Abstract
Trans-acting small interfering RNAs (ta-siRNAs) are plant-specific siRNAs released from TAS precursor transcripts after microRNA-dependent cleavage, conversion into double-stranded RNA, and Dicer-dependent phased processing. Like microRNAs (miRNAs), ta-siRNAs direct site-specific cleavage of target RNAs at sites of extensive complementarity. Here, we show that the DICER-LIKE 4 protein of Physcomitrella patens (PpDCL4) is essential for the biogenesis of 21 nucleotide (nt) ta-siRNAs. In ΔPpDCL4 mutants, off-sized 23 and 24-nt ta-siRNAs accumulated as the result of PpDCL3 activity. ΔPpDCL4 mutants display severe abnormalities throughout Physcomitrella development, including sterility, that were fully reversed in ΔPpDCL3/ΔPpDCL4 double-mutant plants. Therefore, PpDCL3 activity, not loss of PpDCL4 function per se, is the cause of the ΔPpDCL4 phenotypes. Additionally, we describe several new Physcomitrella trans-acting siRNA loci, three of which belong to a new family, TAS6. TAS6 loci are typified by sliced miR156 and miR529 target sites and are in close proximity to PpTAS3 loci.
Keywords: ta-siRNA, miR156, PpDCL3, PpDCL4, development, moss
INTRODUCTION
In plants, different RNA silencing pathways regulate gene expression during development (Du and Zamore, 2005), control genome maintenance (Xie et al., 2004), and act in defense against biotic and abiotic stresses (Borsani et al., 2005; Deleris et al., 2006). For the regulation of these diverse processes, plants evolved complex small non-coding RNA (sRNA) pathways consisting of miRNA, ta-siRNA, natural antisense transcript-derived small interfering RNA (nat-siRNA), and repeat-associated small interfering RNA (ra-siRNA) (Vazquez, 2006). Most plant species possess a set of four Dicer proteins (Margis et al., 2006) with specific functions in the biogenesis of sRNAs from double-stranded RNA precursors. In Arabidopsis thaliana, DCL1 is required for the maturation of miRNAs from hairpin precursors (Park et al., 2002; Reinhart et al., 2002), DCL2 acts in the generation of siRNAs from exogenous RNA sources (Xie et al., 2004), DCL3 guides siRNA formation from heterochromatin-associated regions (Herr et al., 2005; Xie et al., 2004), and DCL4 acts in the formation of ta-siRNAs (Dunoyer et al., 2005; Xie et al., 2005). Although each Dicer protein is associated with a specific pathway, functional redundancies between the individual Dicers do exist (Gasciolli et al., 2005; Henderson et al., 2006; Fahlgren et al., 2009). The Physcomitrella genome also encodes four DCL proteins, but the DCL repertoire varies from that in the flowering plant Arabidopsis. Two Physcomitrella homologs, PpDCL1a and PpDCL1b, show high similarity to AtDCL1, PpDCL3 and PpDCL4 proteins are homologous to AtDCL3 and AtDCL4, respectively, whereas an AtDCL2 ortholog is missing (Axtell et al., 2007). PpDCL1a is essential for miRNA biogenesis and thus the functional equivalent to AtDCL1 whereas PpDCL1b is required for miRNA-directed cleavage of target RNAs (Khraiwesh et al., 2010). Unlike Arabidopsis DCL3, which produces exclusively 24-nt siRNAs (Xie et al., 2004; Gasciolli et al., 2005), PpDCL3 is required for the accumulation of 22–24-nt siRNAs (Cho et al., 2008). In both species, DCL3-dependent siRNAs arise preferentially from intergenic and repetitive genomic regions with dense cytosine methylation (Kasschau et al., 2007; Cho et al., 2008). Moreover, ΔPpDCL3 mutants display accelerated gametophore development, while Arabidopsis dcl3 mutants show no obvious developmental defects. Arabidopsis dcl2/dcl3 or dcl3/dcl4 double mutants show stochastic developmental defects that are more severe in dcl2/dcl3/dcl4 triple mutants, suggesting functional redundancy between these proteins (Gasciolli et al., 2005; Cho et al., 2008).
miRNAs and ta-siRNAs act by base-pairing to their target RNAs, thereby mediating cleavage or translational repression of the target (Qi and Hannon, 2005). Both miRNAs and ta-siRNAs are ∼21 nt in size and are nuclear-encoded, but they differ with respect to their biogenesis from RNA precursors (Vaucheret, 2006). Primary MIRNA transcripts have a characteristic stem-loop structure that is processed into mature miRNA. TAS precursor transcripts contain miRNA binding sites leading to miRNA-directed precursor cleavage, subsequent conversion of the cleavage products into dsRNA by RNA-DEPENDENT RNA POLYMERASE (RDR), and phased dicing of the dsRNA into ta-siRNAs by Dicer proteins (Allen et al., 2005; Talmor-Neiman et al., 2006).
In Arabidopsis, several components of the ta-siRNA biogenesis pathway were identified, including three miRNAs (miR173, miR390, and miR828) that direct site-specific cleavage of distinct TAS transcripts, RDR6 catalyzing the conversion into dsRNA, SUPPRESSOR OF GENE SILENCING 3 (SGS3) protecting the TAS precursors from degradation, ARGONAUTE 1/7 (AGO1/7) for miRNA-directed cleavage of TAS precursors, DICER-LIKE4 (DCL4) for the phased slicing of double-stranded TAS precursors, and DOUBLE-STRANDED RNA BINDING PROTEIN 4 (DRB4) that interacts with DCL4 for proper dicing activity (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Adenot et al., 2006).
In Arabidopsis, four TAS gene families (TAS1–4) have been identified. Ta-siRNA production from TAS1 and TAS2 requires miR173-directed cleavage, miR390 is dedicated to TAS3, and miR828 is assigned for TAS4 (Allen et al., 2005; Rajagopalan et al., 2006). In Arabidopsis, most miRNAs are incorporated into an AGO1-containing RNA-induced silencing complex (RISC) to direct cleavage of their targets, whereas miR390-dependent cleavage of TAS3 requires AGO7 activity (Montgomery et al., 2008). In Physcomitrella, ta-siRNAs originate from four TAS3 precursors (TAS3a–d) that possess miR390-dependent cleavage at two distinct sites spanning the region from which ta-siRNAs are produced (Axtell et al., 2006; Talmor-Neiman et al., 2006). Recently, a fifth TAS family (TAS5) was identified in tomato; sly-TAS5 is triggered by miR482-directed slicing (Li et al., 2012).
Physcomitrella ta-siRNAs regulate transcription factor transcripts encoding a single N-terminal AP2 domain and, similarly to Arabidopsis, mRNAs encoding ARF (Auxin Response Factor) transcription factors (Talmor-Neiman et al., 2006; Axtell et al., 2007). However, to date, a functional analysis of the DCL protein acting in the biogenesis of Physcomitrella ta-siRNAs is missing. The only study on Physcomitrella ta-siRNA biogenesis revealed an essential function of PpRDR6 that is required for the accumulation of ta-siRNAs. ΔPpRDR6 mutant lines lack ta-siRNAs and display an accelerated developmental transition from the juvenile (protonema) to the adult gametophyte (gametophore) (Talmor-Neiman et al., 2006).
Here, we demonstrate that ΔPpDCL4 loss-of-function mutants have dramatic developmental abnormalities. In the ΔPpDCL4 background, PpDCL3 processes TAS substrates into ‘off-sized’ 23–24-nt siRNAs. The ΔPpDCL4 developmental defects are fully ameliorated in ΔPpDCL3/ΔPpDCL4 double mutants, which completely lack TAS-derived sRNAs of all sizes. Therefore, an activity of PpDCL3, not the reduced effectiveness of ta-siRNA function, is responsible for the ΔPpDCL4 developmental phenotypes. We also identified a new family of TAS loci, PpTAS6, characterized by dual miR156 and/or miR529-mediated cleavage sites and close genomic proximity to PpTAS3 loci.
