Abstract
Introduction
Morphea or localized scleroderma is a relatively rare disease whose main symptom is excessive skin fibrosis. Here we focus on the involvement of human endogenous retroviruses (HERVs) in morphea. The HERVs are a vast and intensely growing field in genomics. HERVs are of special interest as far as autoimmune disorders are concerned, yet little effort has been made until now to assess the possible changes of their expression in morphea.
Material and methods
Six sequences of particular interest were chosen for this study. Real-time polymerase chain reaction was performed on samples derived from peripheral blood mononuclear cells (PBMCs) and skin biopsies. The results were normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcription.
Results
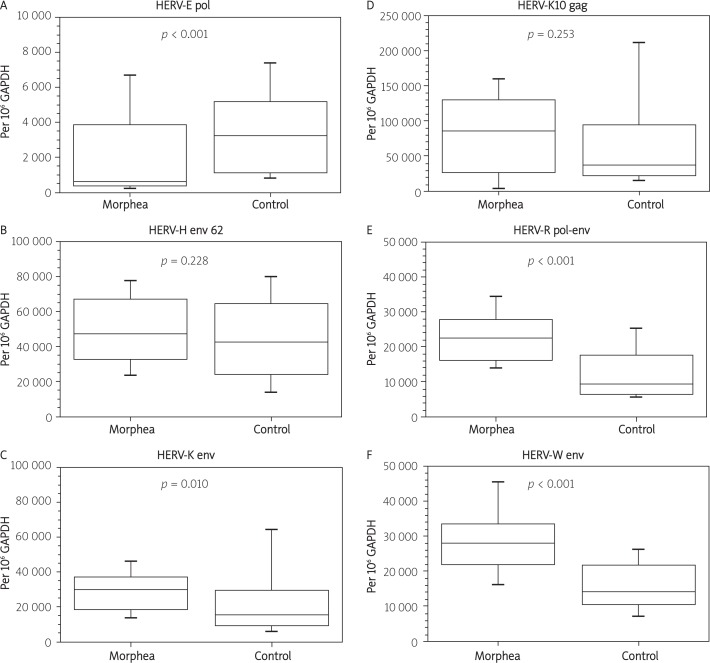
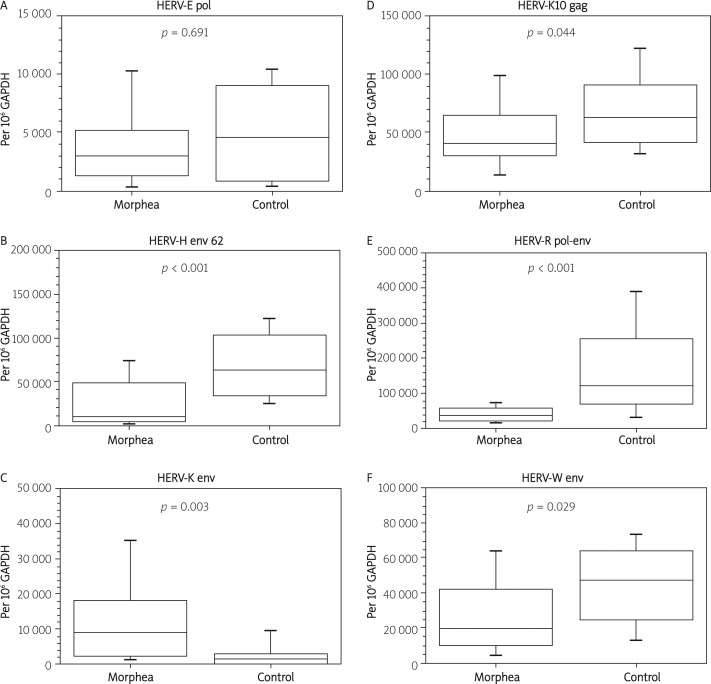
In PBMCs we found a statistically significant decrease of transcription of HERV-E pol, while HERV-K env, HERV-R pol-env, and HERV-W env were found to be up-regulated. In skin biopsies HERV-K env was strongly up-regulated. On the other hand, we noted a decrease of transcription of HERV-H env 62, HERV-K10 gag, HERV-R pol-env, and HERV-W env. In PBMCs we found a statistically significant decrease of transcription of HERV-E pol (–81.8%, p < 0.001), while HERV-K env (+94.1%, p = 0.010), HERV-R pol-env (+140.0%, p < 0.001), and HERV-W env (+97.7%, p < 0.001) were found to be up-regulated. In skin biopsies HERV-K env was strongly up-regulated (+713.0%, p = 0.003). On the other hand, we noted a decrease of transcription of HERV-H env 62 (–83.5%, p < 0.001, HERV-K10 gag (-33.7%, p = 0.044), HERV-R pol-env (–71.3%, p < 0.001), and HERV-W env (–59.3%, p = 0.029).
