Nmyc promoter regulated by tTS, an EGFP cassette was fused in frame into Nmyc exon 2 downstream of the first three amino acid coding sequence (ATGCCCAGC). NmycEGFP positive ES cells were identified by PCR (Figure 2c) and confirmed by DNA sequencing. Three positive ES cell clones were used to generate chimeric NmycEGFP founder mice. The offspring of these founder mice displayed robust EGFP expression in E 12.5d embryos (Figure 2d).
Dox-dependent on/off regulation of EGFP expression
NmycEGFP mice were crossed with tTS transgenic mice, and the offspring were genotyped by PCR (Supplementary Figure S2a). In offspring that developed without exposure to dox, EGFP expression measured by real-time PCR in NmycEGFP:tTS embryos was ∼0.3% of the levels in NmycEGFP embryos, suggesting that the Nmyc promoter activity in NmycEGFP:tTS embryos was tightly repressed by tTS (Figure 3a). In the offspring of pregnant females exposed to dox-containing (2 mg/ml) drinking water for 10 days beginning at E0.5d, there was no significant difference of EGFP expression level between NmycEGFP:tTS embryos and NmycEGFP embryos at E10.5d, indicating that the tTS regulation of Nmyc promoter activity was abolished by dox (Figure 3a). These results were confirmed by western blotting and an EGFP fluorescence assay (Figure 3b and c). In embryos that developed in the presence of dox, western blotting yielded an obvious EGFP fragments, and robust EGFP fluorescence in whole embryos was observed in both NmycEGFP embryos and NmycEGFP:tTS embryos. By contrast, in embryos with no exposure to dox, neither the EGFP band nor the EGFP fluorescence was observed in NmycEGFP:tTS embryos. These results indicated that Nmyc gene promoter could be accurately controlled by tTS–dox system.
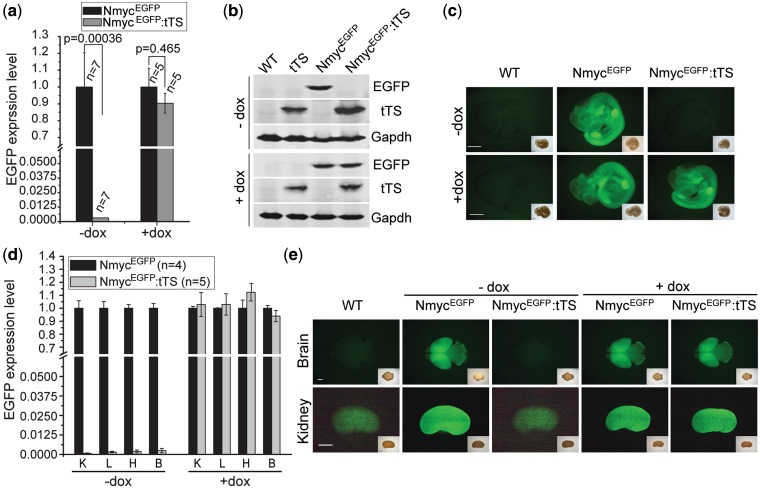
Figure 3.
Dox-dependent on/off regulation of EGFP expression. Embryos were the offspring of a cross of NmycEGFP and tTS transgenic mice. Pregnant mice were reared on drinking water containing 0 mg/ml dox (−dox) or 2 mg/ml dox (+dox). (a) Quantitative real-time PCR of EGFP expression in NmycEGFP and NmycEGFP:tTS E10.5d embryos. (b) Western blots of EGFP expression in NmycEGFP and NmycEGFP:tTS E10.5d embryos. (c) EGFP fluorescence of E10.5d embryos. The insets are bright field images. (d) Real-time PCR quantification of EGFP expression at 2 days postnatal (K: kidney; L: lung; H: heart; B: brain). (e) EGFP fluorescence in brain and kidney of postnatal 2 days mice (a feebly background fluorescence was observed in the kidney of wild-type mice.) The insets are bright field images. Scale bar is 1000 µm.
To test the feasibility of extending this system to other developmental stages and tissues, EGFP expression was analyzed in the brain, heart, lung and kidney of mice at postnatal day 2. In embryos without dox exposure, EGFP expression in these tissues from NmycEGFP:tTS mice were all <0.25% (P < 10−6) of the levels observed in the NmycEGFP mice (Figure 3d), and no EGFP fluorescence was observed in the tissues from NmycEGFP:tTS mice (Figure 3e). In the NmycEGFP:tTS mice exposed to dox beginning at E0.5d, either EGFP mRNA level or EGFP fluorescence intensity was not significantly different in the tissues from both genotypes.
Dox-dependent reversible regulation of EGFP expression
To determine whether the dox-dependent regulation was reversible, pregnant mice were exposed to dox-containing (2 mg/ml) drinking water for 10 days beginning at E0.5d, followed by no exposure to dox for 14 days, and followed by access to dox-containing drinking water again for 7 days (Figure 4a). In the presence of dox, the Nmyc promoter was in the ‘on’ state. After eliminating dox for 2 weeks, EGFP expression in brain, heart, lung and kidney of NmycEGFP:tTS mice measured by real-time PCR were all < 0.7% (P < 10−4) of the expression levels in NmycEGFP mice (Figure 4b). These results indicated that the transcriptional activity of Nmyc in the brain, heart, lung and kidney can switch from the ‘on’ state to the ‘off’ state. The ‘off’ state was reversed back to the ‘on’ state after further exposure to dox-containing drinking water for 7 days (Figure 4b), and the EGFP expression level in NmycEGFP:tTS mice increased to similar levels in NmycEGFP mice. The switch of the on/off status was also observed by changes in EGFP fluorescence in the brain of NmycEGFP:tTS mice (Figure 4c).