RESULTS
Loss of PpDCL4 Results in Severe Developmental Abnormalities
The functional characterization of the PpDCL4 gene was initiated by cloning and sequencing of its full-length cDNA, revealing that it contains all characteristic domains of Dicer proteins (Supplemental Figure 1A). Physcomitrella integrates DNA into its nuclear genome by means of homologous recombination at high frequencies (Strepp et al., 1998) enabling the targeted disruption of genomic loci. Making use of the efficient gene targeting, we generated four independent PpDCL4 null mutants (ΔPpDCL4) by the insertion of a zeocin selection marker cassette into the PpDCL4 genomic locus (Supplemental Figure 1B–1D). Two independent ΔPpDCL4 mutant lines that have integrated a single copy of the PpDCL4 gene disruption construct at the PpDCL4 genomic locus (Supplemental Figure 1E) were used for further analyses.
In Arabidopsis dcl4 mutants, the lack of ta-siRNAs only has minor developmental effects, namely the formation of downward curled and slightly elongated rosette leaf margins and accelerated juvenile-to-adult vegetative phase changes (Gasciolli et al., 2005; Xie et al., 2005). In contrast, Oryza sativa dcl4 mutants display developmental defects in the leaves, are semi-dwarfed, display a severe disturbance of spikelet organ identity, and are sterile (Liu et al., 2007). Comparable to rice dcl4 mutants, we observed severe developmental defects throughout the development of ΔPpDCL4 mutants. Under long-day conditions, ΔPpDCL4 mutants display an aberrant colony morphology characterized by a reduced size and a strongly increased development of filamentous protonema tissue (Figure 1A). Physcomitrella protonema tissue comprises two cell types: chloronema and caulonema cells. Chloronema cells are short, chloroplast-enriched, and characterized by cross walls that are perpendicular to the growth axis. Caulonema cells are elongated and thinner, they contain fewer chloroplasts, and their cross walls are oblique to the growth axis (Reski, 1998). Caulonema development can be triggered by growth in the dark (Vandenbussche et al., 2007). Wild-type plants cultivated for 8 d in complete darkness developed numerous caulonema filaments with a negative gravitropic growth response whereas dark-induced caulonema formation was abolished in the ΔPpDCL4 mutant lines (Figure 1B).
Figure 1.
Phenotypic Analyses of ΔPpDCL4 Mutants.
Both independent ΔPpDCL4 mutants display an identical mutant phenotype. Representative images of one line are shown.
(A) Equal amounts of protonema tissue from wild-type (WT) and the ΔPpDCL4 mutant were spotted onto solid medium and grown under long-day conditions. Colonies were photographed after 4 weeks. Note the increased protonema development in the ΔPpDCL4 mutant. Scale bar: 0.5 cm.
(B) Cultivation for 8 d in darkness results in caulonema formation in WT (black arrows), but not in the ΔPpDCL4 mutant. Scale bar: 0.5 cm.
(C) The ΔPpDCL4 mutant develops a high number of brachycytes (arrows). Scale bar: 0.1 mm.
(D) Under long-day growth conditions, the ΔPpDCL4 mutant develops dwarfed gametophores. Scale bar: 1 mm.
(E) As in (A) grown under short-day conditions. Scale bar: 1 cm.
(F) The gametophores of the ΔPpDCL4 mutant are dwarfed when grown under sporophyte-inducing short-day conditions. Gametophore size in the ΔPpDCL4 mutant is indicated by arrows. Scale bar: 1 mm.
(G) The ΔPpDCL4 mutant develops abnormal spathulate elongated leaves. Scale bar: 0.1 mm.
(H) The ΔPpDCL4 mutant fails to develop sporophytes. Scale bar: 200 μm.
Under various stress conditions or in response to ABA treatment, subapical chloronema cells can form short, thick-walled, rounded brood-cells called brachycytes (Schnepf and Reinhard, 1997; Decker et al., 2006) that act as vegetative spores to ensure plant survival under unfavorable growth conditions. In contrast to the wild-type, ΔPpDCL4 mutants developed a high number of brachycytes even under standard growth conditions (Figure 1C). Further development of protonema tissue is characterized by the formation of three-faced apical cells (buds) that give rise to the leafy gametophores. ΔPpDCL4 mutants are also affected in gametophore development, since gametophores are dwarfed under long- and short-day conditions (Figure 1D and 1F). Furthermore, when grown under short-day conditions, the leaf shape of ΔPpDCL4 mutants deviates severely from wild-type characterized by the development of spatulate elongated leaves with serrate leaf margins (Figure 1E–1G).
Most strikingly, in the ΔPpDCL4 mutant lines, sexual reproduction is abolished, since they are not able to develop the diploid sporophyte. At low temperatures and under inductive conditions, male (antheridia) and female (archegonia) sex organs are formed at the apices of gametophores (Hohe et al., 2002). In the presence of water, spermatozoids reach the archegonia and fertilize the egg cell. After fertilization, the diploid sporophyte develops and, after meiosis, haploid spores are formed within the spore capsule. When wild-type plants were grown under sporophyte-inducing conditions, all 27 analyzed colonies developed maturing sporophytes (Figure 1H). In contrast, sporophyte development was completely abolished in the ΔPpDCL4 mutant lines (27 colonies analyzed from each ΔPpDCL4 mutant line). As the defect in sporophyte development in the ΔPpDCL4 mutants might be caused by defective male and/or female reproductive tissues, we analyzed the morphology of antheridia and archegonia, but did not observe morphological deviations. Consequently, a cross-fertilization experiment was performed with ΔPpDCL4 mutants and Physcomitrella wild-type. To investigate whether the defective sporophyte formation in the ΔPpDCL4 mutants is caused by female sterility, we co-cultivated ΔPpDCL4 mutant lines with wild-type colonies and induced sporophyte formation. In case of male sterility, defects in spermatozoid development in the mutant lines should be complemented by wild-type spermatozoids and should result in the fertilization of egg cells in the archegonia of ΔPpDCL4 mutants. If the development of the female reproductive tissue is affected in the mutant lines, fertilization cannot be compensated by wild-type spermatozoids. Since only wild-type colonies formed sporophytes on mixed plates, but the mutant plants did not, we infer that at least the development of female reproductive tissues is affected in ΔPpDCL4 mutants. However, in addition to that, we cannot exclude defective spermatozoid development in the ΔPpDCL4 mutants as well.
PpDCL4 Is Required for Normal ta-siRNA Accumulation and Function
In order to obtain a genome-wide view on PpDCL4 functions in Physcomitrella sRNA pathways, we compared sRNA profiles from protonema tissue of ΔPpDCL4 mutants and wild-type by smallRNAseq, a commonly used quantitative sRNA profiling method (Lu et al., 2006; Ruby et al., 2006; Kasschau et al., 2007; Lister et al., 2008). Size-fractionated small RNAs (∼10–40 nt) from wild-type and ΔPpDCL4 mutant line 1 harboring a single genomic integration of the ΔPpDCL4 knockout construct at the PpDCL4 locus were sequenced using the SOLiD™ 2 system (Applied Biosystems). Subsequent analysis was restricted to reads with a length of 20–24 nt, resulting in 32 467 reads from wild-type and 106 609 reads from the ΔPpDCL4 mutant. The data were normalized as reads per 10 000 and used to analyze sRNA expression and size distribution from previously described MIR and TAS loci.