Conclusions
The studied HERV sequences generally show an increase of transcription in PBMCs of morphea patients, while being down-regulated in their skin, with some exceptions for both types of tissue.
Keywords: human endogenous retroviruses, real-time polymerase chain reaction, autoimmunity, localized scleroderma
Introduction
Morphea, also known as localized scleroderma (LS), is a relatively rare disease whose main symptom is excessive skin fibrosis. This disorder is believed to be limited to skin, unlike the more thoroughly studied systemic sclerosis (SS), which is characterized by internal organ involvement. The two disorders, although sharing fibrosis, seem to be propelled by different molecular mechanisms. Morphea can be further divided into subgroups: plaque morphea, generalized morphea, linear scleroderma, bullous morphea, and deep morphea [1–3]. Its manifestations can be mimicked by other conditions such as sclerodermiform graft versus host disease, postradiation dermatitis, drug-related sclerodermoid reactions, lichen sclerosus, progerias and many others [3–5]. Although morphea seems under-investigated for the time being, most likely due to good prognosis, there have been ongoing efforts to explain the mechanisms behind such fibrosis [6].
Here we focus on the involvement of human endogenous retroviruses (HERVs) in morphea. HERVs are a vast and intensely growing field in genomics. The group is composed of thousands of elements, many of which are dynamic in the genome and exert innumerable possible interactions with other genes and proteins. The mechanisms through which HERVs influence cells have been investigated for years.
HERVs are remnants of ancient retroviral infections, preserved in the genome and passed through successive generations. Due to mutations, they lack the ability to form fully infectious viral particles, yet have retained their original gene arrangement. With some exceptions such as the HERV-L group, HERVs contain three major genes: Gag, which determines the core structure; Pol, which is responsible for viral replication; and Env, coding for the envelope proteins. HERVs not only can influence the cell directly with the products of those usually truncated and interrupted genes, but also exert certain changes by their own localization near other genes, within promoter regions or even inside introns, which affects splicing of genes. Many other possible mechanisms of interaction have been reported in the literature [7–9]. The HERVs are of special interest as far as autoimmune disorders are concerned, yet little effort has been made until now to assess the possible changes of their expression in morphea.
Here we present the results of a study performed on peripheral blood mononuclear cells (PBMCs) and skin biopsies collected from patients with morphea in 2009-2011. Six HERV sequences of particular interest were chosen for this study. This selection covers HERVs of different taxonomic groups that have been previously studied in other autoimmune disorders.
The goal of this research was to perform a preliminary study of HERV expression in morphea, in order to assess whether such viruses may play any role in the pathomechanisms of the disease, as they do in certain oncogeneses and other autoimmune disorders. This is the first study of this kind showing that indeed certain human endogenous retroviruses do show aberrant expression patterns.
Material and methods
Studied groups
In the study of PBMCs 41 patients with morphea were enrolled (22 women, 19 men, mean age: 44 years). The control group consisted of 47 healthy volunteers (35 women, 12 men, mean age: 33 years).
In the study of skin biopsies 37 patients with morphea were enrolled (18 women, 19 men, mean age: 45 years). The control group consisted of 13 healthy skin samples collected mostly during other surgical procedures (6 women, 7 men, mean age: 52 years).
