Abstract
Organismal response to hypoxia is essential for critical regulation of erythropoiesis, other physiological functions and survival. There is an evidence of individual variation in response to hypoxia as some but not all of the affected individuals develop polycythemia, and or pulmonary and cerebral edema. A significant population difference in response to hypoxia exist as many Tibetans, Ethiopian and Andean natives developed an adaptive mechanisms to extreme hypoxia. A proportion of any non-adapted individuals exposed to high altitude develop pulmonary edema (HAPE), pulmonary hypertension, cerebral edema and extreme polycythemia. The isolation of causative gene(s) responsible for HAPE and other extreme hypoxia complications would provide a rational basis for specific targeted therapy of HAPE, allow its targeted prevention for at-risk populations, and clarification of its, and pathophysiology of other hypoxic maladaptations. As today, the only suggested linkage in unrelated individual with HAPE has been with endothelial nitric oxide synthase (eNOS) gene. Here we describe a family with multiple members affected with HAPE in three generations. Families with multiple affected members with HAPE have not been described. We first ruled out linkage of HAPE with eNOS gene. We then performed analysis of the whole genome using high-density SNP arrays (Affymetrix v5.0) and assuming a single gene causation of HAPE ruled out a linkage with 34 other candidate genes. Only HIF2A haplotype was shared by individuals who exhibit the HAPE phenotype, and the work on their possible causative role in HAPE is in progress. Clearly a small size of our family does not