In Arabidopsis, DCL1 is the essential enzyme for the processing of miRNAs from hairpin-like MIR precursor transcripts (Park et al., 2002; Reinhart et al., 2002), whereas the Arabidopsis DCL4 protein has no effect on miRNA biogenesis (Xie et al., 2005) with the exception of a few irregular, non-conserved miRNAs (Rajagopalan et al., 2006). We observed similar results in Physcomitrella: no apparent differences were observed in the overall accumulation and size distribution of miRNAs between Physcomitrella wild-type and ΔPpDCL4 (Figure 2A and Supplemental Table 1). RNA blots of select miRNAs confirmed that ΔPpDCL4 mutants did not substantially affect miRNA accumulation (Figure 2B). These results are in agreement with our previous studies that identified PpDCL1a to be the essential protein for the maturation of miRNAs in Physcomitrella (Khraiwesh et al., 2010).
Figure 2.
Analyses of miRNAs and ta-siRNAs in ΔPpDCL4 Mutants.
(A) Summed, scaled, repeat-normalized sRNA abundances for all MIRNA-derived sRNAs from wild-type and ΔPpDCL4. Units are normalized reads per 10 000.
(B) Expression analysis of miR390, miR166, miR156, and miR160 in protonema tissue by RNA gel blots. U6 snRNA served as control for normalization. Numbers under blots indicate normalized expression values with respect to wild-type.
(C) Chart indicating the expression of TAS3a-d derived sRNAs inferred from sequencing data of wild-type and ΔPpDCL4.
(D) RNA gel blots for ta-siRNAs derived from TAS3a and TAS3c precursors in wild-type and ΔPpDCL4 mutants. U6snRNA served as control for normalization. Numbers under blots indicate normalized expression values for off-sized ta-siRNAs with respect to the 21-nt wild-type ta-siRNAs.
(E) Transcript levels of ta-siRNA target genes (PpARF; Phypa_203442 and PpAP2; Phypa_65352) in wild-type (WT) and ΔPpDCL4 mutants determined by qRT–PCR. Expression levels were normalized to the constitutively expressed control gene PpEF1α.
(F) RLM 5' RACE products of the miRNA and ta-siRNA target PpARF from WT and ΔPpDCL4 mutants. Arrows mark products of the expected size (1: miRNA-mediated cleavage product; 2: ta-siRNA-mediated cleavage product) that were eluted, cloned, and sequenced. Numbers above miRNA:target and ta-siRNA:target alignments indicate sequenced RACE products with the corresponding 5' end.
The ΔPpDCL4 mutants are strongly affected in the accumulation of TAS3-derived ta-siRNAs. Overall, ta-siRNA accumulation was strongly reduced, and the remaining ta-siRNAs were shifted in size from the wild-type 21–22-nt-dominated profile to a 23–24-nt-dominated profile in the mutant (Figures 2C and 2D). The off-sized ta-siRNAs observed in the ΔPpDCL4 mutant still appeared to be in phase with the flanking miR390 cleavage sites (Supplemental Figure 2), suggesting that an alternative PpDCL protein substituted for PpDCL4 at the TAS3 loci in the ΔPpDCL4 plants. Two known targets of PpTAS3-derived ta-siRNAs accumulated to higher levels in the ΔPpDCL4 mutant, suggesting that ta-siRNA target regulation was compromised in the ΔPpDCL4 background (Figure 2E). One of the two targets analyzed, the Auxin Response Factor (ARF) transcript Phypa_203442, was previously shown to be targeted by both a miRNA (miR1219) and a PpTAS3-derived ta-siRNA, which we refer to as ta-siARF (Axtell et al., 2007). 5'-RACE experiments demonstrated that, while miR1219-mediated cleavage of this ARF transcript persisted in the ΔPpDCL4 mutant, ta-siARF-mediated cleavage was not detectable (Figure 2F).
PpDCL3 Is Responsible for the Phenotypes of ΔPpDCL4 Mutants
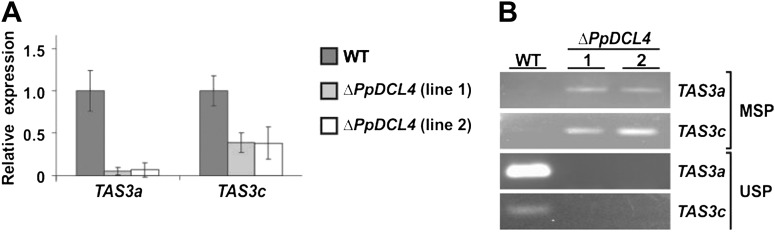
PpDCL3 is required for the accumulation of repeat-associated 22–24-nt siRNAs that derive from densely methylated, intergenic regions of the Physcomitrella genome (Cho et al., 2008). Because the off-sized ta-siRNAs that accumulate in the ΔPpDCL4 mutants are also dominated by 22–24-nt-long species, we hypothesized that, in the absence of PpDCL4, PpDCL3 processes TAS3 dsRNAs, and that the resulting siRNAs are funneled into the PpDCL3-dependent siRNA pathway, perhaps directing inappropriate chromatin modifications at siRNA-generating loci. Consistently with this notion, the accumulation of PpTAS3a primary transcripts was reduced in ΔPpDCL4 mutants (Figure 3A). From bisulfite-treated DNA cytosine methylation-specific primers only amplified PpTAS3-derived PCR products from the ΔPpDCL4 mutant lines, whereas primers specific for non-methylated sequences exclusively generated PCR products from wild-type (Figure 3B). Subsequent cloning and sequencing of the PCR products revealed that PpTAS3a residues were highly methylated in the ΔPpDCL4 mutants (Supplemental Figure 3). We detected cytosine methylation in both symmetrical (CG and CHG) and non-symmetrical (CHH) DNA sequence contexts. Since cytosine methylation of nuclear DNA is well known to cause epigenetic transcriptional silencing (Chan et al., 2005; Goll and Bestor, 2005), we propose that the high degree of cytosine methylation in the PpTAS3a loci of ΔPpDCL4 mutants is directed by PpDCL3-dependent siRNAs and leads to reduced PpTAS3a transcription rates. However, it is important to note that the ectopic DNA methylation of PpTAS3a and associated reduction of the PpTAS3a primary transcript are unlikely to cause the developmental phenotypes observed in ΔPpDCL4, as complete absence of ta-siRNAs in the ΔPpRDR6 mutant does not cause a similar phenotype. To prove this hypothesis, we generated ΔPpDCL3/ΔPpDCL4 double mutants by first establishing a ΔPpDCL3 mutant line that integrated a single PpDCL3 knockout construct at the PpDCL3 locus and was confirmed to be a null mutant by RT–PCR (Supplemental Figure 4A–4E). Subsequently, the PpDCL4 knockout construct was introduced into the ΔPpDCL3 mutant background resulting in the generation of six independent ΔPpDCL3/ΔPpDCL4 double mutants, all of which were confirmed to lack both the PpDCL3 and PpDCL4 transcripts (Supplemental Figure 4F). Strikingly, the developmental defects observed in the ΔPpDCL4 mutants were not present in the ΔPpDCL3/ΔPpDCL4 double mutants (Figures 4A–4D). The colony morphology, protonema shape, size of gametophores, caulonema initiation in the dark, gametangia development, sporophyte formation, and maturation of spore capsules in ΔPpDCL3/ΔPpDCL4 mutants were similar to wild-type. In ΔPpDCL3/ΔPpDCL4 mutants, we only observed an accelerated gametophore production that resembles the phenotype of ΔPpDCL3 mutants (Cho et al., 2008) as well as the ΔPpRDR6 mutants (Talmor-Neiman et al., 2006). RNA blots indicated that ta-siRNAs of all sizes were completely absent in ΔPpDCL3/ΔPpDCL4 mutants, while miRNA accumulation was unaffected (Figure 4E). We therefore conclude that the developmental defects caused by loss of PpDCL4 function cannot be attributed solely to reduction of ta-siRNA accumulation. Instead, the PpDCL4 phenotypes are caused by PpDCL3 activity, which only manifests in the absence of PpDCL4 function. We further characterized the PpDCL3-dependent ta-siRNAs by performing target prediction with the 23 and 24-nt PpTAS3a-derived small RNAs present in the ΔPpDCL4 small RNA library. With maximum stringent conditions of target prediction, we identified 69 target sites from 49 distinct genes that are probably affected in the ΔPpDCL4 mutant (Supplemental Table 2).