The study was approved by the Bioethics Committee of the Poznan University of Medical Sciences. All patients and volunteers gave written consent.
Sample preparation
Peripheral blood mononuclear cells
EDTA-collected blood was separated in a Ficoll gradient (1.077 g/cm3, 800 × g, 45 min, 23°C). The PBMCs were collected, washed in phosphate buffered saline (PBS), spun down and disintegrated in TriPure reagent (Roche Applied Science). RNA was isolated with the use of a modified Chomczynski and Sacchi protocol [10]. 1 µg of each RNA sample was used in a reaction with DNase I to minimize genomic DNA contamination (Ambion, USA). Reverse transcription (RT) was performed with the Transcriptor First Strand cDNA Synthesis Kit from Roche Applied Science with random hexamer primers. cDNA was stored in aliquots at –80°C prior to real-time polymerase chain reaction (PCR). Each RNA sample was assessed through a PCR no-RT negative reaction.
Skin biopsies
Skin biopsies of morphea patients were collected from pathological areas under local anesthetic. Upon collection samples were transferred to an empty Eppendorf tube, which was immediately put into liquid nitrogen. Prior to further processing, skin samples were stored at –80°C. If necessary, frozen samples were cut prior to rotor-stator homogenization that was carried out on ice in TriPure reagent. After removal of debris by centrifugation, RNA was isolated with the use of a modified Chomczynski and Sacchi protocol [10]. Since only in a few cases the total amount of RNA exceeded 1 µg, all of it was used for the following steps of enzymatic genomic DNA removal and cDNA synthesis. The remaining steps were the same as in the case of PBMCs.
Real-time polymerase chain reaction
cDNA samples were amplified by real-time” PCR in a LightCycler 2.0 thermocycler using LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics GmbH, Germany) with additionally added uracil-n-glycosylase (Fermentas, Canada) for carry-over prevention. Standard curves were prepared using amplicon samples of known concentrations. The results of HERV expression were normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcription.
Primers
It is relevant to note that although reference accession data are provided, our primers would usually bind to more than one locus in the human genome, due to the fact that either exact or very similar copies of individual HERVs are spread across it. Sequences of primers for HERV-K10 gag, HERV-R pol-env, HERV-W env, and GAPDH were imported from our previous study [11]. The sequences and accession data are provided in Table I.
Table I.
Sequences of primers used for real-time PCR
| Primer name | Sequence (5’-3’) | Accession/location |
|---|---|---|
| HERV-E pol F | GTC ATT TGT ATT CTA CCG GAG | Retrosearch [12]: |
| HERV-E pol R | AAT ACT GCA AAG TTT GGG AC | Orf ID: 8435 |
| HERV ID: 1109 [13] | ||
| Chr. Xq21.32 | ||
|
| ||
| HERV-H (env62) F | TAT GTC ATC CTC TAC CTC TCC C | Retrosearch: |
| HERV-H (env62) R | CCA GCA GTT GTT CAC TAA GGA | Orf ID: 82113 |
| HERV ID: 10816 | ||
| Chr. 2q24.3 | ||
|
| ||
| HERV-K env F | CAC TTG GGT TAA GAC CAT TGG A | Retrosearch: |
| HERV-K env R | GGA GCT GTT GAG TAC ACC TG | Orf ID: 204173 |
| HERV ID: 29013 | ||
| Chr. 8p23.1 | ||
|
| ||
| HERV-K10 gag F | GTA ATG GCT CAG TCA ACG CA | GenBank: |
| HERV-K10 gag R | GCC CCA TTA ATT CTG GAC CT | M14123 [14] |
|
| ||
| HERV-R pol-env F | GGG CCA ATT ATG CTT ACC AA | GenBank: |
| HERV-R pol-env R | ATG GGC TGA TCT GGC TCT AA | M12140 |
| Chr. 7q11.2 | ||
|
| ||
| HERV-W env F | TCA TAT CTA AGC CCC GCA AC | GenBank: |
| HERV-W env R | GAG GTT GTG ATA CCG CCA AT | AF072506 |
|
| ||
| GAPDH F | CTG CAC CAC CAA CTG CTT AG | Ensembl: ENST00000229239 |
| GAPDH R | TTC TGG GTG GCA GTG ATG | |
Statistical analysis
Normality was assessed with Shapiro-Wilk test using Past v.2.03 software, while significant outliers were detected with Grubbs’ test at α = 0.05 (data not published). Due to the lack of normality and the presence of significant outliers in the samples, median values were taken into consideration, while the two independent samples Wilcoxon-Mann-Whitney U test was performed for the assessment of the statistical significance of the found differences (Past v.2.03). The graphs were generated using QtiPlot v.0.9.7 software.