provide sufficient power for a conclusive analysis of linkage; we hope that collaboration with other investigators referring us more HAPE patients in effort to increase sample size would lead to identification of gene(s) responsible for HAPE and possibly other maladaptive hypoxic complications.
Keywords: Hypoxia, Erythropoietin, High Altitude Pulmonary Edema (HAPE), Endothelial Nitric Oxide (eNOS), Hypoxia Inducible Factor (HIF)
INTRODUCTION
The response to hypoxia is crucial for multiple body functions, including erythropoiesis, energy metabolism, respiration, angiogenesis and other functions.1 The transcription of as many as 3% of the genes in the genome is regulated by ubiquitously expressed transcription factor HIF-1, a master transcription factor discovered from studies of erythropoietin gene regulation.2 In some tissues, HIF-1’s homologue HIF-2 regulates distinct functions (for example renal erythropoietin gene is regulated by HIF-1 but hepatic and cerebral erythropoietin expression is regulated by HIF-2)3 and both HIF-1 and HIF-2 are regulated not only by hypoxia but by other mechanisms including heat shock protein 9-(HSP 90 and RACK1) as well as oxygen radicals.4,5
Hypoxic preconditioning of mammalian cells leads to limited hypoxic response after re-oxygenation and is associated with protective response to ischemic damage.6 Further, normoxic mice with prior hypoxic exposure have an augmented ventilatory response to subsequent acute hypoxia.7 The current concept of hypoxia induced regulation is depicted in Figure 1.8
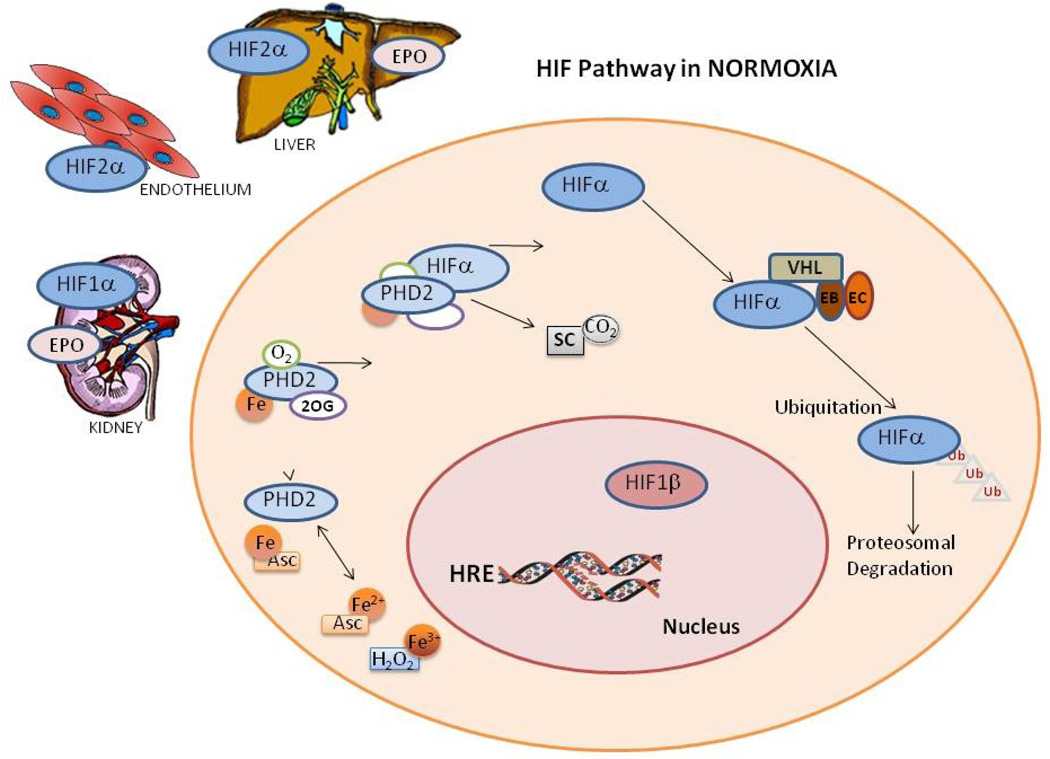
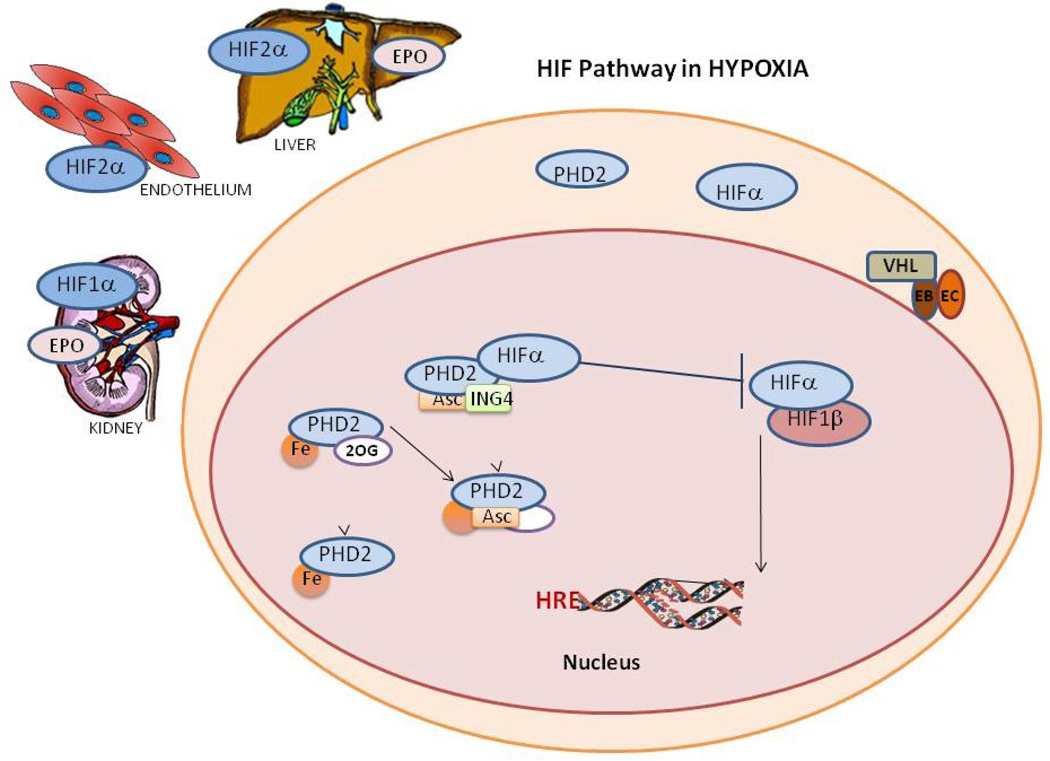
Figure 1. Hypoxia Inducible Factor (HIF) pathways.
