Abstract
Previous studies have suggested that polymorphism in the serotonin transporter gene (5-HTTLPR) influences responses to serotonergic manipulation, with opposite effects in patients recovered from depression (rMDD) and controls. Here we sought to clarify the neurocognitive mechanisms underpinning these surprising results. Twenty controls and 23 rMDD subjects completed the study; functional magnetic resonance imaging (fMRI) and genotype data were available for 17 rMDD subjects and 16 controls. Following tryptophan or sham depletion, subjects performed an emotional-processing task during fMRI. Although no genotype effects on mood were identified, significant genotype*diagnosis*depletion interactions were observed in the hippocampus and subgenual cingulate in response to emotionally valenced words. In both regions, tryptophan depletion increased responses to negative words, relative to positive words, in high-expression controls, previously identified as being at low-risk for mood change following this procedure. By contrast, in higher-risk low-expression controls and high-expression rMDD subjects, tryptophan depletion had the opposite effect. Increased neural responses to negative words following tryptophan depletion may reflect an adaptive mechanism promoting resilience to mood change following perturbation of the serotonin system, which is reversed in sub-groups vulnerable to developing depressive symptoms. However, this interpretation is complicated by our failure to replicate previous findings of increased negative mood following tryptophan depletion.
Keywords: Depression, serotonin transporter polymorphism (5-HTTLPR), acute tryptophan depletion, functional magnetic resonance imaging (fMRI), emotional processing
Introduction
A long-standing model posits that serotonin plays a central role in the pathophysiology of major depressive disorder (MDD). Supporting the serotonin hypothesis, reducing synthesis using the acute tryptophan depletion (ATD) method reinstates depressive symptoms in some remitted MDD (rMDD) subjects (Ruhe et al., 2007). In controls ATD does not commonly alter mood, but does impair cognitive processing of affective stimuli, in particular negatively biasing responses to emotional information (Murphy et al., 2002; Roiser et al., 2008). Such findings have led to a re-evaluation of the role of serotonin in MDD, focusing on its role in the processing of affective stimuli (Harmer et al., 2009).
Further evidence relevant to the serotonin hypothesis comes from genetic association studies, particularly those investigating the serotonin transporter gene. A variable number of tandem repeats (VNTR) polymorphism in the promoter region of this gene, with a 20–23 base-pair imperfect repeat, is known as the serotonin transporter-linked polymorphic region (5-HTTLPR). This results in long (‘L’) and short (‘S’) variants, corresponding to high and low serotonin transporter expression, respectively (Heils et al., 1996). An additional functional A/G polymorphism occurs almost exclusively within the L-allele, resulting in comparable expression to the S-allele (Hu et al., 2006). The S-allele has been associated with both neuroticism (Munafo et al., 2009b), a risk factor for MDD (Kendler et al., 1993), and MDD itself (Clarke et al., 2010), particularly in the context of stressful life events (Caspi et al., 2003), though some studies have reported discrepant results for the latter association (Risch et al., 2009). Moreover, the S-allele has been associated with vulnerability to developing negative mood symptoms under ATD in individuals without MDD (Neumeister et al., 2002), as well as poor response to antidepressant treatment in MDD (Serretti et al., 2007), though again negative results have been reported (Hu et al., 2007).
A number of studies reported that the influence of 5-HTTLPR polymorphism on responses to serotonergic manipulation may depend on an individual's history of psychiatric illness (Lenzinger et al., 1999; Moreno et al., 2002; Pierucci-Lagha et al., 2004). Surprisingly, these studies suggested that carrying either one or two copies of the high-expression 5-HTTLPR allele (L) was associated with a greater risk of ATD-induced mood change in individuals with a history of affective disorders. By contrast, high-expression genotype controls appeared relatively resilient to the ATD procedure (Neumeister et al., 2002; Roiser et al., 2006). Neumeister and colleagues (2006) were the first to compare directly the effects of the 5-HTTLPR on responses to ATD between individuals with and without a history of MDD. They confirmed that rMDD subjects carrying the high-expression allele were more vulnerable to ATD than those of low-expression genotypes, while the opposite pattern was evident in controls. Moreover, using positron emission tomography to assess glucose metabolism, the authors were able to demonstrate significant diagnosis-by-treatment-by-genotype interactions in the subgenual cingulate cortex (SGC) and hippocampus, with a trend in the amygdala (Neumeister et al., 2006), regions implicated in MDD (Drevets et al., 2008). rMDD subjects with high-expression genotypes, who exhibited the greatest mood change following ATD, exhibited increased glucose metabolism in these regions (Neumeister et al., 2006), mirroring the pattern observed during major depressive episodes (Drevets et al., 2008).
We sought to understand the neurocognitive mechanisms underpinning this result, using the Affective Go/No-go test (AGNG), on which depressed subjects exhibit a more negative processing bias than controls (Erickson et al., 2005; Murphy et al., 1999). Previously, we reported significant interacting effects of emotional valence, tryptophan depletion and rMDD diagnosis on behavioral and hemodynamic responses on this task (Roiser et al., 2008, 2009). Since we utilized an identical tryptophan depletion protocol to that of Neumeister and colleagues (2006), though our sample was independent, we were presented with a unique opportunity to assess the neurocognitive mechanisms underpinning the moderation of response to ATD by 5-HTTLPR polymorphism, and how this moderation might differ in subjects with and without a history of MDD. Therefore, we reanalyzed these data including 5-HTTLPR genotype as an additional factor, focusing on the four-way interaction between emotional valence, history of depression, tryptophan depletion and 5-HTTLPR genotype. We hypothesized that the above reciprocal responses to ATD in different 5-HTTLPR sub-groups resulted from biases during the processing of emotional information in prefrontal and subcortical circuits involved in mood regulation (Harmer et al., 2009). Specifically, on the basis of the findings of Neumeister and colleagues (2006), we predicted four-way valence-by-diagnosis-by-treatment-by-genotype interactions in the SGC, hippocampus, and amygdala. These regions form part of a ‘visceromotor’ network thought to subserve the assessment of and response to the emotional salience of sensory stimuli (Ongur and Price, 2000), consistently implicated in the pathophysiology of MDD (Drevets et al., 2008).