Figure 3.
Reduced Transcript Accumulation and Increased DNA Methylation at TAS Loci in ΔPpDCL4 Mutants.
(A) Expression level of the TAS precursors TAS3a and TAS3c determined by qRT–PCR. Expression levels were normalized to the constitutively expressed control gene PpEF1α.
(B) Bisulfite PCR with methylation-specific (MSP) and unmethylation-specific (USP) primers for TAS3a and TAS3c.
Figure 4.
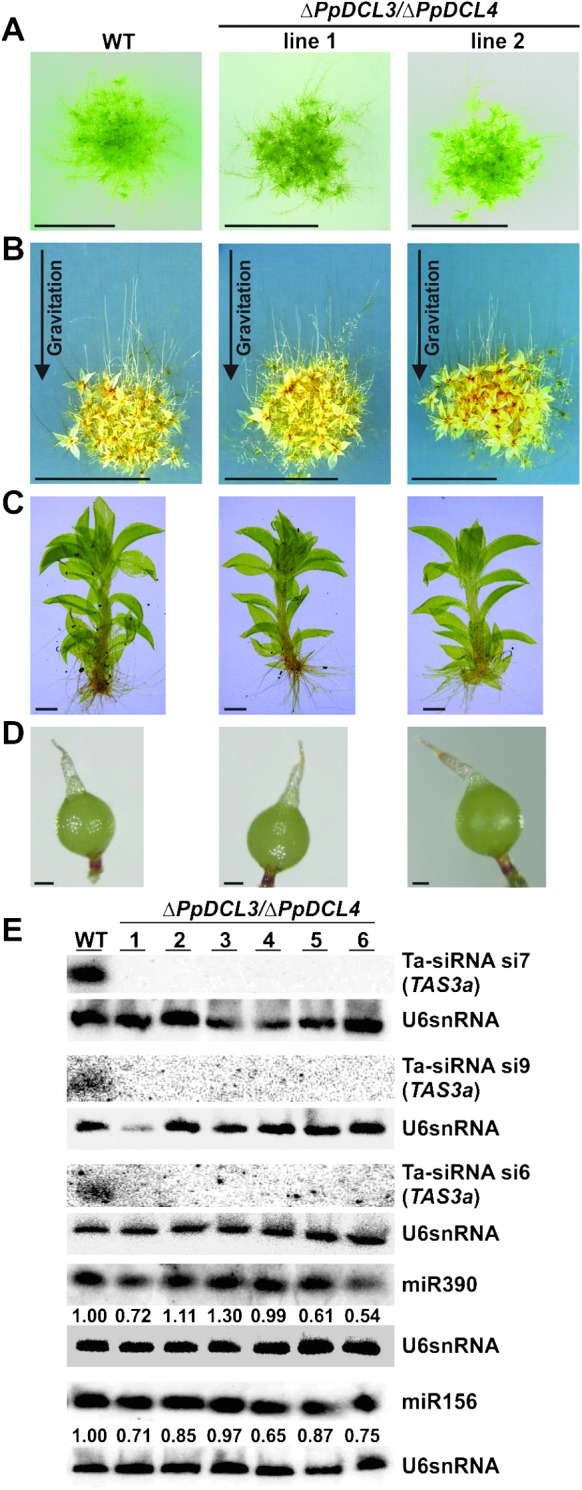
Phenotypic Analyses of ΔPpDCL3/ΔPpDCL4 Double Mutants.
(A) Equal amounts of protonema tissue from wild-type (WT) and ΔPpDCL3/ΔPpDCL4 mutants were spotted onto solid medium and developed colonies were photographed after 3 weeks. ΔPpDCL3/ΔPpDCL4 mutants show accelerated gametophore development. Scale bar: 0.5 cm.
(B) WT and ΔPpDCL3/ΔPpDCL4 mutants display normal caulonema development after 10 d growth in darkness. Scale bar: 0.5 cm.
(C) Gametophore size is similar in WT and ΔPpDCL3/ΔPpDCL4 mutants. Scale bar: 2 mm.
(D) Sporophyte formation is rescued in ΔPpDCL3/ΔPpDCL4 mutants. Scale bar: 200 μm.
(E) RNA gel blots hybridized with antisense probes of miR390, miR156, and ta-siRNAs generated from the PpTAS3a precursor in WT and ΔPpDCL3/ΔPpDCL4 double mutants. An antisense probe for U6 snRNA served as control for normalization. Numbers under blots indicate normalized miRNA expression values with respect to WT.
Identification of Additional DCL4/RDR6-Dependent TAS Loci in Physcomitrella
The four previously known PpTAS3 loci were discovered based upon phased patterns of siRNA accumulation and the presence of miR390 complementary sites (Axtell et al., 2006; Talmor-Neiman et al., 2006). As described above, siRNA accumulation patterns at these four loci change in distinctive patterns in ΔPpDCL4 and ΔPpRDR6 plants; ΔPpDCL4 mutants shift the size profile of the siRNAs from a 21-nt-dominated pattern to a 23–24-nt-dominated pattern, while nearly all siRNA accumulation is lost in ΔPpRDR6 mutants. We therefore used existing Physcomitrella smallRNAseq data to search for additional small RNA loci showing similar changes in siRNA accumulation in ΔPpDCL4 and ΔPpRDR6 plants. Initially, seven loci were identified, four of which overlapped the previously described PpTAS3a-d (Figure 5, Table 1, and Supplemental Figure 5). Two of the three novel loci overlapped previously described hotspots of non-miRNA 21nt-dominated small RNA expression (Pp21SR6 and Pp21SR39; Cho et al., 2008). The third novel locus as well as Pp21SR6 possessed dual miR390 complementary regions and thus we named them PpTAS3e and PpTAS3f, respectively. Degradome data (Addo-Quaye et al., 2009) demonstrated that the four miR390 sites within PpTAS3e and PpTAS3f were sliced (Table 1 and Supplemental Figure 5). Two distinct PpTAS3 ta-siRNA populations have previously been shown to be capable of directing the slicing of targets in trans: one population targets AP2-domain transcripts, while the other directs slicing of ARF-domain transcript (Talmor-Neiman et al., 2006; Axtell et al., 2007; Khraiwesh et al., 2010). PpTAS3e could produce a ta-siARF with reasonable base-pairing to one of the known ta-siARF target sites, while PpTAS3f could produce both ta-siAP2 and ta-siARF (Table 1, Supplemental Figure 5, and Supplemental Table 3). The PpTAS3e siRNA population was typical in that it was largely contained between, and in phase with, the two miR390 complementary regions (Supplemental Figure 5). In contrast, the PpTAS3f siRNA population extended beyond the region bounded by the dual miR390 complementary sites, and was not well phased (Supplemental Figure 5).
Figure 5.
Neighboring miR156- and miR390-Sliced TAS Loci.
(A) An annotated genomic snapshot of PpTAS3a and PpTAS6a. Shaded regions indicate boundaries of indicated TAS loci, with locations and strand orientations of miRNA complementary sites and known functional ta-siRNAs shown. Insets above show the small RNA size distribution in the indicated genotypes, as well as the ‘phasing’ distributions of wild-type small RNAs in 21-nt bins (numbers on periphery indicate percentage of siRNAs in each bin). Browser track small RNA data (blue) and degradome data (magenta) show the 5' end positions from wild-type samples, with positive values indicating Watson-strand mapped reads, and negative values indicating Crick-strand mapped reads. Insets below show miRNAs aligned with miRNA complementary sites. Numbers above alignments are the number of degradome-derived 5' ends mapped to the 10th nucleotide of the alignment.