A. In normoxia HIF hydroxylation and degradation occurs in the cytoplasm which involves, independent oxidation reduction of ascorbic acid (Asc) and iron (Fe). Fe then binds to prolyl hydroxylase (PHD2) forming a complex with 2-oxoglutarate (2OG) and oxygen, this complex in turn hydroxylate HIFA with 2 by products succinate and carbon dioxide. Unbound hydroxylated HIFA binds with the von Hippel-Lindau-Elongin B-Elongin C complex ubiquinating HIFA for proteosomal degradation.
B. While in Hypoxia HIFA enters the nucleus for hydroxylation and escape degradation, PHDs binds to Fe, 2-oxoglutarate (2OG) and ascorbic acid but not oxygen. The protein inhibitor of growth 4 (ING4) binding to PHD2 may regulate HIFA transcriptional activity and blocks HIFA-HIFB complex binding. When HIFA and HIFB binding occurs the HIF dimer can activate the gene in the hypoxia response element (HRE) site, activating HIFA dependent genes where protein levels were upregulated like HIFA and PHD2.
There is clear evidence of individual variation in response to hypoxia as witnessed by the fact that hypoxemia from pulmonary or heart disease, or from exposure to extreme high altitude, leads in some but not all of the affected individuals to an appropriate increase of red cell mass.9 Further, athletes trained in hypobaric chambers have variable responses to improved athletic performance that correlates with commensurate increase of erythropoietin levels.10,11 The individual variation of erythropoietic responses to a hypoxic stimulus suggests that there are likely genetic determinants including genetic polymorphisms underlying the erythropoietic response to hypoxia.
Similarly, there are significant population differences in response to hypoxia. Native Tibetan populations, highlanders in Ethiopia, and populations in the Andes appear to have developed an adaptive mechanism to protect against extreme hypoxia.12 However, a proportion of non-adapted Caucasians and Han Chinese and other populations exposed to extreme hypoxia of high altitude tend to develop significant complications in extreme hypoxia which include high altitude pulmonary edema (HAPE), cerebral edema, pulmonary hypertension13, and extreme polycythemia14, these complications can be fatal.
In non-acclimatized individuals one of these complications, HAPE, can occur at as low as 2,500 meters. This often lethal complication occurs with what appears to be individual predisposition.15,16 The only gene with suggested a linkage with HAPE is endothelial nitric oxide synthase (eNOS). Its polymorphisms in studies of unrelated individuals Japanese and Asian Indian ethnic subjects.16,17 There are reports that carriers of the 27bp VNTR of eNOS gene have lower nitric oxide plasma levels and decreased protein expression.16,17 Lower eNOS mRNA and serum nitric oxide levels were found in individuals with the −786C eNOS variant, this association was strengthen by in vitro reporter gene assays.21 Human in vivo studies suggested that subjects homozygous for the −786C allele have a decreased maximal forearm blood flow response to acetylcholine, a pharmacologic tool to evaluate nitric oxide production in vivo.22,23
However, to our knowledge, families with multiple affected members with HAPE have not been described. In this report, we describe one such family and demonstrate the lack of association of linkage with the eNOS gene. We also performed preliminary analysis of the whole genome using high-density SNP arrays, ruled out a linkage with other candidate genes; we believe extending these studies to other affected families or multiple individuals should lead to identification of gene(s) responsible for HAPE.
MATERIALS AND METHODS
DNA isolation
Blood samples were processed with the approval of Ethics Committee of Institute of High Altitude Medicine of Qinghai University at Xining, China. Genomic DNA was extracted from peripheral blood using the Gentra Puregene Blood Kit (Qiagen, Germantown, MA) following the manufacturer’s instruction, and concentration determined using an ND-1000 spectrophotometer (Thermo Scientific, Nanodrop, Wilmington, DE). Quality and sizing of genomic DNA was determined by agarose gel electrophoresis.