Materials and methods
Participants
Healthy volunteers (N=20) and rMDD patients (N=23) were recruited through the outpatient clinical services of the National Institute of Mental Health (NIMH) and by newspaper advertisement. Current and past psychopathology was assessed using the structured clinical interview for DSM-IV (SCID) (Spitzer et al., 2002). All participants provided informed consent following a full explanation of the procedures, risks and benefits of the study. This study was approved by the NIMH Institutional Review Board. See Roiser et al. (2009) for further details.
The majority of participants were of Caucasian ancestry (78.0%); three rMDD subjects were African American (7.3%); two control subjects were Asian (4.9%); and two control subjects and two rMDD subjects were Hispanic (9.8%). Genotype did not differ according to ethnicity (χ2=5.065, Fisher's exact p=0.1).
Experimental procedure
Participants attended two sessions, in both of which they were administered 70 white capsules containing: L-isoleucine; L-leucine; L-lysine; L-methionine; L-phenylalanine; L-threonine; and L-valine (total amino acid load: 31.5g). They were also administered four pink capsules containing either lactose (tryptophan depletion day) or tryptophan (1.2 g, sham depletion day) (Neumeister et al., 2004), in a double-blind, placebo-controlled crossover design. Mood ratings and blood samples were obtained immediately prior to amino acid ingestion (T0) and 5 h later (T5), just prior to the magnetic resonance imaging (MRI) scan. See Roiser et al. (2008) for further details
Cognitive activation paradigm (Affective Go/No-go task)
Participants performed the AGNG task during functional MRI (fMRI), using a block design. The design of the task was almost identical to that described by Elliott et al. (2000, 2002), using the same conditions, stimuli and stimulus timings. See Roiser et al. (2008) for further details.
MRI scanning
Participants were scanned during task performance using a 3T scanner (GE Signa, Milwaukee, WI, USA). Three runs of 208 functional blood oxygenation level-dependent (BOLD) MRI images were acquired using an EPI pulse sequence. See Roiser et al. (2008) for further details.
Biochemical measures
Plasma total amino acid concentrations were measured by means of high-performance liquid chromatography with fluorescence end-point detection and pre-column sample derivatization adapted from the methods of Furst et al. (1990). See Roiser et al. (2008) for further details.
Genotyping
Genotyping was performed using the two-stage 5′-nuclease assay described in Hu et al. (2006). Stage 1 distinguished the S from L alleles for the VNTR polymorphism. Stage 2 distinguished LA from LG alleles for the rs25531 polymorphism. For both assays the 5-HTTLPR region was amplified (L amplicon 182 bp, and the S amplicon 138 bp) using a common pair of primers [Forward- GCAACCTCCCAGCAACTCCCTGTA, Reverse- GAGGTGCAGGGGGATGCTGGAA]. The S and L alleles were distinguished by the use of a labeled oligonucleotide probe [FAM- TGCAGCCCCCCCAGCATCTCCC-MGB] capable of hybridizing only once to the 43-bp L insertion, and by an internal control probe (ICP) [VIC- TCCCCCCCTTCACCCCTCGCGGCATCC- MGB] directed against a unique sequence, not involved in the insertion/deletion, located within the amplicon. For stage 2, the LA and LG alleles for single nucleotide polymorphism (SNP) rs25531 were identified using allele-specific probes [LA: FAM- CCCCCCTGCACCCCCAGCATCCC-MGB; LG: VIC- CCCCTGCACCCCCGGCATCCCC-MGB]. Polymerase chain reaction (PCR) was performed in a 25 μL volume, with 25–50 ng DNA, PCR amplification primers (1 μM of each), allelic discrimination probes (240 nM-FAM probes, 160 nM VIC-probes) dimethyl sulfoxide (DMSO) (4% by volume), and 1× TaqMan Master Mix (Applied Biosystems – Part No. 4304437). Amplification conditions were 2 min at 50°C, 10 min at 95°C, and then 40 cycles at 96°C for 15 s and at 62.5°C for 90 s. Genotypes were generated using ABI PRISM 7700 Sequence Detection system software.
Stage 1 and stage 2 genotypes were combined to assign samples to one of six genotypes: SS, SLA, SLG, LALA, LALG, and LGLG. On each plate, previously sequenced standards were introduced. Stage 1 standards were SS, LS, and LL, and stage 2 standards were LALA, LALG, and LGLG. SS, SLG and LGLG individuals were defined as ‘low expression’; SLA, and LALG were defined as ‘medium expression’; LALA individuals were defined as ‘high expression’. Genotype distributions did not deviate from those expected under Hardy–Weinberg equilibrium assumptions, assessed using the likelihood ratio test in ExactoHW (Fisher's exact p=0.98; Engels, 2009). This software utilizes a permutation approach, essential for the present analysis, since the rare LG allele and relatively low number of subjects together produce low expected numbers of subjects in several genotype groups, which invalidates the standard approach based on the asymptotic assumption of the chi-square distribution.
Data analysis
Data were analyzed using SPSS 20 (IBM, Armonk, New York, USA) using analysis of variance or non-parametric equivalents if data were not normally distributed. Due to equipment failure, behavioral performance data were not recorded for two rMDD subjects. Genotype data from one healthy volunteer and one rMDD subject (for one of whom behavioral performance data were also not recorded) were unavailable. A threshold of p<0.05 (two-tailed) was adopted throughout. See Roiser et al. (2008) for further details.
Functional magnetic resonance imaging data analysis
Analysis of BOLD fMRI data was performed using the general linear model within SPM5 (Wellcome Trust Centre for Neuroimaging, London, UK), following realignment, spatial normalization and smoothing (see Roiser et al. 2008 for further details). Data from three controls and five rMDD subjects were excluded due to excessive head movement. Therefore analyses relating to hemodynamic responses and genotype were based on data from 16 healthy volunteers and 17 rMDD subjects. Single-subject interaction maps were created by contrasting the difference maps between emotional conditions across the two treatment conditions (e.g. (positive-negative target words following sham depletion)-(positive-negative target words following tryptophan depletion)). At the second level (group analysis), regions showing a significant main effects or interactions were identified through random-effects analysis of the single-subject contrast maps using F-contrasts.