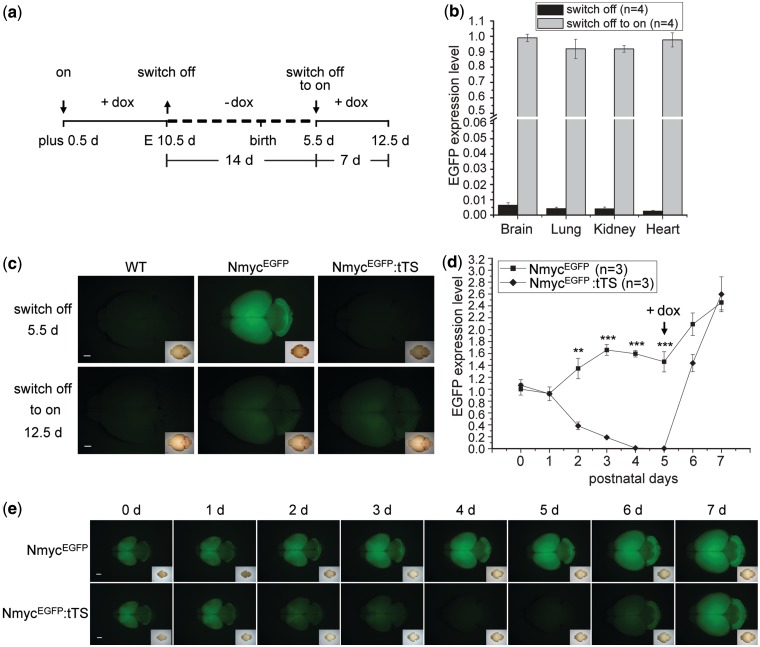
Figure 4.
Dox-controlled reversible regulation of EGFP expression. (a) Schedule of dox introduction to or removal from the drinking water of pregnant mice from the NmycEGFP and tTS cross. The symbols of ‘↓’ and ‘↑’ together with the terms of ‘on’, ‘switch off’ and ‘switch off to on’ indicate the action of adding and removing dox, respectively, at the corresponding time point and its resulting effect. (b) Real-time PCR of the EGFP expression in NmycEGFP:tTS mice of postnatal 5.5d and 12.5d before and after dox retreatment. The EGFP expression level in NmycEGFP:tTS mice was normalized to EGFP expression in NmycEGFP mice. EGFP expression in the NmycEGFP:tTS mice was switched off after dox was removed for 2 weeks and switched off to on after dox was administered for 1 week. (c) Fluorescence of EGFP expression in the brain of NmycEGFP:tTS mice is detected reversibly by removing and adding dox. (d) Time course of dox-controlled reversible EGFP expression by real-time PCR measurement. 0 day means the birth day. Note the expression of EGFP is still on at this time point although the dox was withdraw at E10.5d. (e) Time course of dox-controlled reversible EGFP expression by EGFP fluorescence detection. Scale bar is 1000 µm.
The time-course of dox-induced EGFP expression (Figure 4a) in NmycEGFP:tTS mice was measured by real-time PCR in brain from birth to postnatal 7 days. At postnatal days 0 and 1, EGFP expression in the brain of NmycEGFP:tTS pup mice was similar with that of NmycEGFP mice. At postnatal day 4, EGFP expression sharply decreased to 0.6% relative to that of NmycEGFP mice (Figure 4d). After exposing lactating female mice to dox-containing water at postnatal day 5, EGFP expression in the brain of NmycEGFP:tTS mice recovered to a similar level of NmycEGFP mice in 2 days (Figure 4d). This decrease and recovery was confirmed by EGFP fluorescence in the brain of NmycEGFP:tTS mice (Figure 4e). These experiments showed that the tTS on/off switch regulating EGFP expression in NmycEGFP:tTS mice could be flipped after dox was removed at least 13 days, and EGFP expression could be fully restored after dox was supplied for 2 days.