(B) Evidence for trans-acting slicing directed by the PpTAS6a-derived ta-siZNF. Annota
ted genomic snapshot showing protein-coding transcripts (gray), polyA+ RNAseq data (not strand-specific (Zemach et al., 2010), and degradome data. Inset below-left shows ta-siZNF/target alignment, with the number of degradome reads at the 10th nucleotide of the alignment indicated. Inset below-right shows schematic of the protein (from Phytozome).
Table 1.
Summary of Physcomitrella DCL4/RDR6-Dependent TAS Loci.
| Locus | 5' miR390 site | 3' miR390 site | 5' miR529 site | 5' miR156 site | 3' miR156 site | ta-siAP2 | ta-siARF | ta-siZNF |
| PpTAS3a/PpTAS6a | S | S | S | S | S | + | + | + |
| PpTAS3b | P | P | N/A | N/A | N/A | + | + | N/A |
| PpTAS3c | S | S | N/A | N/A | N/A | – | + | N/A |
| PpTAS3d/PpTAS6c | S | S | P | S | S | + | + | Absent |
| PpTAS3e | S | S | N/A | N/A | N/A | – | + | N/A |
| PpTAS3f/PpTAS6b | S | S | P | P | S | + | + | Absent |
S, degradome-based evidence of slicing; P, predicted miRNA target site w/o degradome evidence; +, ta-siRNA pairs well with one or more target; –, ta-siRNA does not pair well (score ≥ 6) with any targets; N/A, not applicable; absent, site or ta-siRNA not found within locus.
The final novel DCL4/RDR6-dependent siRNA locus that we found in our initial computational screen produced siRNAs from a region bounded by upstream miR156 and miR529 complementary sites and a downstream miR156 site (Figure 5A). Degradome data support slicing at all three complementary sites and the siRNA population is largely in a 21-nt phase with the 3' miR156 cleavage site (Figure 5A). This locus, previously described as Pp21SR39 but heretofore unrecognized as a TAS locus (Cho et al., 2008), did not share detectable similarity with any previously described families of plant TAS loci. Therefore, we renamed it PpTAS6a. Curiously, PpTAS6a is located within ∼0.7 kb of PpTAS3a, and their respective miRNA complementary sites are located on the same strand (Figure 5A). Inspection of the genomic region surrounding PpTAS3f revealed a second nearby cluster of mainly 21-nt siRNAs in the wild-type. We found that this region also contained a high-confidence miR156 complementary site downstream of the siRNA-generating region, which showed degradome-based evidence of slicing (Supplemental Figure 5). In addition, upstream miR156 and miR529 sites could also be observed, albeit with extensive mismatches and no evidence of slicing. Based on this fact, we named this locus PpTAS6b, although there was no detectable sequence similarity outside of the pattern of miR156 and miR529 complementary sites between PpTAS6a and PpTAS6b. Like its neighbor PpTAS3f, siRNA production from PpTAS6b was atypical in that it was not in phase with the miRNA-directed cleavage site(s) and extended for a long distance beyond the complementary sites (Supplemental Figure 5). Finally, we detected sequence similarity between PpTAS6a and a region neighboring PpTAS3d. Although we could find no evidence of small RNA production from this region, we did observe a pattern of miR156 and miR529 complementary sites similar to the other TAS6 loci with some weak evidence of slicing; therefore, we have provisionally named this locus PpTAS6c (Supplemental Figure 5). Phylogenetic analysis demonstrated that the six PpTAS3 loci fall into two distinct clades: PpTAS3a, -d, and -f form one clade, while PpTAS3b, -c, and -e form the other clade (Supplemental Figure 6). All three members of the PpTAS3a/d/f clade have upstream flanking regions that produce miR156 and/or miR529-sliced RNAs, in at least two cases (PpTAS3a/PpTAS6a and PpTAS3f/PpTAS6b) corresponding with the production of phased DCL4/RDR6-dependent siRNAs. Despite the fact that PpTAS6a and PpTAS6b do not share sequence similarity in their siRNA-generating regions, their common arrangement proximal to PpTAS3 loci and their shared presence of sliced miR156 sites strongly suggest a common ancestry that we believe justifies their classification into the same family. We have yet to observe siRNA accumulation from PpTAS6c, so this annotation should be regarded as provisional.
We used all 248 PpTAS6a and PpTAS6b-derived siRNAs from our wild-type sequencing data as queries for ta-siRNA target predictions using a standard scoring matrix (Allen et al., 2005). One hundred and fifty target sites, from 129 distinct transcripts, were predicted with alignment scores of 3.5 or less (Supplemental Table 4). At the present time, we have low confidence in these predictions, as the cohort of transcripts did not seem to have a coherent functional theme, and none of these predictions was supported by robust evidence of slicing in the available degradome data. We did identify robust degradome data supporting the slicing of a zinc-finger domain transcript by a PpTAS6a-derived siRNA with a higher alignment score (5.5; Figure 5B). While this alignment score is relatively high, we note that the siRNA target mismatches are concentrated on the 3' end of the complementary region and that the degradome data strongly support the in vivo accumulation of the predicted slicing remnant. We conclude that at least one PpTAS6a ta-siRNA, which we name ta-siZNF, is functional in slicing a zinc-finger domain transcript in trans, and that other trans targets may exist for other PpTAS6-derived siRNAs.
DISCUSSION
The most striking features of the ΔPpDCL4 mutants are the pleiotropic defects throughout vegetative development and sterility. The defects in the vegetative development include developmental programs such as gametophore elongation, protonema and leaf development, as well as specific growth programs linked to altered physiological conditions. ΔPpDCL4 mutants fail to develop caulonema filaments with a negative gravitropic growth in darkness and they form numerous brachycytes that usually only develop in response to abiotic stress conditions in Physcomitrella wild-type. The enhanced brachycyte development could either be due to the constitutive activation of the underlying developmental program or could result from an altered plant physiology causing abiotic stress symptoms that result in the onset of stress-associated pathways and brachycyte formation. Even though we did not detect morphological differences of antheridia and archegonia in ΔPpDCL4 mutants, the cross-fertilization experiment with wild-type plants indicated at least female sterility.
At the molecular level, we found that PpDCL4 is essential for the accumulation of 21-nt ta-siRNAs from TAS transcripts corresponding to the function of DCL4 orthologs in Arabidopsis and rice (Gasciolli et al., 2005; Liu et al., 2007). Loss of PpDCL4 function allowed PpDCL4 substrates to be processed instead by PpDCL3, leading to the accumulation of off-sized, 23–24-nt siRNAs in the ΔPpDCL4 mutant. Such conditional DCL redundancy upon TAS substrates has also been observed in rice and Arabidopsis dcl4 mutants (Gasciolli et al., 2005; Liu et al., 2007). In all cases, a DCL3 homolog becomes competent for ta-siRNA biogenesis upon the loss of DCL4 function. Concerning this, Physcomitrella differs from rice and Arabidopsis only in that Physcomitrella lacks the DCL2 homolog that in rice and Arabidopsis also contributes to ta-siRNA production upon loss of DCL4 function. Our evidence suggests that the PpDCL3-dependent ta-siRNAs that arise in the ΔPpDCL4 plants are functional in cis: TAS primary transcripts are reduced concomitant with an increase in DNA methylation at TAS chromatin.