We limit the discussion of the fMRI results to maxima reaching the p<0.05 (family-wise error corrected) threshold, using small volume correction (SVC) at the voxel level across three primary regions of interest (ROIs), about which we had strong a priori hypotheses. These ROIs were: amygdala; hippocampus and subgenual cingulate (Neumeister et al., 2006). ROIs were defined using the Wake Forest University Pickatlas toolbox for the amygdala and hippocampus (Maldjian et al., 2003), and using an 8 mm sphere around the coordinates (x=1; y=25; z=−6) (Drevets et al., 1997) for the SGC. These SVC procedures were carried out separately for each of the three regions in order to minimize the risk of Type II error. Post-hoc analysis of significant interactions was performed by calculating the simple main or interaction effects of interest. To construct these contrasts, we used the pooled mean squared error term and degrees of freedom from the relevant interaction term in the omnibus ANOVA, since in no case was there a departure from the assumption of homogeneity of covariance. F-values were square root transformed to t-values, which are reported in the text.
Power analysis
The contrast of primary interest in this study was the comparison of the effects of ATD on neural responses in high-expression versus low-expression unmedicated rMDD patients. The only previous study to report such an effect, though utilizing a different neuroim-aging method, was that of Neumeister and colleagues (2006), where this contrast was the main contributor to the diagnosis-by-treatment-by-genotype interaction identified (see Figure 2 of that paper).
Based on the means, standard errors and numbers of participants reported, we were able to calculate effect sizes for this contrast in our primary ROIs (calculation available from the authors on request). The effect sizes (Cohen's d) varied across the ROIs, but were with one exception between ∼1.6 and ∼2.2. With the numbers of participants in our high-expression and low-expression rMDD sub-groups (four and five, respectively) our analysis had power equivalent to minimum 48% (assuming effect size=1.6) and maximum 74% (assuming effect size=2.2) at p=0.05 (two-tailed) in order to detect effects of this magnitude.
For completeness, where significant four-way interactions were identified in our fMRI analysis, we report effect sizes comparing the high-expression and low-expression genotypes at the peak voxel of the four-way interaction in the control and rMDD groups separately. Unlike the analyses of the simple main and interaction effects described above, these effect size calculations used the error terms and numbers of subjects of the two groups in question for comparability with previous studies. It should be noted that effect sizes based on the peak voxel almost certainly yield inflated values relative the true effect size (Kriegeskorte et al., 2010) and as such these values represent over-estimates.
Results
Demographic and clinical data
There were no differences between the control and rMDD groups on demographic variables (see Table 1 and Roiser et al., 2009), and there was no effect of genotype on clinical variances in the rMDD group. However, there was a significant main effect of genotype on age: medium-expression subjects were significantly older than high-expression and low-expression subjects (F(2,35)=4.8, p=0.015; post-hoc: medium vs. high expression p=0.009; medium vs. low expression p=0.038). There was also a significant main effect of genotype on IQ: the medium and low-expression subjects were lower in IQ (F(2,33)=4.7, p=0.015; post-hoc: high vs. medium expression p=0.069; high vs. low expression p=0.005). The group×genotype interactions were non-significant for both measures. Gender was well matched between the genotype groups (χ2<1). Including these variables as covariates in analyses of behavior on the AGNG and hemodynamic responses did not affect the results, and in no case was any covariate significant in the model. Therefore, all results presented are without covariates.
Table 1.
Demographic and clinical data by genotype.
| Controls (N=19) | rMDD (N=22) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| High expression | Medium expression | Low expression | High expression | Medium expression | Low expression | |
| N (male) | 5 (3) | 9 (1) | 5 (3) | 6 (1) | 9 (3) | 7 (1) |
| Age | 26.4 (5.4) | 36.3 (5.1) | 25.0 (5.2) | 27.7 (6.9) | 34.7 (10.7) | 32.0 (10.2) |
| IQ | 127.6 (10.1) | 124.4 (10.2) | 117.8 (10.5) | 135.6 (7.5) | 127.8 (8.0) | 120.1 (10.9) |
| Ethnicity | 3C, 2H | 9C | 3C, 2As | 4C, 2Af | 8C, 1H | 5C, 1Af, 1H |
| Number of first degree relatives with depression | - | - | - | 1.2 (0.75) | 0.89 (0.78) | 1.3 (0.95) |
| Age of onset of depression (years) | - | - | - | 19.3 (7.3) | 19.9 (7.3) | 20.4 (11.8) |
| Number of depressive episodes | - | - | - | 2.7 (1.6) | 2.0 (1.3) | 3.6 (2.9) |
| Time in remission (weeks) | - | - | - | 27.3 (18.1) | 49.8 (55.6) | 17.0 (8.3) |
| Time since last medication (weeks) | - | - | - | 25.2 (13.3) | 75.9 (65.6) | 20.4 (48.7) |
| Number (%) with past SSRI use | - | - | - | 5 (83.3) | 7 (77.8) | 5 (71.4) |
| Number (%) hospitalized for depression | - | - | - | 1 (16.7) | 2 (22.2) | 2 (28.6) |
| Number (%) with past suicide attempt | - | - | - | 2 (33.3) | 2 (22.2) | 1 (14.3) |
IQ, intelligence quotient; C, Caucasian; Af, African American; As, Asian; H, Hispanic; SSRI, selective serotonin reuptake inhibitor. Figures represent means (standard deviations).
In addition, since population stratification can influence the results of genetic association analyses in ethnically heterogeneous samples, we repeated all analyses including only subjects of Caucasian ancestry. In no case did this change the results, and hence all analyses reported include both Caucasian and non-Caucasian subjects.