Median values are expressed as the number of transcript copies per million copies of GAPDH transcripts. In all graphs the middle-box line indicates the median, boxes cover the 25-75 percentile range, while whiskers cover 10-90 percentiles.
Results
Real-time PCR was optimized for each amplicon to reach appropriate electrophoresis bands and single melting curve analysis peaks (data not shown). Melting curve analysis was performed for each of over eight hundred PCR reactions and the obtained melting temperatures were compared to standards obtained during optimization.
Approximately 16% of all reaction batches were duplicated for pipetting error analysis. The duplicated batches were chosen randomly. In such cases, mean values of the two reactions were taken for further analysis (Tables II, III, Figures 1, 2).
Table II.
Expression of selected HERV sequences in PBMCs
| HERV sequence | Morphea median1 | H. control median1 | Median change | U test p - value |
|---|---|---|---|---|
| HERV-E pol | 586 | 3219 | –81.8% | < 0.001 |
| HERV-H env 62 | 47315 | 42727 | 10.7% | 0.228 |
| HERV-K env | 30000 | 15459 | 94.1% | 0.010 |
| HERV-K10 gag | 86058 | 38220 | 125.2% | 0.253 |
| HERV-R pol-env | 22529 | 9388 | 140.0% | < 0.001 |
| HERV-W env | 28103 | 14214 | 97.7% | < 0.001 |
Table III.
Expression of selected HERV sequences in skin biopsies
| HERV sequence | Morphea median1 | H. control median1 | Median change | U test p-value |
|---|---|---|---|---|
| HERV-E pol | 2996 | 4557 | –34,2% | 0.691 |
| HERV-H env 62 | 10406 | 63125 | –83,5% | < 0.001 |
| HERV-K env | 9064 | 1115 | 713,0% | 0.003 |
| HERV-K10 gag | 41336 | 62385 | –33,7% | 0.044 |
| HERV-R pol-env | 34455 | 119879 | –71,3% | < 0.001 |
| HERV-W env | 19271 | 47292 | –59,3% | 0.029 |
Median – values are expressed as the number of transcript copies per million copies of GAPDH gene transcripts of the median patient
Figure 1.
Expression of selected HERV sequences in PBMCs. Expression levels are normalized to GAPDH reference gene
Figure 2.
Expression of selected HERV sequences in skin biopsies. Expression levels are normalized to GAPDH reference gene
In PBMCs we found a statistically significant decrease of transcription of HERV-E pol, while HERV-K env, HERV-R pol-env, and HERV-W env were found to be up-regulated. In skin biopsies HERV-K env was strongly up-regulated. On the other hand, we noted a decrease of transcription of HERV-H env 62, HERV-K10 gag, HERV-R pol-env, and HERV-W env. In general, the level of transcription of the studied sequences tends to increase in PBMCs, while it decreases in skin.
Discussion
HERV-E pol
In PBMCs there is a statistically significant, roughly 82% down-regulation of HERV-E pol in morphea, compared to healthy control (Table II). This is quite interesting, as all other sequences in this study appear to be somehow up-regulated in PBMCs, along with the fact that most other studies claim HERV expression to be generally increased in autoimmune disorders. In skin the median value for morphea is lower by over 34%. However, there seems not to be any statistical significance at this point, especially taking into consideration the relatively small control group (Table III). We have reported this sequence to be down-regulated by roughly half in psoriasis vulgaris in PBMCs [15]; thus a wider perspective for the down-regulation of this particular HERV-E pol begins to emerge.