Dox-induced reversible regulation of endogenous Nmyc gene
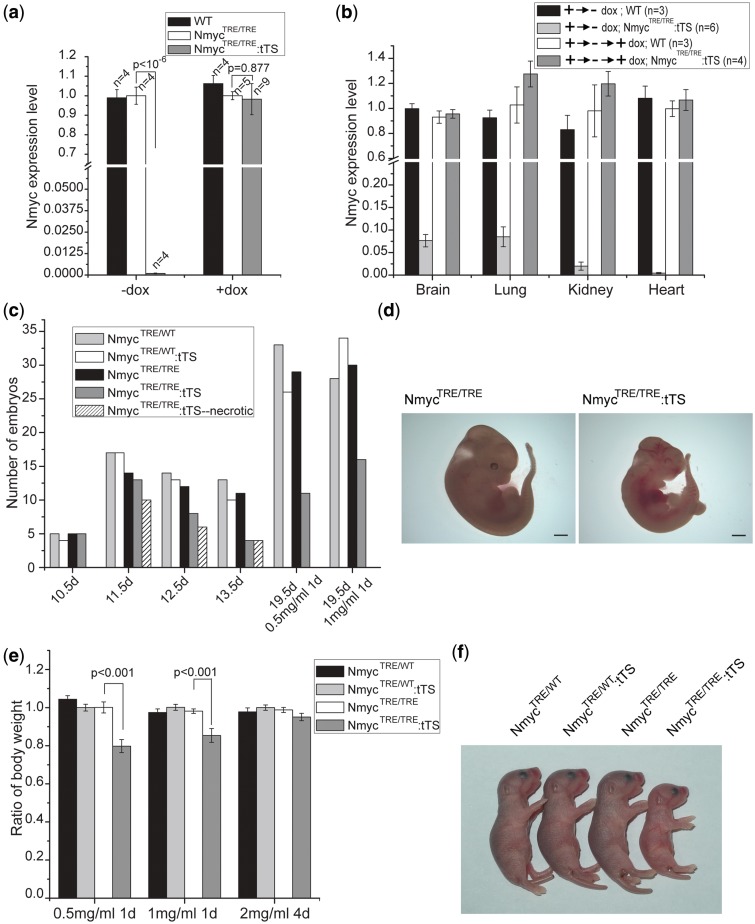
When NmycEGFP mouse was crossed with EIIA-Cre mouse, the Nmyc first intron-EGFP-FRT-NeoR-FRT fragment between the two loxP sites was excised, and NmycTRE/WT mice were generated. NmycTRE/WT mice were crossed with NmycTRE/WT:tTS mice to generate NmycTRE/TRE and NmycTRE/TRE:tTS mice. Genotypes were confirmed by PCR (Supplementary Figure S2b). In embryos without exposure to dox, Nmyc expression in NmycTRE/TRE:tTS E10.5d embryos was ∼0.1% of the levels in NmycTRE/TRE E10.5d embryos (Figure 5a). In embryos grown with prenatal exposure to dox, Nmyc expression was similar in both types of embryos (Figure 5a). Regardless of dox exposure, Nmyc expression in NmycTRE/TRE embryos was not significantly different from wild-type embryos (Figure 5a), indicating that the knockin of the TRE site did not affect Nmyc gene expression. Using the same dox cycle previously described (as in Figure 4a), the reversibility of Nmyc regulation was confirmed (Figure 5b). But some leaky expression of Nmyc in the brain and lung of NmycTRE/TRE:tTS mice was observed after removing dox for 2 weeks, which were, respectively, 7.6% and 8.5% of the levels in NmycTRE/TRE mice.
Figure 5.
Dox-controlled inducible and reversible regulation of endogenous Nmyc expression. (a) Real-time PCR quantification of endogenous Nmyc expression in E10.5d embryos of wild-type, NmycTRE/TRE and NmycTRE/TRE:tTS whose mothers reared on drinking water containing 0 mg/ml dox (−dox) or 2 mg/ml dox (+dox). (b) Real-time PCR measurement shows dox-dependent reversible regulation of endogenous Nmyc expression in NmycTRE/TRE:tTS mice. The pregnant mice were exposed to dox-containing (2 mg/ml) drinking water for 10 days beginning at E0.5d, followed by no exposure to dox for 14 days (+→− dox group), and followed by access to dox-containing drinking water again for 7 days (+→-→+ dox group). Nmyc expression was normalized to NmycTRE/TRE levels. (c) NmycTRE/TRE:tTS embryos whose mothers without dox exposure died at E11.5d. However, prenatal exposure to dox for at least 1 day led to NmycTRE/TRE:tTS embryo survival. (d) NmycTRE/TRE and NmycTRE/TRE:tTS embryo from same litter at E11.5d from a mother without dox exposure. (e) Differences in body weight were observed in NmycTRE/TRE:tTS mice depended on the different dose and duration of dox treatment on their mother. Body weight was normalized to NmycTRE/WT:tTS (n = 13–24 for each genotype in the different exposure duration groups). (f) NmycTRE/TRE:tTS mice were smaller at 2 days postnatal than littermates of other genotypes. The mother had been exposed to drinking water containing 0.5 mg/ml dox for 1 day at E0.5d. Scale bar is 1000 µm.
Blocking of Nmyc expression results in embryonic lethality in NmycTRE/TRE:tTS mice
It is known that the loss of Nmyc results in embryonic lethality and Nmyc−/− embryos die prenatally at E11.5d (15,16). To verify this functional effect, NmycTRE/WT:tTS mice were interbred and the genotypes of newborn pups were analyzed (Table 1). As expected (Supplementary Figure S3a), in the absence of prenatal dox exposure, no NmycTRE/TRE:tTS mice were obtained in a total of 162 live births, which included 6.8% NmycTRE/TRE mice. In the presence of dox, the expected proportion of NmycTRE/TRE:tTS mice were observed. These results demonstrated that in the absence of dox, the Nmyc expression is blocked in NmycTRE/TRE:tTS embryo, thus resulting the embryo death.
Table 1.