Plasma amino acid concentrations
TRP- (tryptophan depleted amino acid mixture) administration reduced the ratio of plasma tryptophan to the other large neutral amino acids by 90%, while TRP+ (tryptophan containing amino acid mixture) induced a 35% reduction in this ratio (treatment×time interaction: F(1,39)=50.7, p<0.001; see also Roiser et al., 2009). Critically, all genotype interactions with treatment and time were non-significant. However, there was a significant diagnosis×genotype interaction for the plasma ratio of tryptophan to the other large neutral amino acids (trypophan:ΣLNAA) (F(2,34)=4.6, p=0.017), as well as a main effect of genotype (F(2,34)=3.5, p=0.041). Post-hoc analysis revealed that the effect of genotype was significant in the controls (F(2,15)=5.3, p=0.018) but not in rMDD subjects (F(2,19)=1.3, p=0.3). In the controls the overall tryptophan:ΣLNAA ratio was lower in the medium-expression controls than high-expression (p=0.018) and low-expression (p=0.017) controls.
Mood
There were no significant main effects or interactions with genotype on the HAM-D or HAM-A (p>0.1 for all).
Hemodynamic responses
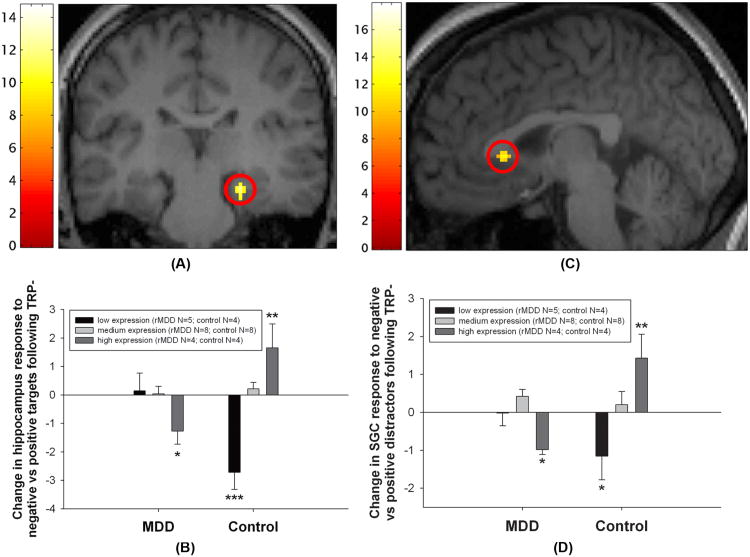
There was a significant valence×diagnosis×treatment×genotype interaction in the right hippocampus for the positive-negative targets contrast ((x=24, y=−28, z=−12), Z=3.60, p<0.05 (SVC); Figure 1A). Post-hoc analysis indicated significant, but opposite, effects of 5-HTTLPR polymorphism in the two groups (Figure 1B). In high-expression rMDD subjects (t(27)=2.43, p=0.022) and low-expression controls (t(27)=5.18, p<0.0001), TRP- decreased hemo-dynamic responses to negative relative to positive targets. However, in high-expression controls, TRP- increased hemodynamic responses to negative relative to positive targets (t(27)=3.16, p=0.004). Further analysis revealed a significant difference between high-expression and low-expression controls (t(27)=6.60, p<0.0001; Cohen's d at peak voxel: 2.06), and between high-expression and low-expression rMDD patients (t(27)=2.14, p=0.042; Cohen's d at peak voxel: 1.18). Significant valence×diag nosis×treatment×genotype interactions were not detected in the positive-negative targets contrast in the SGC, left hippocampus or left/right amygdala.
Figure 1.
Valence×diagnosis×treatment×genotype interactions in hippocampus and subgenual cingulate (SGC) cortex. (A) Four-way interaction in the right hippocampus for positive vs. negative target words. The peak voxel was significant at p<0.05 (two-tailed, corrected), restricting the search volume on the basis of an anatomical mask (see Methods for details). For display purposes, the image is thresholded at p<0.001 (two-tailed, uncorrected) without applying any anatomical mask. The color bar indicates F-values. (B) Plot of contrast estimates at the peak voxel in the hippocampus for the positive relative to negative target words under sham (TRP+) relative to tryptophan (TRP−) depletion contrast (i.e. each bar represents a two-way valence×treatment interaction in each of the genotype sub-groups). In high-expression rMDD subjects and low-expression controls, TRP− increased hemodynamic responses to positive relative to negative targets, while in high-expression controls TRP− increased hemodynamic responses to negative relative to positive targets. (C) Four-way interaction in the SGC for positive vs. negative distractor words. The peak voxel was significant at p<0.05 (two-tailed, corrected), restricting the search volume on the basis of an anatomical mask (see Materials and methods for details). For display purposes, the image is thresholded at p<0.001 (two-tailed, uncorrected) without applying any anatomical mask. The color bar indicates F-values. (D) Plot of contrast estimates at the peak voxel in the SGC for the positive relative to negative distractor words under TRP+ relative to TRP− contrast (i.e. each bar represents a two-way valence×treatment interaction in each of the genotype sub-groups). In high-expression rMDD subjects and low-expression controls, TRP− increased hemodynamic responses to positive relative to negative distractors, while in high-expression controls TRP− increased hemodynamic responses to negative relative to positive distractors. Error bars represent one standard error of the mean. * p<0.05; ** p<0.01; *** p<0.001 (two-tailed, one-sample t-test relative to zero, equivalent to a valence×treatment interaction).
There was a significant valence×diagnosis×treatment×geno type interaction in the SGC for the positive-negative distractors contrast ((x=−4, y=21, z=1), Z=3.63, p<0.05 (SVC); Figure 1C). Post-hoc analysis indicated significant, but opposite, effects of 5-HTTLPR polymorphism in the two groups (Figure 1D). In high-expression rMDD subjects (t(27)=2.26, p =0.032) and low-expression controls (t(27)=2.67, p=0.013), TRP-decreased hemodynamic responses to negative relative to positive distractors. However, in high-expression controls, TRP- increased hemodynamic responses to negative relative to positive distractors (t(27)=3.31, p=0.003). Further analysis revealed a significant difference between high-expression and low-expression controls (t(27)=4.72, p=0.0001, Cohen's d at peak voxel: 1.66), with a trend towards a difference between high-expression and low-expression rMDD subjects (t(27)=1.76, p=0.090; Cohen's d at peak voxel: 1.62). Significant valence×diagnosis×treatment×genotype interactions were not detected in the positive-negative distractors contrast in the left or right hippocampus or amygdala. See supplementary material Tables S1–S4.