HERV-H env 62
A 62 kDa protein encoded by this particular sequence was shown to be immunosuppressive in vivo [16]. This human protein seems to have the capacity to allow tumor cells to escape immune rejection in transgenic mice, although the mechanism of this phenomenon is yet to be revealed. It is apparently not involved in morphea in PBMCs, as no statistically valid difference was found (Table II). In psoriasis vulgaris and pemphigus vulgaris, however, we have reported a 2-3-fold increase in transcription of HERV-H env 62 in PBMCs [17, 18]. On the other hand, a strong down-regulation by over 80% in skin may suggest its weakened immunosuppressive properties through this mechanism (Table III). This can translate to an intensified immune response within the morpheic skin. At the moment it is unknown whether the transcription of this HERV-H env is intentionally inhibited for tissue protection. On the other hand, it may be just a side effect of different processes that result in the altered immunologic response in morphea. The relation between this particular tumor inducing protein and the inhibition of its gene in morphea needs a further and thorough analysis.
HERV-K env
Both PBMCs and skin biopsies show a statistically significant increase of expression of HERV-K env in morphea; the latter however is a strong 7-fold increase (Tables II, III). The increase of HERV-K transcription have been reported in many studies, e.g. in breast cancer [19], which seems to be somewhat similar to the case of the aforementioned HERV-H env. Both envelope proteins are reported to be involved in tumorigenesis. Although one could try to draw some analogies between a turgid breast tumor and morphea, the reason for such increase cannot be explained without further studies. It has to be emphasized that HERV groups contain many representatives and sequences studied in breast cancer may not be functionally linked to this one. The studied HERV-K env seems to be particularly interesting also due to the fact that this is the only strongly up-regulated HERV transcript in skin in our study, opposite to all others showing relatively weaker but decreasing trends. Notwithstanding some similarities between HERV-H and HERV-K envelope proteins, it remains a mystery why the expression of the former is apparently inhibited in morpheic skin, while the latter is promoted.
HERV-K10 gag
Elevated HERV-K10 expression has been detected in placenta, embryonic tissue, and cell lines. Much focus has been placed on unraveling the apparent influence of this sequence in testicular and breast cancers [20, 21].
In both PBMCs and biopsy samples the expression of the HERV-K10 gag sequence shows a particularly large spread around the median. The median normalized expression level in our healthy control group of PBMCs is strikingly similar to a study of exactly the same sequence using the same primers by Namysł et al. (38220 vs. 33692 copies per million GAPDH) [11], although that group consisted of only ten volunteers. Our results indicate an increase of that transcript in morphea by over 125%, which is however not statistically significant, most likely due to a significant difference between mean and median values within our healthy control group. Regardless of that, this increase is substantially lower than the ones found by Namysł et al. in systemic sclerosis and psoriasis [11].
The expression of this sequence in skin samples is statistically lower in morphea just by approximately 34%. However, the significance level of that decrease is just below the p = 0.05 threshold and may turn out to be insignificant for a larger healthy control group, especially taking into consideration the fact that the values for the skin of morphea and PBMCs of healthy control are quite similar. Therefore, the apparent decrease of the HERV-K10 gag sequence in the skin of morphea must be approached with caution.
HERV-R (ERV-3) pol-env
ERV-3 is one of the most studied HERVs. There is strong evidence indicating that it plays an important role in placenta. This retroviral copy was integrated in the genome approximately 30-40 million years ago; thus it has apparent positive functions, otherwise it would have been mutated or deleted [22].
We found that healthy skin produces a lot more of this transcript than PBMCs (over a 12-fold increase), being the highest level of transcription of all HERV sequences in this study. There is also a strong and significant increase of expression of this HERV-R sequence in PBMCs along with an intense significant decrease in the skin in morphea. Although the level of transcription is decreased in the skin of morphea patients, it is still higher than in the PBMCs of morphea, where an increase was noted.