Evaluation of eNOS gene polymorphism, its linkage to HAPE, and whole genome SNP study
We examined the 894G/T, −922A/G, −786T/C polymorphisms and 27bp-VNTRs in eNOS gene. The genotype of these polymorphisms were identified with a polymerase chain reaction (PCR) followed by sequencing of both strands. The primers applied to the current PCR experiments were as follows: eNOS894-F: 5'-AAGGCAGGAGACAGTGGATGGA-3', eNOS894-R: 5'-CCCAGTCAATCCCTTTGGTGCTCA-3' for 894G/T amplification; eNOS27-F: 5'-AGGCCCTATGGTAGTGCCTT-3', eNOS27-R: 5'-TCTCTTAGT GCTGTGGTCAC-3' for 27bp-VNTRs amplification; 5UTR-F1: 5'-CTCTGAGCACA GCCCGTTCC-3', 5UTR-F2: 5'-CCCACCCCAACCTTATCCTC-3', 5UTR-R: 5'-ATG GCAGCCCACACTCTCAG-3' for −922A/G, −786T/ C polymorphisms detection.; The reactions were performed using standard protocol. The PCR product of 27bp-VNTRs was detected by 3% agarose gel electrophoresis. Other products were electrophoresed on a 1% agarose gel and purified with QIAquick Gel Extraction Kit (Qiagen, Germantown, MA). Sequencing of the PCR products was done on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Affymetrix GenomeWide SNP 5.0 arrays were used following the protocol supplied by the manufacturer (Affymetrix, Santa Clara, CA). Briefly, 250 ng genomic DNA were digested for Nsp and Sty enzymes, followed by ligation-specific adaptors and amplified by PCR using the kit primer. The amplicons were purified and quantified. The products were fragmented and labeled, following by hybridization to the array chips at 48°C for 16–18 hours. Excess unhybridized products were washed, followed by scanning with a GeneChip Scanner 3000 (Affymetrix, Santa Clara, CA). Genotypes were called using the Affymetrix BRLMM-p algorithm as implemented in the Genotyping Console software (Affymetrix, Santa Clara, CA). All samples had BRLMM call rates greater than the 95% cutoff.
We conducted a genome-wide scan of single nucleotide polymorphisms (SNPs) to determine whether we could exclude candidate genes associated with hypoxia sensing, assuming a single-gene dominant mode of inheritance. We examined patterns of haplotype inheritance using the pedigree analysis software package MERLIN Abecasis (www.sph.umich.edu/csg/abecasis/merlin). The most likely SNP haplotypes were estimated for 100KB regions spanning 35 candidate gene regions (Supplementary Table 1) by assessing informative meioses in the pedigree. We also used nonparametric linkage analysis to determine LOD scores for each region (Kong and Cox 1997), although pedigree size limits the statistical power to detect linkage.
RESULTS
Clinical presentation and description of phenotype
An 11-year-old boy, ethnic Han Chinese, was brought by ambulance to the emergency department of Lhasa Hospital (altitude 3,648 m). As related by history, upon arrival to Lhasa airport he experienced severe headache, nausea, and breathlessness. A few hours latter, he rapidly developed tachypnea and labored breathing and expectorated foamy red-tinged sputum. He was taken to Lhasa hospital. Physical examination revealed a marked cyanosis, tachypnea, and tachycardia, slight right ventricular heave and a loud second heart (pulmonary) sound. Rales were present extending from lung bases to the middle lung fields; the arterial oxygen saturation (SaO2) was 65%. The chest radiography showed moderate infiltrates in the both lung fields and slight prominence of the pulmonary artery and its branches, cardiac size was normal. He was treated with oxygen and pulmonary vasodilators. The clinical signs and symptoms rapidly improved, and SaO2 was normalized. He was diagnosed with high-altitude pulmonary edema (HAPE).
The propositus was born and raised in Chendu (altitude 500 m), Sichuan province of China, and had experienced three previous episodes of HAPE while visiting his father in Lhasa, Tibet. After the propositus’ hospital discharge and when fully recovered, his mean pulmonary arterial pressure measured by Doppler echocardiography after breathing 10% O2 was increased from 15mmHg to 25mmHg consistent with pulmonary hypertension. His plasma concentration of nitrite and nitrate were markedly lower (12.93 µmol/L) when compared with age- and sexmatched mean controls (33.80 µmol/L).