Behavior on the AGNG
Behavioral data are listed in Table 2. For commission errors (inappropriate responses), there was a significant valence-by-genotype interaction (F(2,34)=5.7, p=0.007). Over both testing sessions, subjects with low-expression (t(34)=5.23, p<0.0001) and high-expression (t(34)=3.34, p=0.002) genotypes showed a significant positive bias (more inappropriate responses to positive than negative distractors), while subjects with medium expression did not (t(34)=1.13, p=0.27). All higher-order interactions were non-significant. No significant effects of genotype were detected for reaction times or omission errors.
Table 2.
Mood, plasma amino acid and behavioral data by genotype.
| Controls (N=19) | rMDD (N=22) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| High expression | Medium expression | Low expression | High expression | Medium expression | Low expression | |
| HAM-D TRP+ T0 | 0.40 (0.55) | 0.89 (2.0) | 0.40 (0.55) | 1.0 (1.5) | 1.2 (1.5) | 1.4 (1.7) |
| HAM-D TRP+ T5 | 1.0 (1.7) | 1.0 (1.3) | 3.2 (2.7) | 3.2 (5.3) | 3.2 (2.5) | 2.6 (2.7) |
| HAM-D TRP- T0 | 0.0 (0.0) | 0.22 (0.67) | 0.80 (0.84) | 1.7 (1.6) | 1.9 (1.5) | 2.0 (2.2) |
| HAM-D TRP- T5 | 1.0 (0.71) | 1.0 (1.9) | 2.6 (2.7) | 2.5 (1.9) | 2.1 (2.8) | 3.0 (2.3) |
| TRP:ΣLNAA ratio TRP+ T0 | 0.11 (0.088) | 0.075 (0.025) | 0.16 (0.053) | 0.14 (0.031) | 0.13 (0.053) | 0.096 (0.037) |
| TRP:ΣLNAA ratio TRP+ T5 | 0.11 (0.093) | 0.043 (0.019) | 0.092 (0.044) | 0.10 (0.049) | 0.072 (0.033) | 0.063 (0.016) |
| TRP:ΣLNAA ratio TRP- T0 | 0.16 (0.084) | 0.087 (0.026) | 0.13 (0.033) | 0.13 (0.042) | 0.13 (0.046) | 0.12 (0.069) |
| TRP:ΣLNAA ratio TRP- T5 | 0.023 (0.023) | 0.0072 (0.0020) | 0.023 (0.0050) | 0.0095 (0.0066) | 0.014 (0.011) | 0.010 (0.0045) |
| Happy RT TRP+ | 614.5 (29.7) | 617.4 (97.3) | 609.3 (45.5) | 670.6 (65.3)* | 657.3 (50.4) | 658.3 (74.9) |
| Sad RT TRP+ | 601.5 (57.4) | 633.6 (76.5) | 610.3 (40.6) | 669.7 (76.1)* | 648.1 (45.0) | 647.2 (72.9) |
| Happy RT TRP- | 632.9 (53.9) | 645.3 (67.4) | 637.9 (49.0) | 648.7 (63.6)* | 662.0 (67.5) | 655.3 (64.2) |
| Sad RT TRP- | 612.0 (49.3) | 668.2 (65.8) | 615.0 (44.4) | 627.5 (67.7)* | 652.8 (74.1) | 649.3 (48.1) |
| Happy CE TRP+ | 1.7 (1.9) | 1.7 (0.87) | 2.0 (1.3) | 1.7 (1.4)* | 1.2 (0.91) | 1.6 (1.1) |
| Sad CE TRP+ | 0.87 (0.69) | 1.4 (1.0) | 0.80 (0.69) | 1.0 (0.62)* | 1.3 (1.42) | 0.67 (1.1) |
| Happy CE TRP- | 1.4 (0.70) | 1.4 (1.1) | 1.7 (0.43) | 1.6 (1.4)* | 1.8 (1.1) | 1.7 (0.49) |
| Sad CE TRP- | 0.63 (0.53) | 1.8 (1.3) | 1.5 (2.3) | 0.69 (0.75)* | 1.2 (0.91) | 0.81 (0.69) |
| Happy OE TRP+ | 0.87 (1.2) | 1.4 (2) | 1.1 (0.64) | 1.2 (1.4)* | 1.3 (1.3) | 1.4 (1.2) |
| Sad OE TRP+ | 0.93 (0.89) | 0.96 (1.2) | 0.73 (0.72) | 1.0 (1.0)* | 0.96 (1.3) | 0.9 (0.69) |
| Happy OE TRP- | 0.73 (0.89) | 1.5 (1.5) | 1.7 (0.49) | 0.61 (0.57)* | 1.1 (1.3) | 1.0 (0.94) |
| Sad OE TRP- | 0.70 (0.77) | 1.2 (1.5) | 0.73 (0.95) | 0.83 (0.55)* | 0.81 (1.4) | 0.43 (0.32) |
behavioral data from the Affective Go/No-go task were not recorded for one female high-expression rMDD subject
HAM-D, Hamilton Depression Rating Scale; TRP+, sham depletion; TRP-, tryptophan depletion TRP:ΣLNAA, tryptophan to sum of large neutral amino acid; RT, reaction time; CE, commission error; OE, omission error. Figures represent means (standard deviations).
Correlations
There were no significant correlations between plasma amino acid levels, mood ratings, behavior on the AGNG or neural responses to emotional words.