In this particular case the decrease of transcription in the skin may be directly linked to the substantially lowered number of sebaceous glands. It was reported that the level of transcription of this ERV-3 is quite high in these glands in healthy people [23]. Since morphea generally involves a decrease in the number of sebaceous glands, it might be responsible for the lowered expression of this sequence in the entire skin sample. The report was not quantitative, so that hypothesis cannot be confirmed at the moment.
HERV-W env
This sequence was also investigated by Namysł et al. [11]. In PBMCs, however, our normalized median values were generally an order of magnitude higher than in that work. On the other hand, our results indicate a roughly 2-fold and statistically significant increase of the expression of that sequence in morphea, in comparison to over a 4-fold increase in SS and an even bigger up-regulation in psoriasis found by Namysł et al.
Quite surprising again is the apparent down-regulation of that sequence in the skin of morphea, although both patients with morphea and healthy volunteers show greater expression of that sequence in skin than in PBMCs.
In conclusion, we have shown that the expression of certain HERVs is altered in morphea. The question concerning the contribution of this phenomenon to its pathogenesis is yet to be answered.
The studied HERV sequences generally show an increase of transcription in PBMCs of morphea patients, while being down-regulated in their skin, with some exceptions for both types of tissue. Direct skin assessment seems to have a greater scientific value over PBMCs, as being the target pathological area of morphea.
The vast majority of HERV orientated research has been focused on the detection of human endogenous retroviruses, by different means such as Western blotting, ELISA, hybridization of tissues with labeled probes, and PCR. Only a margin of these works try to elucidate the underlying mechanisms behind HERVs. Most of the studies concerning HERVs are focused on their influence on oncogenesis, although recently a slightly wider scope of approach can be observed, which includes autoimmune diseases as well.
We are aware that this analysis only pinpoints certain paths that might be followed in the future, e.g. the up-regulation of HERV-K env in morpheic skin, while it does not directly answer the most important question, which is the influence of HERVs in the pathogenesis of morphea.
Sclerotic skin seem to alter its gene expression quite substantially; thus one needs to be aware that GAPDH may turn out not to be necessarily the best choice for normalization of expression in this case. Studies concerning the use of GAPDH as a reference gene in real-time PCR assessment of the skin can easily be misinterpreted, as some authors suggest that it is appropriate, while others state the contrary [24–26], yet none of these studies seem to fully fit our model. The best choice is to assess the stability of the reference gene for each studied model separately and choose the best one, or to use more than one of such genes. Further study, with a different normalization protocol, seems a good choice. Notwithstanding normalization issues, the results do show interesting patterns, yet the mechanisms underlying them remain to be determined.
References
- 1.Szymanek M, Chodorowska G, Kowal M, Pietrzak A, Miturska R, Krasowska D. Serum soluble Fas levels in patients with systemic sclerosis. Postep Derm Alergol. 2010;27:406–14. [Google Scholar]
- 2.Peterson LS, Nelson AM, Su WP. Classification of morphea (localized scleroderma) Mayo Clin Proc. 1995;70:1068–76. doi: 10.4065/70.11.1068. [DOI] [PubMed] [Google Scholar]
- 3.Kreuter A, Krieg T, Worm M, et al. AWMF Guideline no. 013/066. Diagnosis and therapy of circumscribed scleroderma [German] J Dtsch Dermatol Ges. 2009;6(7 Suppl):S1–14. doi: 10.1111/j.1610-0387.2009.07178.x. [DOI] [PubMed] [Google Scholar]
- 4.Reich A, Heisig M, Baran E. Werner syndrome – a premature aging syndrome that can mimic scleroderma [Polish] Postep Derm Alergol. 2010;27:490–4. [Google Scholar]
- 5.Nedoszytko B. Genetically determined syndromes associated with premature aging [Polish] Postep Derm Alergol. 2010;27:282–90. [Google Scholar]