Observed and expected genotypic frequency from a cross between NmycTRE/WT:tTS individuals reared in the presence and absence of dox
| Group (No. of progeny) | Genotypic frequency of surviving offspring (%) |
|||||
|---|---|---|---|---|---|---|
| NmycWT/WT |
NmycTRE/WT |
NmycTRE/TRE |
||||
| tTS (−) | tTS (+) | tTS(−) | tTS(+) | tTS(−) | tTS(+) | |
| Expected (%) | 6.25 | 18.75 | 12.5 | 37.5 | 6.25 | 18.75 |
| −Dox (N = 162) | 8 (4.9) | 39 (24.1) | 23 (14.2) | 81 (50) | 11 (6.8) | 0 (0) |
| +Dox (N = 126) | 11 (8.7) | 22 (17.5) | 18 (14.3) | 48 (38.1) | 6 (4.8) | 21 (16.7) |
To further investigate if the NmycTRE/TRE:tTS embryos died at the same stage as Nmyc−/− embryos, we genotyped embryos from E10.5d to E13.5d from the pregnant mice mating by NmycTRE/TRE and NmycTRE/WT:tTS mice in the absence of prenatal dox exposure (Figure 5c). At E10.5d, no dead embryos were found, and the genotypic ratio was consistent with Mendelian patterns (Supplementary Figure S3b). Beginning at E11.5d, 16.4% of the embryos were dead or resorbed. Genotyping confirmed that they were all NmycTRE/TRE:tTS embryos (Figure 5d). By E13.5d, no live NmycTRE/TRE:tTS embryos were found. Thus, in the absence of prenatal exposure to dox, NmycTRE/TRE:tTS mice exhibited the embryonic lethality at same stage observed in conventional Nmyc−/− knockout mice.
Dox administration rescues the embryonic lethality of NmycTRE/TRE:tTS mice
We investigated how long dox administration could rescue the embryonic lethality of NmycTRE/TRE:tTS mice. Living NmycTRE/TRE:tTS pups could be found when the pregnant mice were fed drinking water containing dox (0.5 or 1 mg/ml) only for a single day beginning at E0.5d (Figure 5c). However, 31% or 21% NmycTRE/TRE:tTS pups died within 2 days after birth for prenatal exposure to 0.5 or 1 mg/ml dox at E0.5d, respectively, the surviving ones were significantly smaller by body mass compared with offspring from other three genotypic classes (Figure 5e and f). No obvious reduced body weight or postnatal death of NmycTRE/TRE:tTS mice was observed if the pregnant mice were fed drinking water containing 2 mg/ml dox for 4 days beginning at E0.5d (Figure 5e).
Limited Dox administration results in cerebellar developmental defects and syndactyly
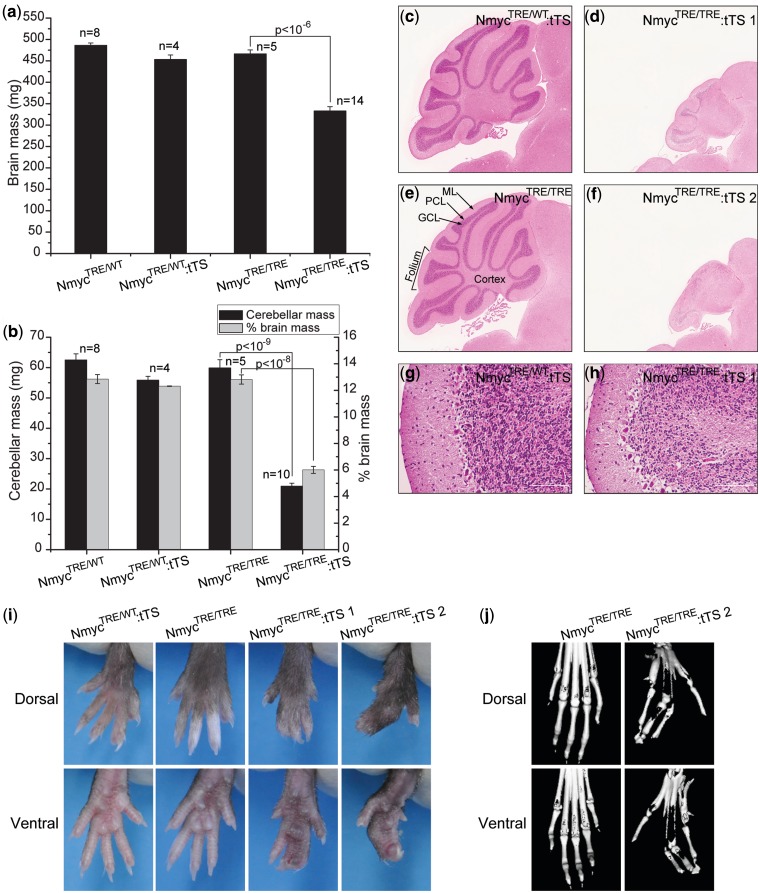
To determine whether mice generated with this method would present the same phenotypes observed in the Nmyc conditional knockout model, we monitored the phenotypes of surviving NmycTRE/TRE:tTS mice whose dam was exposed to drinking water containing 1 mg/ml dox for 1 day at E0.5d. At 12 weeks, NmycTRE/TRE:tTS mice exhibited microencephaly with a significant reduction in absolute brain mass (28.6%) and brain mass relative to total body weight (14.4%) compared with NmycTRE/TRE mice (Figure 6a). This reduction was most severe in the cerebellum, which had a significant reduction in absolute mass (65%) and the ratio relative to brain mass (53.1%) (Figure 6b). Histological analysis of these cerebellums revealed defects in foliation and reduced granular cell density (Figure 6c–h). In addition, NmycTRE/TRE:tTS mice showed different degrees of syndactyly (Figure 6i). Based on micro-CT scanning of limbs, the digit skeleton of limbs in NmycTRE/TRE:tTS mice kept individualization (Figure 6j), indicating that the syndactyly was due to the failure of separation of soft tissue between adjacent fingers.
Figure 6.