Discussion
These data provide insight into the cognitive and functional neu-roanatomical mechanisms underpinning the effects of 5-HTTLPR genotype on response to ATD. Consistent with previous reports (Neumeister et al., 2006), neural responses following ATD were strongly influenced by genotype in SGC and medial temporal regions, but in opposite directions in rMDD subjects and controls. The high-expression rMDD subjects and low-expression controls exhibited qualitatively similar changes in hemodynamic responses to emotional words following ATD. Specifically, ATD decreased responses to negative relative to positive words in these groups, who were reported to be more vulnerable to mood change following tryptophan depletion in previous studies (Lenzinger et al., 1999; Moreno et al., 2002; Neumeister et al., 2002, 2006; Pierucci-Lagha et al., 2004). By contrast, in high-expression controls ATD increased hemodynamic responses to negative relative to positive words. These neural effects were unconfounded by changes in mood, raising the possibility that alterations in the processing of emotional stimuli following changes in 5-HT transmission may occur in the absence of mood change, in line with a recent theory of antidepressant drug action (Harmer et al., 2009).
The pattern of results that we observed may be relevant to understanding two effects of the 5-HTTLPR that have received considerable attention in recent years. First, there have been a number of reports that carriers of low-expression alleles at the 5-HTTLPR are more vulnerable to developing depression, particularly following stressful life events (Caspi et al., 2003, 2010; Clarke et al., 2010; Uher and McGuffin, 2010). This finding is corroborated by a frequently described and consistently replicated increase in amygdala responsivity in these individuals (Munafo et al., 2008), as well as reports of increased vulnerability to mood and motivational disturbance following tryptophan depletion in low-expression healthy volunteers (Neumeister et al., 2002; Roiser et al., 2006).
These observations suggest the hypothesis that the increased hemodynamic responses in high-expression controls to negative versus positive words following tryptophan depletion might constitute an adaptive response that confers resilience, and that this response is reversed in more vulnerable low-expression controls. The fact that these interactions, which were of similar shape, were identified in different regions for the target words (medial temporal) and distractor words (SGC) contrasts may be related to the different roles that these interconnected regions play in cognitive processing (emotional memory and emotional inhibition, respectively: Silbersweig et al., 2007; Strange and Dolan, 2006). This finding is also consistent with evidence that 5-HTTLPR variation influences functional connectivity of both the SGC (Pezawas et al., 2005) and hippocampus (Canli et al., 2006), particularly in individuals who had suffered from stressful life events. Interestingly, a recent study reported also reduced hippocampal volumes in MDD patients carrying the S-allele who had suffered from childhood adversity, suggesting that this region may also play a role in mediating resilience (Frodl et al., 2010). However, it should also be noted that a number of studies have failed to confirm the gene×environment interaction described above (Munafo et al., 2009a; Risch et al., 2009), and as such this hypothesis remains speculative and requires testing in future, larger studies.
Second, there have been several reports that carriers of the high-expression 5-HTTLPR allele are more responsive to antide-pressant medication (Kato and Serretti, 2010; Serretti et al., 2007), although again this pharmacogenetic interaction has received inconsistent support (Taylor et al., 2010). Conversely, other studies reported that the same high-expression allele increases vulnerability to tryptophan depletion-induced mood change in rMDD patients (Neumeister et al., 2006), although we were unable to replicate this effect. Nonetheless, our interpretation of tryptophan depletion-induced altered hemodynamic responses to negative relative to positive stimuli outlined above might also apply to rMDD subjects: only high-expression rMDD subjects exhibited decreased hemodynamic responses to negative vs. positive words, while hemodynamic responses in our ROIs to emotionally valenced words were apparently unchanged by tryptophan depletion in the low-expression rMDD subjects.
We failed to replicate the finding of increased negative mood following tryptophan depletion in high-expression rMDD patients and low-expression controls (Neumeister et al., 2006). This could reflect low statistical power, since the magnitude of the effect of the 5-HTTLPR on behavior was lower than the effect on regional metabolism in the prior study of Neumeister and colleagues (indeed the treatment×group×genotype×time interaction narrowly missed statistical significance in that study). In addition, the sample we recruited may have been unusually resilient to tryptophan depletion for some reason: notably, we did not detect any significant mood changes following tryptophan depletion relative to sham depletion, even when considering the rMDD group as a whole (Roiser et al., 2009). One possible explanation is the relatively high IQ of our rMDD sample (mean ∼136 in the high-expression subgroup). Previous studies have reported that high IQ depressed patients respond better to selective serotonin reuptake inhibitor (SSRI) treatment (Fournier et al., 2009); possibly, high IQ individuals are also less vulnerable to serotonin depletion. However, this hypothesis has not been tested empirically to our knowledge. Alternatively, different demographic and clinical characteristics may explain this discrepancy: on average, our sample of rMDD subjects was younger and had suffered fewer depressive episodes than that tested by Neumeister and colleagues (2006). This failure to replicate also raises the issue of why we were able to detect effects of the 5-HTTLPR on hemodynamic responses, but not on mood responses, following tryptophan depletion. Although a detailed examination of this issue, which is widespread in fMRI research, is beyond the scope of this paper (see for example Murray et al., 2010 for a thorough discussion), it may be that objectively measured hemodynamic responses are more sensitive to pharmacological manipulation than mood rating scales (see also Harmer et al., 2009).
A number of limitations of our study merit comment. First, the small number of participants we tested raises the possibility of Type II error, although even with this small number of participants we had between 48% and 74% power to detect effects of the magnitude reported by Neumeister and colleagues (2006). It should also be noted that while the Type I error rate within our study was controlled, the ratio of false positive to true positive findings is raised across studies if several have low statistical power (Sterne and Davey Smith, 2001). Second, in light of the small number of subjects we were able to test, we adopted strict inclusion/exclusion criteria in order to decrease heterogeneity and consequently to reduce the chance of Type II error. However, this aspect of our study also limits the generalizability of our findings; for example, the effects of genotype on response to tryptophan depletion could potentially be quite different in medicated rMDD subjects. Third, we restricted our analysis to a priori anatomically defined ROIs, improving our sensitivity to detect effects. However, it is likely that we may have missed effects smaller than those reported previously, especially outside these regions, which our study was not powered to detect. Fourth, the number of independent contrasts between conditions we performed and the number of regions examined also raises the possibility of Type I error. However, the relatively stringent correction for multiple comparisons we employed, controlling the family-wise error rate, should ensure that the number of false positives per contrast remained relatively low. Moreover, the similar shape of the significant interactions identified and the replication of previous results (Neumeister et al., 2006) encourages greater confidence in our findings. Finally, while the majority of our participants reported that they were of Caucasian ancestry, and all results remained significant when only Caucasians were included in the analyses, we did not use informative markers to corroborate ancestry. Therefore there is a very small chance that our results could be confounded by population stratification, although such stratification would be unlikely to vary between the control and rMDD groups in a systematic fashion.