The dramatic morphological defects seen in ΔPpDCL4 plants cannot be attributed solely to reduced ta-siRNA accumulation. ΔPpRDR6 plants completely lack all known ta-siRNAs (Talmor-Neiman et al., 2006; Cho et al., 2008) but nonetheless have a very mild phenotype consisting of an accelerated rate of transition from the filamentous protonematal growth stage to the leafy gametophore stage. The morphological phenotypes apparent in ΔPpDCL4 plants require the activity of PpDCL3, as the ΔPpDCL3/ΔPpDCL4 double-mutant plants resemble the comparatively mild growth changes seen in ΔPpRDR6. Thus, the role that PpDCL3 plays in the ΔPpDCL4 background appears to be specifically detrimental to normal Physcomitrella development. A similar observation has been made in Arabidopsis where ta-siRNAs of aberrant sizes are made by DCL2 and DCL3 in the absence of DCL4, and dcl1/dcl4 double mutants exhibit pleiotropic developmental defects absent in the dcl1/dcl2/dcl4 triple mutant (Bouche et al., 2006). These data imply that, as in the case of Physcomitrella DCL3, ectopic DCL2 activity in Arabidopsis can trigger dramatic phenotypic consequences. However, compensatory or ectopic DCL activities do not always cause major phenotypes. The phase-change and polarity defects of Arabidopsis dcl4 mutants are enhanced, not eliminated, in dcl3/dcl4 mutants and these defects are identical to those observed in rdr6 mutants (Gasciolli et al., 2005). Oryza dcl3dcl4 double mutants have not been reported. However, the strong phenotypes observed in OsDCL4 (SHO1) mutants are identical to those seen in plants defective in the rice RDR6 homolog, SHL2 (Nagasaki et al., 2007), strongly suggesting that reduction of ta-siRNA accumulation, not the effects of compensatory DCLs, underlies the phenotypes observed in OsDCL4 mutant plants.
Why might PpDCL3 activity have such dramatic effects in the PpDCL4 background? The distinct phenotype seems likely to be explained by the regulatory activities of the PpDCL3-dependent off-sized ta-siRNAs that arise solely in this background. These off-sized ta-siRNAs may target RNAs that are not under sRNA control in wild-type, and/or regulate normal ta-siRNA targets with an unusual strength or via different molecular mechanisms. Representatives of two known ‘normal’ ta-siRNA targets were up-regulated in the ΔPpDCL4 background, suggesting that perhaps other targets of the ‘off-sized’ ta-siRNAs cause the ΔPpDCL4 phenotype. To address this question, we performed target predictions for all 23–24-nt ta-siRNAs arising from the PpTAS3a precursor (Supplemental Table 2). Among the predicted targets, we find transcripts encoding proteins that were previously shown to control developmental processes and the misregulation of which could account for the developmental defects in the ΔPpDCL4 mutant. For example, auxin triggers specific developmental steps in P. patens (Ludwig-Muller et al., 2009) and we identified putative off-sized ta-siRNA target transcripts encoding proteins that are involved in auxin transport and signaling as well as auxin-induced downstream transcripts. One putative off-sized ta-siRNA target (Pp1s42_203V6.1) encodes a homolog of A. thaliana BIG, a calossin-like protein required for polar auxin transport (Gil et al., 2001). Two other predicted targets (Pp1s74_86V6.1 and Pp1s5_432V6.1) are homologs of auxin-regulated AP2/EREBP genes, which control cell division patterning at early stages of root primordium development in A. thaliana (Hirota et al., 2007). Another putative off-sized ta-siRNA target (Pp1s220_89V6.1) encodes a homolog of the A. thaliana N-MYC down-regulated-like 2 (NDL2) protein that acts in the auxin signaling pathway and down-regulation of members of the NDL family was shown to alter root architecture in A. thaliana (Mudgil et al., 2009). Additional targets were predicted that do not act in auxin-responsive pathways but have an impact on the development of plants. For example, one of the predicted targets (Pp1s60_37V6.1) encodes a protein homologous to MOS1 (modifier of snc1) from A. thaliana that controls the expression of SUPPRESSOR OF npr1-1 CONSTITUTIVE1 (SNC1). mos1 mutants show retarded growth and it was suggested that MOS1 regulates gene expression at the chromatin level (Li et al., 2010). Another predicted target (Pp1s642_3V6.1) encodes a A-type cyclin (CYCA1) homolog that is involved in meiotic division in A. thaliana. cyca1 mutants fail to enter meiosis II and consequently produce diploid spores (d’Erfurth et al., 2010). In addition, two homologs (Pp1s42_258V6.1 and Pp1s336_16V6.1) of HAP6 from A. thaliana were predicted as off-sized ta-siRNA targets that act in pollen grain development, since pollen grains of A. thaliana hap6 mutants cannot fertilize properly (Johnson et al., 2004). The sterility defects observed in ΔPpDCL4 mutants may be attributed to the down-regulation of the PpCYCA1 and/or PpHAP6 homologs.
An alternative possibility is that off-sized, DCL3-dependent miRNAs from MIRNAs normally processed by PpDCL4 are responsible for the phenotypes of ΔPpDCL4 plants. Although we cannot rule out this possibility completely, our RNA blot and smallRNAseq analyses found no evidence for PpDCL4-dependent miRNAs nor evidence for off-sized miRNA accumulation in the ΔPpDCL4 background. Future experiments should seek to understand the molecular basis for the PpDCL3-dependent phenotypes observed in the ΔPpDCL4 background.
In Arabidopsis (Howell et al., 2007) and Physcomitrella genome, only a limited number of endogenous DCL4/RDR6-dependent small RNA loci have been identified. Our analysis of DCL4/RDR6-dependent loci in Physcomitrella has revealed several interesting findings. All of the Physcomitrella DCL4/RDR6-dependent siRNAs appear to derive from primary transcripts sliced at least once, and often several times, by a miRNA(s). We describe a new family of TAS loci that are associated with miR156- and miR529-directed slicing. miR156 is an ancient plant miRNA that has a cohort of conserved, protein-coding targets independently of TAS loci. In all plant species examined, including Physcomitrella, miR156 targets mRNAs encoding Squamosa Promoter Binding Like (SBP/SPL) transcription factors (Poethig, 2009). In angiosperms, miR156-mediated regulation of SBP/SPL targets is critical for many distinct aspects of developmental timing (Wu and Poethig, 2006; Wu et al., 2009; Nodine and Bartel, 2010). However, miR156-associated TAS loci have not been previously described in any plant species. miR529 is related in sequence to miR156, and also widely conserved. At least one PpTAS6-derived siRNA appears to be a true ta-siRNA, in that we could identify a sliced zinc-finger domain target. The PpTAS6 family is also unique in its very close proximity to miR390-targeted PpTAS3 loci; the three TAS3/TAS6 pairs are separated by only ∼0.7 kb, and their miRNA complementary sites are all on the same strands. This suggests that these PpTAS6/PpTAS3 pairs could share single common primary transcripts, and that miR156-, miR529-, and miR390-mediated activities may be interrelated in Physcomitrella.
Our findings demonstrate ta-siRNA biogenesis mechanisms in Physcomitrella and the existence of additional TAS precursors. This indicates that ta-siRNA-regulated mechanisms are much more complex than thought before, and opens new avenues for investigating the molecular mechanisms that control small RNA-directed developmental processes.
METHODS
Plant Material and Phenotypic Analysis
Physcomitrella cultivation, sporophyte induction, protoplast isolation, PEG-mediated transformation, and regeneration of stably transformed plants were performed according to standard procedures (Frank et al., 2005). The lines reported in this study are deposited in the International Moss Stock Center (IMSC, www.moss-stock-center.org) with the accession numbers IMSC 40436–40437 (ΔPpDCL4 mutants), IMSC 40433 (ΔPpDCL3 mutant), and IMSC 40440, 40441, 40552, 40553, 40555, 40556 (ΔPpDCL3/ΔPpDCL4 double mutants). Pictures of plants were taken with either a KAISER Scan Do scanner (Buchen, Germany) or a Zeiss Axioplan 2 microscope fitted with an Axiocam MRc5 (Zeiss, Jena). For caulonema induction, 5-μl droplets from freshly dispersed liquid cultures with an adjusted density of 100 mg L−1 dry weight were spotted onto solid growth medium. The plates were sealed and cultivated under standard growth conditions for 3 weeks. Subsequently, the plates were wrapped in aluminum foil, positioned vertically, and cultured for another 8 days.