Dox-controlled switch off Nmyc leads to cerebellar developmental defects and syndactyly. (a) Absolute brain mass in four genotypic classes of mice. (b) Absolute cerebellar mass and ratio of cerebellar mass to total brain mass in four genotypic classes of mice. (c–h) Representative hematoxylin and eosin (H&E) staining of cerebellum from 12-week mice from three genotypic classes. c, e: control cerebellums. ML, molecular layer; PCL, Purkinje cell layer; GCL, granular cell layer. d, f: cerebellums from NmycTRE/TRE:tTS mice. g, h: Folia of NmycTRE/WT:tTS mouse and NmycTRE/TRE:tTS mouse. (i) Dorsal and ventral views of hindlimbs of 12-week mice from different genotypic classes. (j) Dorsal and ventral 3D micro-CT reconstructions of hindlimbs. Panels c–f were magnified 4×. Scale bar is 100 µm.
Dox adminstration regulates the severity of cerebellar developmental defects
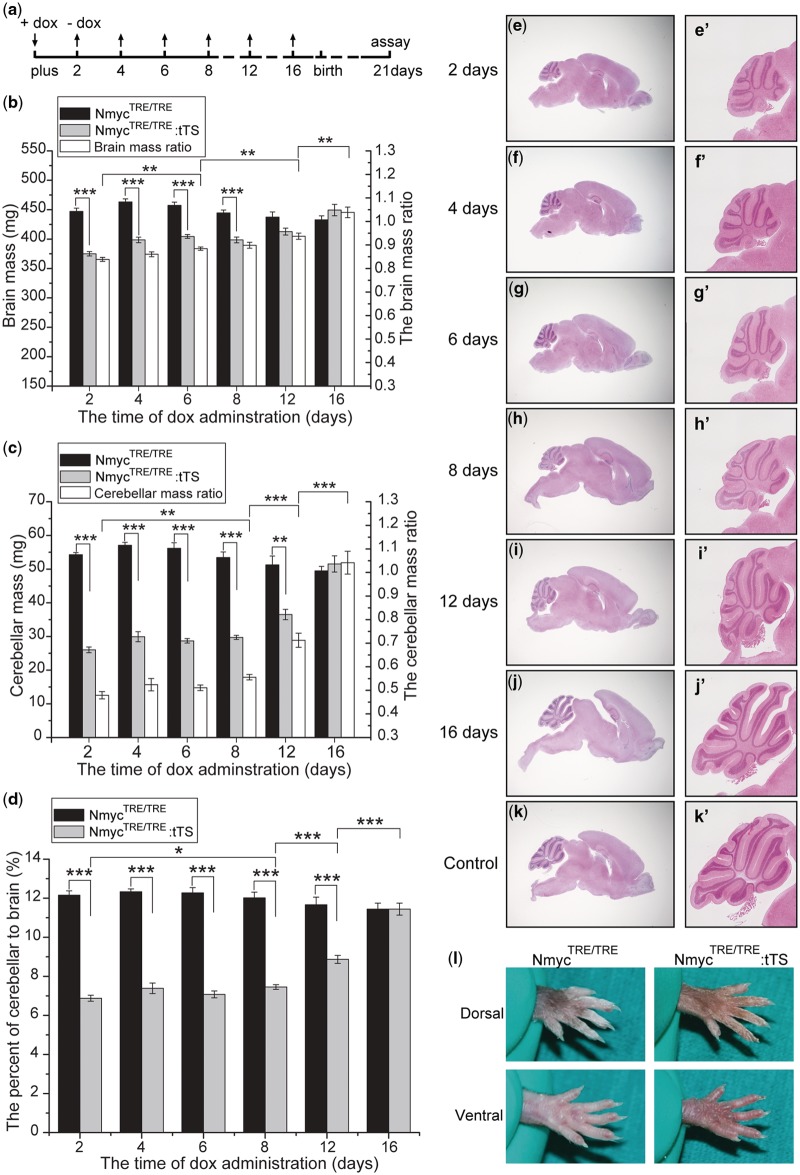
To determine if this method could be used to explore Nmyc gene function at different developmental stages by changing the exposure time to dox, NmycTRE/TRE:tTS mice were mated with NmycTRE/TRE mice and the pregnant mice were exposed to 2 mg/ml dox-containing drinking water for different durations (Figure 7a) beginning at E0.5d. At postnatal day 21, absolute brain mass was significantly reduced in NmycTRE/TRE:tTS mice whose dam was exposed to dox for <8 days compared with littermates (Figure 7b). Absolute cerebellar mass and the ratio of cerebellar mass to total brain mass were significantly reduced in NmycTRE/TRE:tTS mice whose dam was exposed to dox for <12 days (Figure 7c and d). The absolute brain mass, cerebellar mass and the ratio of cerebellar mass to total brain mass increased significantly along with prolonged dox exposure time. These differences were confirmed by histological analysis of the brain (Figure 7e–k, e′–k′). These results suggest that Nmyc plays an important role in cerebellar formation during late development. Although no obvious syndactyly phenotype in offspring was observed, when the dam were feeded by 2 mg/ml dox in drinking water just for 2 days (Figure 7l). Our results suggest that this method can be used to study the time window of Nmyc function for a specific developmental event.
Figure 7.