Family Studies
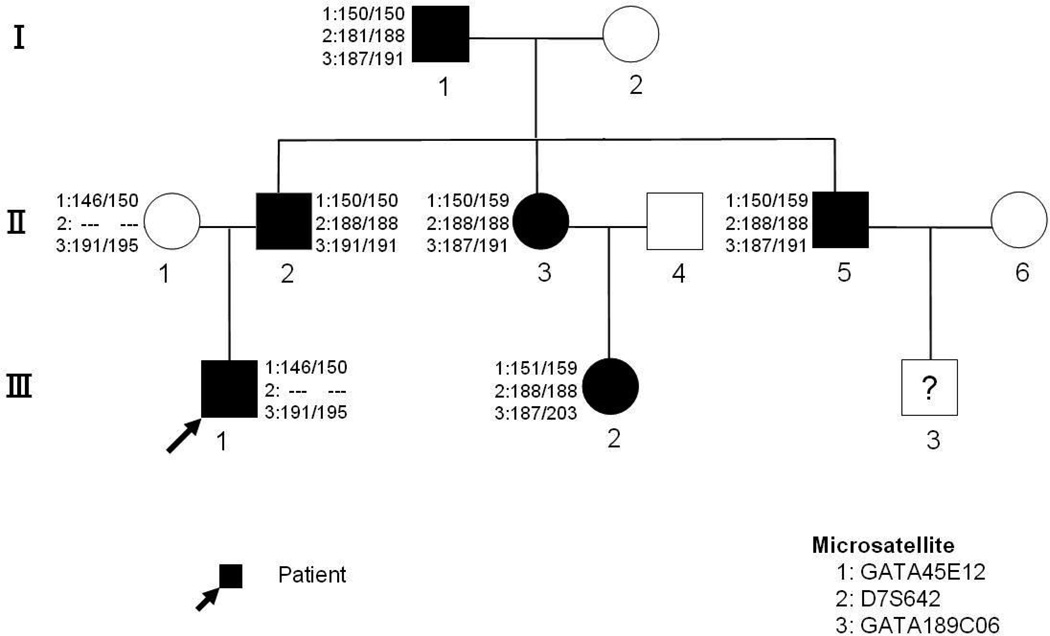
The detailed interview revealed other family members with documented HAPE, all requiring hospitalizations (confirmed by review of medical records). These affected relatives included an 8-year-old first cousin (III-2), the boy’s father (II-2), both his siblings (II-3 and II-5) and parental grandfather (I-1), all living in Lhasa while the members of the third generation and paternal grandmother (I-2) live in low altitude and visit Lhasa on rare occasions (Figure 2).
Figure 2. Pedigree chart of HAPE affected family.
Shaded and unshaded figures indicate affected and unaffected individual respectively. Numeric paired values represent the genotype alignment of the 3 eNOS microsatellite markers used to rule out association with HAPE.
While the propositus had a typical presentation of recurrent HAPE 24,25 he also had a clear genetic susceptibility to HAPE as evidenced by multiple affected first-degree relatives. HAPE is a rare life-threatening condition characterized by marked pulmonary vasoconstriction, which normally occurs in healthy persons after rapid exposure to altitudes in excess of 2,500 m. above sea level. The pathogenesis of the recurrent HAPE and familial HAPE has not yet been delineated.
eNOS
In view of these reports, we analyzed the 4 polymorphisms of eNOS gene in our HAPE family and found that only the patient harbors the following eNOS3 polymorphisms: 27bp-VNTRs 4a/b, T−786→C, A−922→G. Thus linkage of HAPE with eNOS could not be established in this family using these particular polymorphisms. We then employed 3 microsatellites in the vicinity eNOS3 gene (GATA45E12--Chromosome 7:162.33cM, D7S642--Chromosome 7: 162.62cM, GATA189C06--Chromosome 7:163.03cM) to analyze all family members and found no linkage of HAPE with eNOS (Figure 2. microsatellite in HAPE pedigree). Thus eNOS cannot account for the HAPE familial phenotype in this Chinese family.