In conclusion, consistent with previous reports (Lenzinger et al., 1999; Moreno et al., 2002; Neumeister et al., 2004, 2006; Pierucci-Lagha et al., 2004), 5-HTTLPR polymorphism modulated hemodynamic responses to emotional stimuli following tryptophan depletion in an opposite manner in rMDD patients and healthy volunteers. While these paradoxical reciprocal effects require confirmation and remain incompletely understood, they underscore the importance of studying pharmacogenetic interactions in samples of patients with depression, since these interactions may be qualitatively different from those identified healthy volunteer populations. Such strategies may ultimately aid the identification of specific sub-groups of patients that might show better response to treatment.
Acknowledgments
We thank Judy Starling for preparation of the amino acid mixtures, Mike Franklin for analysis of the plasma amino acids, Harvey Iwamoto for programming the AGNG, Rebecca Elliott and Judy Rubinsztein for providing the original word lists for the AGNG, Jeanette Black and Renee Hill for technologist support, the nurses of ward 5 South-West for their clinical support and Karl Friston for guidance regarding fMRI analysis. We thank Joan Williams, Michele Drevets and Paul Carlson for help with recruitment and clinical support. We thank Longina Akhtar for assistance with tissue culture and Gary Jenkins for assistance with genotyping. We thank Ruth Waxman for help with manuscript preparation. Finally, we thank all the volunteers who participated.
Funding: This research was supported by the Intramural Research Program of the NIMH. JPR was supported by the NIH-Cambridge Health Science Scholars Program.
Footnotes
Conflict of interest: JPR and BJS have received compensation for consultancy work from Cambridge Cognition Ltd., who now own the behavioral version of the AGNG. The other authors have no conflict of interest to declare.
References
- Canli T, Qiu M, Omura K, et al. Neural correlates of epigenesis. Proc Natl Acad Sci U S A. 2006;103:16033–16038. doi: 10.1073/pnas.0601674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, et al. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am J Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Clarke H, Flint J, Attwood AS, et al. Association of the 5- HTTLPR genotype and unipolar depression: a meta-analysis. Psychol Med. 2010;40:1767–1778. doi: 10.1017/S0033291710000516. [DOI] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Simpson JR, Jr, et al. Subgenual prefrontal cortex abnormalities in mood disorders. Nature. 1997;386:824–827. doi: 10.1038/386824a0. [DOI] [PubMed] [Google Scholar]
- Elliott R, Rubinsztein JS, Sahakian BJ, et al. Selective attention to emotional stimuli in a verbal go/no-go task: an fMRI study. Neuroreport. 2000;11:1739–1744. doi: 10.1097/00001756-200006050-00028. [DOI] [PubMed] [Google Scholar]
- Elliott R, Rubinsztein JS, Sahakian BJ, et al. The neural basis of mood-congruent processing biases in depression. Arch Gen Psychiatry. 2002;59:597–604. doi: 10.1001/archpsyc.59.7.597. [DOI] [PubMed] [Google Scholar]
- Engels WR. Exact tests for Hardy-Weinberg proportions. Genetics. 2009;183:1431–1441. doi: 10.1534/genetics.109.108977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson K, Drevets WC, Clark L, et al. Mood-congruent bias in affective go/no-go performance of unmedicated patients with major depressive disorder. Am J Psychiatry. 2005;162:2171–2173. doi: 10.1176/appi.ajp.162.11.2171. [DOI] [PubMed] [Google Scholar]
- Fournier JC, DeRubeis RJ, Shelton RC, et al. Prediction of response to medication and cognitive therapy in the treatment of moderate to severe depression. J Consult Clin Psychol. 2009;77:775–787. doi: 10.1037/a0015401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodl T, Reinhold E, Koutsouleris N, et al. Childhood stress, serotonin transporter gene and brain structures in major depression. Neuropsychopharmacology. 2010;35:1383–1390. doi: 10.1038/npp.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst P, Pollack L, Graser TA, et al. Appraisal of four pre-column derivatization methods for the high-performance liquid chromatographic determination of free amino acids in biological materials. J Chromatogr. 1990;499:557–569. doi: 10.1016/s0021-9673(00)97000-6. [DOI] [PubMed] [Google Scholar]
- Harmer CJ, Goodwin GM, Cowen PJ. Why do antidepressants take so long to work? A cognitive neuropsychological model of antidepressant drug action. Br J Psychiatry. 2009;195:102–108. doi: 10.1192/bjp.bp.108.051193. [DOI] [PubMed] [Google Scholar]
- Heils A, Teufel A, Petri S, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66:2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XZ, Rush AJ, Charney D, et al. Association between a functional serotonin transporter promoter polymorphism and citalopram treatment in adult outpatients with major depression. Arch Gen Psychiatry. 2007;64:783–792. doi: 10.1001/archpsyc.64.7.783. [DOI] [PubMed] [Google Scholar]
- Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry. 2010;15:473–500. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Kessler RC, Neale MC, et al. The prediction of major depression in women: toward an integrated etiologic model. Am J Psychiatry. 1993;150:1139–1148. doi: 10.1176/ajp.150.8.1139. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Lindquist MA, Nichols TE, et al. Everything you never wanted to know about circular analysis, but were afraid to ask. J Cereb Blood Flow Metab. 2010;30:1551–1557. doi: 10.1038/jcbfm.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzinger E, Neumeister A, Praschak-Rieder N, et al. Behavioral effects of tryptophan depletion in seasonal affective disorder associated with the serotonin transporter gene? Psychiatry Res. 1999;85:241–246. doi: 10.1016/s0165-1781(99)00009-8. [DOI] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, et al. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003;19:1233–1239. doi: 10.1016/s1053-8119(03)00169-1. [DOI] [PubMed] [Google Scholar]