Dox-regulated severity of cerebellar developmental defects. (a) Dox was added or removed from the drinking water of pregnant female mice at the indicated time points. (b–d) Absolute brain mass, absolute cerebellar mass and the ratio of cerebellar mass to total brain mass in mice exposed to dox for different durations (n = 5–9 for each genotype in the different exposure duration groups). Ratios in b and c are the ratios of total brain and cerebellar mass in NmycTRE/TRE:tTS mice to the NmycTRE/TRE mice with the same dox exposure. (e–k) Representative H&E staining of brain in mice exposed dox for different durations; (e′–k′) magnification of the cerebellum in e–k. e–j: brains from NmycTRE/TRE:tTS mice; k: the brain of NmycTRE/TRE mice. (l) Dorsal and ventral views of hindlimbs of 24-week mice from different genotypic classes with exposure to 2 mg/ml dox for 2 days since pregnancy (ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001).
The expression of N-myc during the postnatal day 2–11 determines postnatal cerebellar development
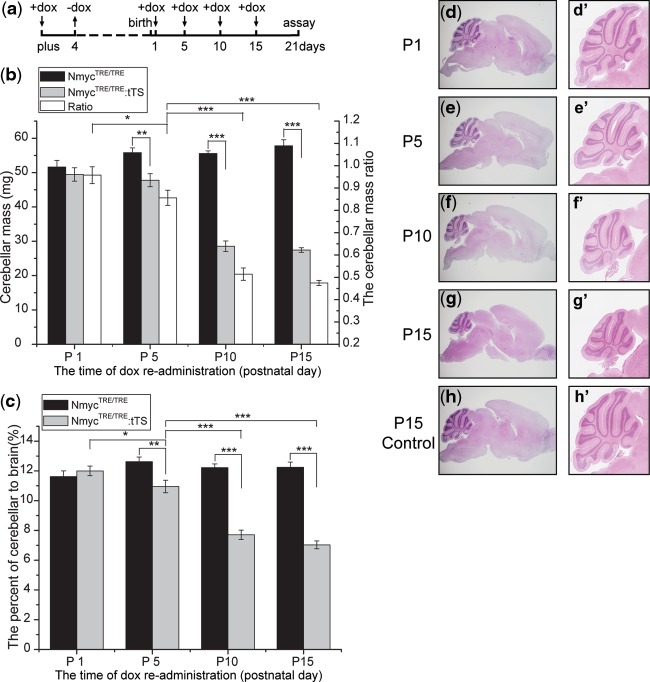
To determine the time window of Nmyc functions in postnatal cerebellar development, a series of reversible regulation of Nmyc were performed at different time points after birth. After mating between NmycTRE/TRE:tTS mice and NmycTRE/TRE mice, the pregnant mice were exposed to 2 mg/ml dox-containing drinking water at E0.5d for 4 days followed by removing the drug for a certain time (Figure 8a), and then exposed to dox again until postnatal day 21. At postnatal day 21, the absolute cerebellar mass and the ratio of cerebellar mass to total brain mass had no significant reduction in NmycTRE/TRE:tTS mice whose dam was exposed to dox again since postnatal day 1,while reduction of cerebella mass was observed if the dam was exposed to dox again since postnatal day 5 (Figure 8b and c). However, much serious abnormality was found in cerebellar of NmycTRE/TRE:tTS mice if their dam was not exposed to dox again, and such a deficiency cannot be rescued if the dox was given at postnatal day 10. These data were confirmed by histological analysis of the brain (Figure 8d–h, d′–h′). Considering that there were 1–2 days interval from dox readministration to Nmyc re-expression, our results indicated that Nmyc plays an important role in postnatal cerebellar development, and the functional time window located between postnatal days 2 and 11.
Figure 8.
Analysis of Nmyc functional time window in postnatal cerebellar development. (a) Dox was added or removed from the drinking water of pregnant female mice at the indicated time points. (b–c) Absolute cerebellar mass and the ratio of cerebellar mass to total brain mass in mice exposed to dox for different durations (n = 4–6 for each genotype in the different exposure duration groups). Ratios in b are the ratios of cerebellar mass in NmycTRE/TRE:tTS mice to the NmycTRE/TRE mice with the same dox exposure. (d–h) Representative H&E staining of brain in mice exposed dox for different durations; (d′–h′) magnification of the cerebellum in d–h. d–g: brains from NmycTRE/TRE:tTS mice; h: the brain of NmycTRE/TRE mice (ANOVA; *P < 0.05; **P < 0.01; ***P < 0.001).
DISCUSSION
We developed a genetic modification strategy based on the tTS–dox system to inducibly and reversibly regulate endogenous genes in mice. To test the efficiency of the system, Nmyc was selected as the target gene. The insertion of a TRE site into the first intron of Nmyc gene was proved not to affect the normal phenotype of the mice, as Nmyc expression in NmycTRE/TRE mice was the same as that in wild-type mice. When these mice were crossed with tTS transgenic mice, Nmyc expression was turned off by the tTS in NmycTRE/TRE:tTS embryos. Loss of Nmyc expression resulted in embryonic lethality at E11.5d, which has been observed in conventional Nmyc knockout mice (15,16). The lethality phenotype was rescued when the dam was exposed to drinking water containing dox at the beginning of the pregnancy. Variation in the duration and dosage of the dox administration led to different degrees of severity of phenotypes, including cerebellar development defects and syndactyly. These phenotypes have also been observed in conditional Nmyc knockout mice (17,18). It is worth to note that increasing dox administration converted the embryonic lethal phenotype into a deficient phenotype spatially restricted to the brain and limbs and finally to a normal phenotype. Although we did not investigate the development situation of all other organs very carefully, the deficiency in brain and limbs is no doubt the most obvious. Intriguingly, the syndactyly phenotype could be rescued when the dam was exposed to dox just 2 days from the beginning of pregnancy. However, the expression of Nmyc during the postnatal days 2–11 is also important for the cerebellar development. This finding is in agreement with previous reports that a massive proliferation undergoes at the external granule layer of cerebellar cortex after birth with the peak at postnatal days 5–8 and then declined (21,22). These results suggested that the essential role of Nmyc to determine digit individualization took place at early stage of development and in a relative sharp time window while the expression of Nmyc during late stage of development after birth played key role to determine cerebellar formation. Thus, this method can be used to evaluate and compare the sensitivity of different organ development to the target gene dosage. Moreover, it also can be applied to explore the role of target gene within a specific time window. These advantages are not with the strategy of conditional knockout and other traditional methods.