Based on SNP analysis, the eNOS gene is not linked to the disease phenotype. Assuming a single gene causes HAPE, we ruled out linkage with 34 other candidate genes. Only HIF2A haplotype was shared by individuals who exhibit the HAPE phenotype (Supplementary Table 1). Therefore, this gene is not excluded based upon patterns of inheritance and will require further investigation. The LOD scores calculated for each of these regions are limited by the small pedigree size and are not reported here (Supplementary Table 1). Although this initial assessment does not produce enough evidence for linkage, the observed patterns provide insight for future analyses.
DISCUSSION
Clearly, the isolation of causative gene (s) responsible for HAPE would provide a rationale basis for specific targeted therapy and allow its targeted prevention for at-risk populations. Because of absence of linkage with the eNOS gene, we initiated a whole genome screen using Affymetrix SNP5.0 chips these analysis also ruled out linkage with eNOS gene. While the relatively small family size does not provide sufficient power for conclusive linkage analysis, these data can be utilized as a background for determining the region of the genome associated with HAPE and provide a basis for isolating the causative gene and elucidating the molecular basis and pathophysiology of HAPE. The low incidence of familial HAPE will require collaboration with other investigators with access to HAPE families, or HAPE affected individuals that should result in identification of causative gene(s) responsible for HAPE. When this is accomplished, this new knowledge should improve the prevention of HAPE by identification of individuals at risk, lead to development of HAPE targeted therapy, and clarification of HAPE pathophysiology.
Supplementary Material
ACKNOWLEDGEMENT
The presented study was supported by R01HL50077-11, VAH Merit review grant (JTP) NIH grant GM-59290 (LJ).
This work was supported by “National Basic Research Program of China, No.2006CB504100”, National Natural Science Foundation of China, No. 30393133. and R01HL50077-14 (NHLBI) PI Prchal, Molecular Biology of Primary Polycythemia and VAH Merit review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hirota K, Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Critical reviews in Oncology/Hematology. 2006;59(1):15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Manalo DJ, Rowan A, Lavoie T, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 3.Rankin EB, Biju MP, Liu Q, et al. Hypoxia-Inducible Factor (HIF)-2 regulates hepatic EPO in vivo. J Clin Invest. 2007;117(4):1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu YV, Baek JH, Zhang H, et al. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell. 2007;26(25(2)):207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda R, Zhang H, Kim JW, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 6.Kline D, Peng Y, Manalo D, Semenza G, Prabhakar N. Defective carotid body function and impaired ventilatory responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1 alpha. Proc. Natl. Acad. Sci. U. S. A. 2002;99:821–826. doi: 10.1073/pnas.022634199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelo G, Duplan E, Boyer N, Vigne P, Frelin C. Hypoxia up-regulates prolyl hydroxylase activity: a feedbackmechanism that limitsHIF-1 responses during reoxygenation. J. Biol. Chem. 2003;278:38183–38187. doi: 10.1074/jbc.M302244200. [DOI] [PubMed] [Google Scholar]
- 8.Qutub A, Popel A. Three autocrine feedback loops determine HIF1A expression in chronic hypoxia. BBA. 2007;1773:1511–1525. doi: 10.1016/j.bbamcr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mejía OM, Prchal JT, León-Velarde F, Hurtado A, Stockton DW. Genetic association analysis of chronic mountain sickness in an Andean high-altitude population. Haematologica. 2005;90(1):13–19. [PubMed] [Google Scholar]
- 10.Ge RL, Witkowski S, Zhang Y, et al. Determinants of Epo release in response to short term, hypobaric hypoxia. Appl Physiol. 2001;92(6):2361–2367. doi: 10.1152/japplphysiol.00684.2001. [DOI] [PubMed] [Google Scholar]