Isolation of PpDCL3 and PpDCL4 Full-Length cDNA
PpDCL3 and PpDCL4 were initially identified by TBLASTN searches in Physcomitrella EST sequences (Rensing et al., 2002) using Arabidopsis DCL1-4 protein sequences (accession numbers P84634, Q9SP32, NP_189978, NP_566199) as queries. The isolation of full-length cDNA sequences was performed by 5'RACE-PCR, 3'RACE-PCR, and RT-PCR using primers deduced from Physcomitrella genomic sequence data. To confirm that PCR products were derived from the same cDNA, all PCR and RACE primers were selected to produce overlapping PCR fragments. The sequences were assembled and full-length PpDCL3 and PpDCL4 cDNA sequences were submitted to GenBank with accession numbers EF670437 and EF670438, respectively. The Pfam database (Finn et al., 2008) was used for protein domain predictions.
Generation of ΔPpDCL4 Mutant Lines
A zeocin selection marker cassette was cloned into a single restriction site present in a PpDCL4 genomic fragment. The gene disruption construct was transfected into Physcomitrella protoplasts and zeocin-resistant lines were analyzed by PCR and genomic DNA blot to confirm precise integration into the PpDCL4 locus. Loss of PpDCL4 transcript was confirmed by RT–PCR. The generation of ΔPpDCL4 mutants is described in detail in Supplemental Material and Methods.
Generation of ΔPpDCL3/ΔPpDCL4 Double Mutants
An nptII (neomycin phosphotransferase II) expression cassette (Strepp et al., 1998) was cloned into the PAZ domain encoding region of PpDCL3, transfected into wild-type protoplasts, and G418-resistant lines were analyzed by PCR and genomic DNA blot to confirm precise integration into the PpDCL3 locus. Loss of PpDCL3 transcript was confirmed by RT–PCR. ΔPpDCL3 mutant protoplasts were subsequently transfected with the PpDCL4 knockout construct where the zeocin selection cassette was replaced by hpt (hygromycin phosphotransferase) resistance cassette (Huether et al., 2005). Antibiotic resistant lines were analyzed by PCR for proper insertion of the PpDCL4 knockout construct. RT–PCR analyses of these lines confirmed the loss of PpDCL3 and PpDCL4 transcripts (ΔPpDCL3/ΔPpDCL4). The generation of ΔPpDCL3/ΔPpDCL4 double mutants is described in detail in Supplemental Material and Methods.
Small RNA Sequencing and Data Analysis
Total RNA was isolated from 10-day-old protonema from ΔPpDCL4 mutants and wild-type plants as previously described (Cho et al., 2008). Small RNAs (∼10–40 nt) were recovered by PAGE fractionation. Small RNA sequencing libraries for the Applied Biosystems SOLiD™ 2 system were prepared using the SOLiD™ small RNA expression kit (Ambion) according to the manufacturer’s instructions. Barcoded libraries were sequenced using a SOLiD™ 2 system. Barcoded color space data were first parsed to separate different samples mixed in the same run. Barcode-sorted color space reads (35 colors) were then parsed to identify the first five nucleotides of the constant 3' adapter. The adapter sequences were trimmed. Only reads with sizes of 20–24 nt were retained and were directly translated to DNA space. The processed and raw data are available from the Gene Expression Omnibus (GEO: Accession GSE18466). The processed reads were mapped to the Physcomitrella genome as previously described (Cho et al., 2008), retaining only exact matches. Custom PERL scripts were used to analyze small RNA expression from MIRNA and TAS loci.
Identification of Additional TAS Loci
SmallRNAseq data from Physcomitrella wild-type (GSM115095, GSM115096, GSM115097, GSM313212, and GSM313213), dcl3 (GSM313214, GSM313215), and rdr6 (GSM313216, GSM313217) was previously described (Axtell et al., 2006; Cho et al., 2008) and the smallRNAseq library from dcl4 mutant current study (GSM459911) was used in the analysis. 20–24-nt small RNAs from each of the 10 libraries were mapped to the Physcomitrella genome (v1.1) using bowtie (Langmead et al., 2009), v0.12.7, with settings ‘-v 1 -a –best –strata -f -p 7 -k 100’ as well as ‘-C’ for color space data. Therefore, mappings with either zero or one mismatch were accepted and, for reads that had more than 100 optimal mapping positions, only the first 100 were reported. Reads mapping to rRNA, tRNA, or MIRNA hairpin positions were then discarded. The total small RNA abundance in 200-nt non-overlapping bins was calculated genome-wide, and scaled to reads per million to facilitate comparisons between different datasets. The top 1000 most abundant bins in the wild-type datasets were then screened to find bins meeting the following criteria: (1) symmetry (defined as the proportion of small RNAs mapped to one or the other strand) of 0.25–0.75, (2) rdr6 total <0.2 × wild-type total, (3) percentage 21–22-nt RNAs in wild-type and dcl3 > 70%, and (4) percentage 23–24-nt RNAs in dcl4 > 50%. These parameters were inferred from the characteristics of the previously known PpTAS3a–d loci. The three novel bins that passed this filter were then further examined by searching for miRNA complementary sites (based on BLASTN) within a centered 1000-nt window. The final annotations of these three novel loci, PpTAS3e, PpTAS3f, and PpTAS6a, were based on manual curation and the positions of the miRNA complementary sites. PpTAS6b did not pass the initial computational filter but was identified serendipitously based upon its proximity to PpTAS3f. Similarly, PpTAS6c lacked corresponding siRNAs in our dataset but was identified based on proximity to PpTAS3d and the presence of miR156 target sites. The siRNA population may be generated from PpTAS6c under different physiological conditions or different tissue types. Degradome sequencing data from Physcomitrella have been previously described (Addo-Quaye et al., 2009). CleaveLand 2 (Ma et al., 2010) was used for degradome analysis. For siRNA target predictions, we used axtell_targetfinder.pl (Ma et al., 2010). Phylogenetic analysis of PpTAS3a-f utilized a MUSCLE alignment (Edgar, 2004) of the regions between the miR390 complementary sites as input to the Maximum Likelihood method in MEGA5 based on the Tamura-Nei model (Tamura and Nei, 1993). The bootstrap consensus tree, based on 500 replicates, was shown. A .bed file has been created (Supplemental Table 5) that indicates genomic locations of Physcomitrella TAS loci (genome assembly version 1.1) as well as important features within the TAS loci.
Detection of Small RNAs by RNA Gel Blots
Total RNA was separated in a 13% polyacrylamide gel containing 8.3 M urea in 1 TBE buffer and electro-blotted onto nylon membranes for 1 h at 400 mA. Oligonucleotides complementary to miRNA, ta-siRNA and U6snRNA sequences were radiolabeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas). Blot hybridization was carried out in buffer containing 0.05 M sodium phosphate (pH 7.2), 1 mM EDTA, 6 SSC, 1 Denhardt’s and 5% SDS. Blots were washed two to three times with 2 SSC, 0.2% SDS, and one time with 1 SSC, 0.1% SDS. Blots were hybridized and washed at temperatures 10°C below the Tm of the oligonucleotide. Sequences of oligonucleotides used for the detection of small RNAs are listed in Supplemental Table 6.
Detection of RNA Cleavage Products
RNA ligase mediated 5' RACE PCR was performed with a modified GeneRacer Kit (Invitrogen) protocol (Llave et al., 2002). PCR reactions were performed using the GeneRacer forward primer 5'-CGACTGGAGCACGAGGACACTGA-3' and the PpARF (Phypa_203442) gene-specific primer 5'-CACTCGACACGTCGTTGCTGAGAGTT-3' followed by a PCR with the nested GeneRacer forward primer 5'-GGACACTGACATGGACTGAAGGAGTA-3' and the nested gene-specific primer 5'-ATGAGGAGGTCCGGGAGGATTCGATA-3'. Amplification products corresponding to the size of expected cleavage products were excised from the gel, cloned, and sequenced.