- Moreno FA, Rowe DC, Kaiser B, et al. Association between a serotonin transporter promoter region polymorphism and mood response during tryptophan depletion. Mol Psychiatry. 2002;7:213–216. doi: 10.1038/sj.mp.4000962. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Brown SM, Hariri AR. Serotonin transporter (5-HTTLPR) genotype and amygdala activation: a meta-analysis. Biol Psychiatry. 2008;63:852–857. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR, Durrant C, Lewis G, et al. Gene = environment interactions at the serotonin transporter locus. Biol Psychiatry. 2009a;65:211–219. doi: 10.1016/j.biopsych.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Munafo MR, Freimer NB, Ng W, et al. 5-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. Am J Med Genet B Neuropsychiatr Genet. 2009b;150B:271–281. doi: 10.1002/ajmg.b.30808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FC, Sahakian BJ, Rubinsztein JS, et al. Emotional bias and inhibitory control processes in mania and depression. Psychol Med. 1999;29:1307–1321. doi: 10.1017/s0033291799001233. [DOI] [PubMed] [Google Scholar]
- Murphy FC, Smith KA, Cowen PJ, et al. The effects of tryptophan depletion on cognitive and affective processing in healthy volunteers. Psychopharmacology (Berl) 2002;163:42–53. doi: 10.1007/s00213-002-1128-9. [DOI] [PubMed] [Google Scholar]
- Murray GK, Corlett PR, Fletcher PC. The neural underpinnings of associative learning in health and psychosis: how can performance be preserved when brain responses are abnormal? Schizophr Bull. 2010;36:465–471. doi: 10.1093/schbul/sbq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumeister A, Hu XZ, Luckenbaugh DA, et al. Differential effects of 5-HTTLPR genotypes on the behavioral and neural responses to tryptophan depletion in patients with major depression and controls. Arch Gen Psychiatry. 2006;63:978–986. doi: 10.1001/archpsyc.63.9.978. [DOI] [PubMed] [Google Scholar]
- Neumeister A, Konstantinidis A, Stastny J, et al. Association between serotonin transporter gene promoter polymorphism (5HTTLPR) and behavioral responses to tryptophan depletion in healthy women with and without family history of depression. Arch Gen Psychiatry. 2002;59:613–620. doi: 10.1001/archpsyc.59.7.613. [DOI] [PubMed] [Google Scholar]
- Neumeister A, Nugent AC, Waldeck T, et al. Neural and behavioral responses to tryptophan depletion in unmedicated patients with remitted major depressive disorder and controls. Arch Gen Psychiatry. 2004;61:765–773. doi: 10.1001/archpsyc.61.8.765. [DOI] [PubMed] [Google Scholar]
- Ongur D, Price JL. The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb Cortex. 2000;10:206–219. doi: 10.1093/cercor/10.3.206. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Meyer-Lindenberg A, Drabant EM, et al. 5-HTTLPR polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. Nat Neurosci. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Feinn R, Modesto-Lowe V, et al. Effects of rapid tryptophan depletion on mood and urge to drink in patients with co-morbid major depression and alcohol dependence. Psychopharmacology (Berl) 2004;171:340–348. doi: 10.1007/s00213-003-1588-6. [DOI] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiser JP, Blackwell AD, Cools R, et al. Serotonin transporter polymorphism mediates vulnerability to loss of incentive motivation following acute tryptophan depletion. Neuropsychopharmacology. 2006;31:2264–2272. doi: 10.1038/sj.npp.1301055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiser JP, Levy J, Fromm SJ, et al. The effects of tryptophan depletion on neural responses to emotional words in remitted depression. Biol Psychiatry. 2009;66:441–450. doi: 10.1016/j.biopsych.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiser JP, Levy J, Fromm SJ, et al. The effect of acute tryptophan depletion on the neural correlates of emotional processing in healthy volunteers. Neuropsychopharmacology. 2008;33:1992–2006. doi: 10.1038/sj.npp.1301581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe HG, Mason NS, Schene AH. Mood is indirectly related to serotonin, norepinephrine and dopamine levels in humans: a meta-analysis of monoamine depletion studies. Mol Psychiatry. 2007;12:331–359. doi: 10.1038/sj.mp.4001949. [DOI] [PubMed] [Google Scholar]
- Serretti A, Kato M, De Ronchi D, et al. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry. 2007;12:247–257. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- Silbersweig D, Clarkin JF, Goldstein M, et al. Failure of frontolimbic inhibitory function in the context of negative emotion in borderline personality disorder. Am J Psychiatry. 2007;164:1832–1841. doi: 10.1176/appi.ajp.2007.06010126. [DOI] [PubMed] [Google Scholar]
- Spitzer MB, Gibbon M, Williams JBW. Stuctured Clinical Interview for DSM-IV-TR, Research Version, Patient Edition (SCID-I/P) New York: Biometrics Research, New York State Biometrics Institute; 2002. [Google Scholar]
- Sterne JA, Davey Smith G. Sifting the evidence-what's wrong with significance tests? BMJ. 2001;322:226–231. doi: 10.1136/bmj.322.7280.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Dolan RJ. Anterior medial temporal lobe in human cognition: memory for fear and the unexpected. Cogn Neuropsychiatry. 2006;11:198–218. doi: 10.1080/13546800500305096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Sen S, Bhagwagar Z. Antidepressant response and the serotonin transporter gene-linked polymorphic region. Biol Psychiatry. 2010;68:536–543. doi: 10.1016/j.biopsych.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the etiology of depression: 2009 update. Mol Psychiatry. 2010;15:18–22. doi: 10.1038/mp.2009.123. [DOI] [PubMed] [Google Scholar]