To address whether or not the tTS–dox switch could repeatedly be turned on and off, we inserted an EGFP expression cassette immediately downstream of Nmyc translation initiation site in exon 2 in the NmycEGFP:tTS mice. With EGFP as the reporter of Nmyc expression, we evaluated how EGFP was regulated by the tTS–dox system. The fluorescence distribution of EGFP was consistent with the previous reports of Nmyc expression by in situ hybridization (23–26). In NmycEGFP:tTS mice, tTS inhibited EGFP expression in multiple tissues with an efficiency between 99% and 100%. The difference in inhibition may reflect a difference in tTS expression between tissues. Exposure to dox rescued the EGFP expression levels. However, the expression levels can be reduced to pre-exposure levels by discontinuing dox exposure. For dox-induced EGFP expression changes, the switch from ‘on’ to ‘off’ required longer time than that from ‘off’ to ‘on’. The on-to-off time likely reflects the time required to fully metabolize all the dox in body, but the detailed mechanism needs to be explored further. For relative long time to switch from ‘on’ to ‘off’, there are some limitations in application of this system to reversible regulation of gene expression in early development cases. We are trying to develop rtTS system, utilizing the rtTS to replace tTS. This replacement may allow us to keep gene normal expression in the absence of dox, switch off gene expression in the presence of dox (6,27). The rtTS may permit us to switch off gene by adding dox and apply to early development cases.
Although tTS regulated Nmyc expression as expected, it is important to understand how the tTS binding to the TRE in Nmyc affected nearby genes. As a transcriptional repressor, tTS regulated gene expression by an epigenetic mechanism. When tTS binds to DNA, the tTS KRAB domain recruits a multimolecular complex that deacetylates and methylates histones and binds to heterochromatin protein1β (HP1β). As a result, a local heterochromatin state extends a radius of 2–3 kb around binding sites (6,28,29). In vitro, KRAB and its corepressor KAP1 can silence promoters that are tens of kilobases away from their DNA binding sites (30). Here, Rpl36, the closest gene to Nmyc, did not exhibit significant differences in expression level between exposure and no exposure to dox (Supplementary Figure S4). Due to the distance effect of tTS on the transcription suppression, the insertion of TRE site was limited to a radius of 2–3 kb around transcription start site, which always included 2- to 3-kb promoter region, the first exon and intron. DNase I hypersensitive site mapping analysis indicated that nearly half of the most open sites to DNase I were in promoters and first exons of known genes, which were the location rich of genetic regulatory elements (31). To reduce the risk of interference with the normal activity of promoter, we analyzed the distribution of transcription factors around the promoter region of Nmyc gene. The first intron was chose in our case to insert TRE site. It is worth to note that intron was a choice but not the absolute option. With increasing of high-throughput DNase I-Seq data and the completion of ENCODE project (32), a comprehensive knowledge of regulatory elements in genome will be built, which will give us more guidelines for the choice of TRE insert site. The risk of insertion element to interfere the promoter activity was also existed in traditional knockout and conditional knockout strategy.
While this article was in preparation, two other studies based on the tTS system were reported by Tanaka et al. (12) and Richardson et al. (13). Comparing with their reports, our study provided a more detailed evaluation of the system. We also successfully used the same strategy to regulate Hprt (a house keeping gene) and Kif18a (a kinesin-like gene) gene expression in different tissues and different development stage including adult (data not shown). So our data and together with previous reports suggested that tTS system could be a general method to inducibly and reversibly regulate an endogenous gene in mouse. We also noticed a powerful system to tissue-specific and inducible control of gene expression in transgenic mice reported by Ko et al., to control gene expression in a tissue-specific and inducible manner in transgenic mice, which was based on the Cre-LoxP system and RNA interference (RNAi) (7).This system relied on the Cre recombinase to tissue-specific expression of short-hairpin RNA and used Tet-on system to control the inducible expression. However, this system also possessed the limitations of transgene and RNAi system, such as laborious screening of ideal founder, shRNA offtarget effect and toxicity. Comparing the currently used methods such as CreER2T and the system based on RNAi, each method strategy had advantages and limitations. tTS system can act as a complementarity and give us more choices to select a suitable method in life science research.
In summary, the genetic manipulation method based on tTS–dox system in this article provides a very powerful strategy to regulate a selected endogenous gene in mouse. In our case, the efficiency to inhibit the target gene Nmyc is nearly the same as the strategy of conditional gene knockout while our method has the advantage over it in some applications, especially, when reversible regulation or temporary shutdown of the target gene is needed in the research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4 and Supplementary Material and Methods.