- 11.Jedlickova K, Stockton DW, Prchal JT. Search for Genetic Determinants of Individual Variability of the Epo Response to High Altitude. ExpHematol 30:44, 2002 and Blood Cells Mol Dis. 2003;31(2):175–182. doi: 10.1016/s1079-9796(03)00153-0. [DOI] [PubMed] [Google Scholar]
- 12.Beall CM. Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integrative and Comparative Biology. 2006;46(1):18–24. doi: 10.1093/icb/icj004. [DOI] [PubMed] [Google Scholar]
- 13.Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. PNAS. 2007;104:8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jefferson JA, Escudero E, Prchal JT, et al. Excessive erythrocytosis, chronic mountain sickness, and serum cobalt levels. Lancet. 2002;359(9304):407–408. doi: 10.1016/s0140-6736(02)07594-3. [DOI] [PubMed] [Google Scholar]
- 15.Kawashima A, Kubo K, Kobayashi T, et al. Hemodynamic responses to acute hypoxia, hypobaria, and exercise in subjects susceptible to high-altitude pulmonary edema. J Appl Physiol. 1989;67(5):1982–1989. doi: 10.1152/jappl.1989.67.5.1982. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzawa Y, Fujimoto, Kobayashi T, et al. Blunted hypoxic ventilatory drive in subjects susceptible to high-altitude pulmonary edema. J Appl Physiol. 1989;66(3):1152–1157. doi: 10.1152/jappl.1989.66.3.1152. [DOI] [PubMed] [Google Scholar]
- 17.Ahsan A, Mohd G, Norboo T, Baig MA, Pasha MA. Heterozygotes of NOS3 polymorphisms contribute to reduced nitrogen oxides in high-altitude pulmonary edema. Chest. 2006;130(5):1511–1519. doi: 10.1378/chest.130.5.1511. [DOI] [PubMed] [Google Scholar]
- 18.Droma Yunden, Hanaoka Masayuki, Ota Masao, Katsuyama Yoshihiko, Koizumi Tomonobu, Fujimoto Keisaku, Kobayashi Toshio, Kubo Keishi. Positive Association of the Endothelial Nitric Oxide Synthase Gene Polymorphisms With High-Altitude Pulmonary Edema. Circulation. 2002;106(7):826–830. doi: 10.1161/01.cir.0000024409.30143.70. [DOI] [PubMed] [Google Scholar]
- 19.Tsukada T, Yokoyama K, Arai T, et al. Evidence of association of the eNOS gene polymorphism with plasma NO metabolite levels in humans. Biochem Biophys Res Commun. 1998;245(1):190–193. doi: 10.1006/bbrc.1998.8267. [DOI] [PubMed] [Google Scholar]
- 20.Wang XL, Sim AS, Wang MX, et al. Genotype dependent and cigarette specific effects o n endothelial nitric oxide synthase gene expression and enzyme activity. FEBS Lett. 2000;471(1):45–50. doi: 10.1016/s0014-5793(00)01356-9. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto Y, Saito Y, Nakayama M, et al. Replication protein A1 reduces transcription of the endothelial nitric oxide synthase gene containing a −786T/C mutation associated with coronary spastic angina. Hum Mol Genet. 2000;9(18):2629–2637. doi: 10.1093/hmg/9.18.2629. [DOI] [PubMed] [Google Scholar]
- 22.Tesauro M, Thompson WC, Rogliani P, et al. Intracellular processing of endothelial nitric oxide synthase isoforms associated with differences in severity of cardiopulmonary diseases: cleavage of proteins with aspartate vs. glutamate at position 298. Proc Natl Acad Sci U S A. 2000;97(6):2832–2835. doi: 10.1073/pnas.97.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattaruzza M, Guzik TJ, Slodowski W, et al. Shear stress insensitivity of endothelial nitric oxide synthase expression as a genetic risk factor for coronary heart disease. Circ Res. 2004;95(8):841–847. doi: 10.1161/01.RES.0000145359.47708.2f. [DOI] [PubMed] [Google Scholar]
- 24.Maggiorini Marco. High altitude-induced pulmonary oedema. Cardiovascular Research. 2006;72(1):41–50. doi: 10.1016/j.cardiores.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Gluecker T, Capasso P, Schnyder P, Gudinchet F, Schaller MD, Revelly JP, Chiolero R, Vock P, Wicky S. Clinical and radiologic features of pulmonary edema. Radio graphics. 1999;19(6):1507–1531. doi: 10.1148/radiographics.19.6.g99no211507. [DOI] [PubMed] [Google Scholar]
- 26.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genetics. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 27.Kong A, Cox. NJ. Allele-Sharing Models: LOD Scores and Accurate Linkage Tests. The American Journal of Human Genetics. 1997;61(5):1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Affymetrix. BRLMM: an improved genotype calling method for the Gene Chip Human Mapping 500K array set. 2006 http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.