- 6.Zulian F. New developments in localized scleroderma. Curr Opin Rheumatol. 2008;20:601–7. doi: 10.1097/BOR.0b013e328309a5eb. [DOI] [PubMed] [Google Scholar]
- 7.Urnovitz HB, Murphy WH. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwolińska K. Retroviruses-derived sequences in the human genome. Human endogenous retroviruses (HERVs) [Polish] Postepy Hig Med Dosw. 2006;60:637–52. [PubMed] [Google Scholar]
- 9.Larek-Rąpała A, Żaba R, Kowalczyk MJ, Szramka-Pawlak B, Schwartz RA. Herpes simplex virus infection as a possible modulator of autoimmune diseases facilitated by human endogenous retroviruses. Postep Derm Alergol. 2011;28:313–6. [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Namysł J, Osmola A, Prokop J. Human endogenous retroviruses sequences expression in patients with progressive systemic sclerosis and different clinical forms of psoriasis. Postep Derm Alergol. 2005;22:99–104. [Google Scholar]
- 12.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu RZ, Zheng S. Identification of a novel human retroviral RNA genome associated with human chronic myeloid leukemia; Unpublished. GenBank: AY208746.1. [Google Scholar]
- 14.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–98. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowalczyk MJ, Szramka-Pawlak B, Zaba R, Silny W. Downregulation of HERV-E pol gene in psoriasis vulgaris; Proceedings of the 19th Congress of the European Academy of Dermatology an Venereology; Gothenburg, Sweden: 2010. Oct 6-10, [Google Scholar]
- 16.Mangeney M, de Parseval N, Thomas G, Heidmann T. The full-length envelope of an HERV-H human endogenous retrovirus has immunosuppressive properties. J Gen Virol. 2001;82:2515–8. doi: 10.1099/0022-1317-82-10-2515. [DOI] [PubMed] [Google Scholar]
- 17.Kowalczyk MJ, Szramka-Pawlak B, Zaba R, Silny W. Proceedings of the 40th Annual Meeting of the European Society for Dermatological Research (ESDR) Helsinki, Finland: 2010. Sep 8-11, Upregulation of HERV-H env gene in psoriasis vulgaris. http://www.esdr2010.org/pdf/2010ESDRabastractbookweb.pdf; 2011. [Google Scholar]
- 18.Kowalczyk M, Gornowicz J, Szramka-Pawlak B, Dmochow ski M, Zaba R. Transcription of three HERV sequences in pemphigus vulgaris; Proceedings of the 8th Congress of the Baltic Association of Dermatovenerologists; Vilnius, Lithuania: 2009. Sep 17-19, Abstract Book: 75. [Google Scholar]
- 19.Wang-Johanning F, Frost AR, Johanning GL, et al. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–60. [PubMed] [Google Scholar]
- 20.Sauter M, Schommer S, Kremmer E, et al. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–21. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goedert JJ, Sauter ME, Jacobson LP, et al. High prevalence of antibodies against HERV-K10 in patients with testicular cancer but not with AIDS. Cancer Epidemiol Biomarkers Prev. 1999;8:293–6. [PubMed] [Google Scholar]
- 22.Kato N, Pfeifer-Ohlsson S, Kato M, et al. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J Virol. 1987;61:2182–91. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersson AC, Merza M, Venables P, et al. Elevated levels of the endogenous retrovirus ERV3 in human sebaceous glands. J Invest Dermatol. 1996;106:125–8. doi: 10.1111/1523-1747.ep12329612. [DOI] [PubMed] [Google Scholar]
- 24.Zainuddin A, Makpol S, Chua KH, Abdul Rahim N, Yusof YA, Ngah WZ. GAPDH as housekeeping gene for human skin fibroblast senescent model. Med J Malaysia. 2008;63(Suppl A):73–4. [PubMed] [Google Scholar]
- 25.Turabelidze A, Guo S, DiPietro LA. Importance of housekeeping gene selection for accurate reverse transcription-quantitative polymerase chain reaction in a wound healing model. Wound Repair Regen. 2010;18:460–6. doi: 10.1111/j.1524-475X.2010.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira JG, Prados RZ, Guedes AC, Ferreira PC, Kroon EG. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase is inappropriate as internal control in comparative studies between skin tissue and cultured skin fibroblasts using Northern blot analysis. Arch Dermatol Res. 1999;291:659–61. doi: 10.1007/s004030050471. [DOI] [PubMed] [Google Scholar]