Expression Analysis by Quantitative Real-Time PCR
RNA was extracted from three biological replicates of Physcomitrella wild-type and two ΔPpDCL4 mutant lines with TRIzol reagent (Invitrogen), treated with DNase I (Fermentas), and reverse-transcribed into first-strand cDNA by Taq Man Reverse Transcription Reagents (Applied Biosystems). Real-time PCR was performed on a Roche 480 Light Cycler using gene-specific primers and Light Cycler 480 SYBR Green I Master (Roche) according to the manufacturer’s instructions. The constitutively expressed gene PpEF1α was used as reference gene for normalization. Expression levels of PpAP2 (Phypa_65352) and PpARF (Phypa_203442) were calculated relative to transcript abundance in wild-type employing relative quantification with efficiency correction (Livak and Schmittgen, 2001). Primer sequences are reported in Supplemental Table 6.
DNA Methylation Analysis
Two μg of genomic DNA from wild-type and two ΔPpDCL4 mutant lines were treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen). TAS3a and TAS3c sequences were analyzed with the MethPrimer program (Li and Dahiya, 2002) to deduce methylation-specific (MSP) and unmethylation-specific primers (USP) for PCR analysis of bisulfite-treated DNA. PCR products were cloned using the CloneJET PCR Cloning Kit (Fermentas) and sequenced. Primer sequences are reported in Supplemental Table 6.
Target Prediction
The targets of 23–24-nt ta-siRNAs generated from PpTAS3a precursor were predicted against the Physcomitrella mRNAs (from Phytozome 7.0; dataset ‘Ppatens_152_transcript.fa’) using the Carrington Lab script ‘targetfinder.pl’ (http://carringtonlab.org/resources/targetfinder). It is based on Allen’s parameters and only targets that gave a maximum score of 4.0 were considered as true targets (Allen et al., 2005; Fahlgren et al., 2007).
Phylogenetic Analysis
The sense-strand sequences between the two miR390 complementary sites for each of the six PpTAS3 loci were aligned using MUSCLE 3.8 (Edgar, 2004) using default parameters. The resulting FASTA-formatted alignment used for phylogenetic analysis is listed in Supplemental Table 7. Phylogenetic analysis was performed using MEGA 5.05 for Mac OSX (Tamura et al., 2011). The bootstrap consensus tree from a maximum likelihood reconstruction (Nucleotide substitution model (Tamura and Nei, 1993); Uniform rates among sites; gaps treated as complete deletion; NNI ML heuristic method with automatic initial tree) using 1000 bootstrap replicates is shown in Supplemental Figure 6. Nodes with less than 100% bootstrap support are annotated with percent support. Scale bar: substitutions per site (n = 103 informative sites).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by Landesstiftung Baden-W ü rttemberg (P-LS-RNS/40 to W.F. and R.R.), the German Federal Ministry of Education and Research (FRISYS 0313921 to R.R. and W.F.), the German Academic Exchange Service (DAAD; Ph.D. fellowship to M.A.A. and I.F.), the graduate school GRK1305 (Ph.D. fellowship to A.K.B.), and the US National Institutes of Health (R01GMR084051 to M.J.A.).
Supplementary Material
Acknowledgments
We thank Jo Ann Snyder for assistance with moss propagation; Jim Carrington and Blake Meyers for discussion on TAS nomenclature; and Jeff Duckett and Jan-Peter Frahm for discussions on the morphology of mutant lines. Author contributions: M.A.A. generated and analyzed ΔPpDCL4 and ΔPpDCL3/ΔPpDCL4 mutants; I.F. and A.K.B. helped in the analysis of ΔPpDCL4 mutants; M.J.A. and S.H.C. performed sRNA sequencing; M.J.A. and Z.M. analyzed sRNA sequencing data and annotated novel TAS loci; W.F. designed research; M.A.A., M.J.A., and W.F. analyzed the data; and M.A.A. and W.F. wrote the paper with support from R.R., A.K.B., S.H.C., and M.J.A. No conflict of interest declared.
References
- Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–2121. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis . Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana . Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Cho SH, et al. Physcomitrella patens DCL3 is required for 22–24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 2008;4:e1000314. doi: 10.1371/journal.pgen.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth I, et al. The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 2010;6:e1000989. doi: 10.1371/journal.pgen.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker EL, Frank W, Sarnighausen E, Reski R. Moss systems biology en route: phytohormones in Physcomitrella development. Plant Biol. Stuttg. 2006;8:397–405. doi: 10.1055/s-2006-923952. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. Plos ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W, Decker EL, Reski R. Molecular tools to study Physcomitrella patens . Plant Biol. Stuttg. 2005;7:220–227. doi: 10.1055/s-2005-865645. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Gil P, et al. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis . Genes Dev. 2001;15:1985–1997. doi: 10.1101/gad.905201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis . Plant Cell. 2007;19:2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohe A, Rensing SA, Mildner M, Lang D, Reski R. Day length and temperature strongly influence sexual reproduction and expression of a novel MADS-box gene in the moss Physcomitrella patens . Plant Biol. 2002;4:595–602. [Google Scholar]
- Howell MD, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huether CM, et al. Glyco-engineering of moss lacking plant-specific sugar residues. Plant Biol. 2005;7:292–299. doi: 10.1055/s-2005-837653. [DOI] [PubMed] [Google Scholar]
- Johnson MA, et al. Arabidopsis hapless mutations define essential gametophytic functions. Genetics. 2004;168:971–982. doi: 10.1534/genetics.104.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, et al. Transcriptional control of gene expression by microRNAs. Cell. 2010;140:111–122. doi: 10.1016/j.cell.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Orban R, Baker B. SoMART, a web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.04922.x. doi: 10.1111/j.1365–313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Li Y, Tessaro MJ, Li X, Zhang Y. Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 2010;153:1425–1434. doi: 10.1104/pp.110.156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis . Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell. 2007;19:2705–2718. doi: 10.1105/tpc.107.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Lu C, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J, Decker EL, Reski R. Dead end for auxin conjugates in Physcomitrella? Plant Signal Behav. 2009;4:116–118. doi: 10.4161/psb.4.2.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Coruh C, Axtell MJ. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell. 2010;22:1090–1103. doi: 10.1105/tpc.110.073882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis R, et al. The evolution and diversification of Dicers in plants. FEBS Lett. 2006;580:2442–2450. doi: 10.1016/j.febslet.2006.03.072. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell. 2009;21:3591–3609. doi: 10.1105/tpc.109.065557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl Acad. Sci. U S A. 2007;104:14867–14871. doi: 10.1073/pnas.0704339104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–2692. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana . Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis . Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Small RNAs and developmental timing in plants. Curr. Opin. Genet. Dev. 2009;19:374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Hannon GJ. Uncovering RNAi mechanisms in plants: biochemistry enters the foray. FEBS Lett. 2005;579:5899–5903. doi: 10.1016/j.febslet.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Rombauts S, Van de Peer Y, Reski R. Moss transcriptome and beyond. Trends Plant Sci. 2002;7:535–538. doi: 10.1016/s1360-1385(02)02363-4. [DOI] [PubMed] [Google Scholar]
- Reski R. Development, genetics and molecular biology of mosses. Bot. Acta. 1998;111:1–15. [Google Scholar]
- Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C . elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Schnepf E, Reinhard C. Brachycytes in Funaria protonemate: induction by abscisic acid and fine structure. Plant Physiol. 1997;151:166–175. [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl Acad. Sci. U S A. 1998;95:4368–4373. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmor-Neiman M, Stav R, Klipcan L, Buxdorf K, Baulcombe DC, Arazi T. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 2006;48:511–521. doi: 10.1111/j.1365-313X.2006.02895.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Van Der Straeten D. Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol. 2007;7:65. doi: 10.1186/1471-2229-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis . Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana . Proc. Natl Acad. Sci. U S A. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.