FUNDING
The National Key Project [2010CB945501, 2010CB912604]; the National Natural Science Foundation of China [81171188]; the Science and Technology Commission of Shanghai Municipality [08140900700, 08140901900, 10DZ2251500] the E-Institutes of Shanghai Municipal Education Commission [E03003]. Funding for open access charge: Science and Technology Commission of Shanghai Municipality [08140900700].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Huimin Yan for the assistance on micro CT scanning, Chen Chen for the help to take photos and Wenting Wu for the help to draw the schematic pictures.
REFERENCES
- 1.Zeng H, Horie K, Madisen L, Pavlova MN, Gragerova G, Rohde AD, Schimpf BA, Liang Y, Ojala E, Kramer F, et al. An inducible and reversible mouse genetic rescue system. PLoS Genet. 2008;4:e1000069. doi: 10.1371/journal.pgen.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71–80. doi: 10.1006/meth.2001.1159. [DOI] [PubMed] [Google Scholar]
- 3.Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 4.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat. Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson JD, Chen YN, Cornell-Kennon SA, Dorsch M, Kim S, Leszczyniecka M, Sellers WR, Lengauer C. Validating cancer drug targets. Nature. 2006;441:451–456. doi: 10.1038/nature04873. [DOI] [PubMed] [Google Scholar]
- 6.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat. Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 7.Ko JK, Choi KH, Zhao X, Komazaki S, Pan Z, Weisleder N, Ma J. A versatile single-plasmid system for tissue-specific and inducible control of gene expression in transgenic mice. FASEB J. 2011;25:2638–2649. doi: 10.1096/fj.11-181560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 9.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z, Ma B, Homer RJ, Zheng T, Elias JA. Use of the tetracycline-controlled transcriptional silencer (tTS) to eliminate transgene leak in inducible overexpression transgenic mice. J. Biol. Chem. 2001;276:25222–25229. doi: 10.1074/jbc.M101512200. [DOI] [PubMed] [Google Scholar]
- 11.Mallo M, Kanzler B, Ohnemus S. Reversible gene inactivation in the mouse. Genomics. 2003;81:356–360. doi: 10.1016/s0888-7543(03)00032-6. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka KF, Ahmari SE, Leonardo ED, Richardson-Jones JW, Budreck EC, Scheiffele P, Sugio S, Inamura N, Ikenaka K, Hen R. Flexible Accelerated STOP Tetracycline Operator-knockin (FAST): a versatile and efficient new gene modulating system. Biol. Psychiatry. 2010;67:770–773. doi: 10.1016/j.biopsych.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson-Jones JW, Craige CP, Guiard BP, Stephen A, Metzger KL, Kung HF, Gardier AM, Dranovsky A, David DJ, Beck SG, et al. 5-HT1A autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron. 2010;65:40–52. doi: 10.1016/j.neuron.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurlin PJ. N-Myc functions in transcription and development. Birth Defects Res. C Embryo Today. 2005;75:340–352. doi: 10.1002/bdrc.20059. [DOI] [PubMed] [Google Scholar]
- 15.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 16.Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 17.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ota S, Zhou ZQ, Keene DR, Knoepfler P, Hurlin PJ. Activities of N-Myc in the developing limb link control of skeletal size with digit separation. Development. 2007;134:1583–1592. doi: 10.1242/dev.000703. [DOI] [PubMed] [Google Scholar]
- 19.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon JW, Ruddle FH. Gene transfer into mouse embryos: production of transgenic mice by pronuclear injection. Methods Enzymol. 1983;101:411–433. doi: 10.1016/0076-6879(83)01031-9. [DOI] [PubMed] [Google Scholar]
- 21.Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Curr. Opin. Neurobiol. 1997;7:40–47. doi: 10.1016/s0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- 22.Vaillant C, Monard D. SHH pathway and cerebellar development. Cerebellum. 2009;8:291–301. doi: 10.1007/s12311-009-0094-8. [DOI] [PubMed] [Google Scholar]
- 23.Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE, et al. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- 24.Mugrauer G, Alt FW, Ekblom P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. J. Cell Biol. 1988;107:1325–1335. doi: 10.1083/jcb.107.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Downs KM, Martin GR, Bishop JM. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989;3:860–869. doi: 10.1101/gad.3.6.860. [DOI] [PubMed] [Google Scholar]
- 26.Hirning U, Schmid P, Schulz WA, Rettenberger G, Hameister H. A comparative analysis of N-myc and c-myc expression and cellular proliferation in mouse organogenesis. Mech. Dev. 1991;33:119–125. doi: 10.1016/0925-4773(91)90078-k. [DOI] [PubMed] [Google Scholar]
- 27.Markusic D, Seppen J. Doxycycline regulated lentiviral vectors. Methods Mol. Biol. 2010;614:69–76. doi: 10.1007/978-1-60761-533-0_4. [DOI] [PubMed] [Google Scholar]
- 28.Moosmann P, Georgiev O, Thiesen HJ, Hagmann M, Schaffner W. Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Kruppel-type zinc finger factor. Biol. Chem. 1997;378:669–677. doi: 10.1515/bchm.1997.378.7.669. [DOI] [PubMed] [Google Scholar]
- 29.Senatore B, Cafieri A, Di Marino I, Rosati M, Di Nocera PP, Grimaldi G. A variety of RNA polymerases II and III-dependent promoter classes is repressed by factors containing the Kruppel-associated/finger preceding box of zinc finger proteins. Gene. 1999;234:381–394. doi: 10.1016/s0378-1119(99)00182-1. [DOI] [PubMed] [Google Scholar]
- 30.Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.