Abstract
Cotton (Gossypium hirsutum L.) fibers are single-celled trichomes that synchronously undergo a phase of rapid cell expansion, then a phase including secondary cell wall deposition, and finally maturation. To determine if there is coordinated regulation of gene expression during fiber expansion, we analyzed the expression of components involved in turgor regulation and a cytoskeletal protein by measuring levels of mRNA and protein accumulation and enzyme activity. Fragments of the genes for the plasma membrane proton-translocating ATPase, vacuole-ATPase, proton-translocating pyrophosphatase (PPase), phosphoenolpyruvate carboxylase, major intrinsic protein, and α-tubulin were amplified by polymerase chain reaction and used as probes in ribonuclease protection assays of RNA from a fiber developmental series, revealing two discrete patterns of mRNA accumulation. Transcripts of all but the PPase accumulated to highest levels during the period of peak expansion (+12–15 d postanthesis [dpa]), then declined with the onset of secondary cell wall synthesis. The PPase was constitutively expressed through fiber development. Activity of the two proton-translocating-ATPases peaked at +15 dpa, whereas PPase activity peaked at +20 dpa, suggesting that all are involved in the process of cell expansion but with varying roles. Patterns of protein accumulation and enzyme activity for some of the proteins examined suggest posttranslational regulation through fiber development.
Plant cell expansion occurs through the interaction of multiple influences, including cell wall-yield properties; the opposing force of turgor pressure; the biosynthesis of new membrane lipids, cell wall components, and proteins; and proper trafficking of these newly synthesized materials to their final cellular destination. During cell expansion the force of turgor pressure is related to the osmotic potential and to the transport coefficient for water uptake (Cosgrove, 1986). A vital component of rapid and sustained cell expansion is the maintenance of sufficient osmoticum to compensate for dilution effects resulting from the influx of water. In plant cells, osmoticum and water accumulate primarily in the vacuoles, although osmoticum in the cytoplasm also contributes to cellular osmotic potential. The driving force for the transport and accumulation of ions in the vacuole is provided by two types of electrogenic, proton-translocating pumps, one that hydrolyzes ATP (V-ATPase) and another that hydrolyzes PPi (PPase). Membrane potential across the PM can be generated by a H+-ATPase, which is structurally quite different from the V-ATPase. Movement of anions, which serve as osmoticum, across the tonoplast and the PM occurs through various carriers and channels (Barkla and Pantoja, 1996). Experimental and theoretical approaches to gain a better understanding of the factors regulating cell expansion and tissue growth are often confounded by the varying properties of different cell types in a tissue (Silk, 1984; Cosgrove, 1986). Thus, dissection of the mechanisms controlling cell expansion in a single cell type may provide a foundation for understanding the regulation of tissue growth.
The fiber cells of cotton (Gossypium hirsutum L.) represent a single cell type that undergoes a period of very rapid elongation. Cotton fibers also represent an agricultural commodity of major economic importance in the United States. Fiber cells are single-celled trichomes, which arise in near synchrony from the epidermis of the ovule and may elongate at peak rates in excess of 2 mm d−1 during the rapid polar-expansion phase of development (Schubert et al., 1973; Basra and Malik, 1984). Ultrastructural evidence indicates that expansion occurs through a diffuse growing mechanism, albeit with some bias for deposition of newly synthesized cell wall materials at the tip (Tiwari and Wilkins, 1995). To gain insight into the processes responsible for rapid fiber growth, we initially focused on characterizing the V-ATPase in cotton fiber cells at the molecular level (Wilkins, 1993; Wan and Wilkins, 1994a; Hasenfratz et al., 1995).
In this work we have assayed the expression of several components, outlined below, involved in cell expansion at the levels of RNA accumulation, protein accumulation, and enzymatic activity throughout fiber development. The V-ATPase and PPase, as mentioned above, are responsible for driving solute movement into the vacuole, which is important for maintaining the osmotic potential necessary to generate turgor pressure. The PM H+-ATPase translocates protons out of the cytosol, acidifying the apoplast, which is theorized to effect a change in cell wall extensibility (Rayle and Cleland, 1992). Proteins in the MIP superfamily have been shown to act as aquaporins (Maurel et al., 1993), which may reduce the resistance to water transport across the tonoplast and/or the PM. Malate, synthesized in the cytoplasm through the activity of the highly regulated enzyme PEPCase, accumulates as osmoticum in the vacuole (Barkla and Pantoja, 1996). Finally, α-tubulin is a protein subunit of microtubules, which appear to function by coordinating the organellar organization in the cytoplasm (Goddard et al., 1994) and are related to the orientation of cellulose microfibrils in the cell wall (Cyr and Palevitz, 1995). By comparing the patterns and peak levels of expression of these various components, we hope to gain insight into the role of each through fiber expansion. Our results, especially when considered together with data describing expression of other genes in fibers, suggest that there are at least three discrete patterns of gene expression that can be related to the period of organellar biogenesis, a period of rapid expansion, and primary and secondary cell wall deposition. In addition to coordination of transcript accumulation for vacuolar, PM, and cytoplasmic components during fiber expansion, we present evidence supporting the posttranslational regulation of enzyme activity throughout fiber development.
MATERIALS AND METHODS
Growth Conditions
Cotton (Gossypium hirsutum L. cv Acala SJ-2) was grown in greenhouse conditions as previously described (Hasenfratz et al., 1995). Staged ovules were collected based on the phyllotactic arrangement of flowering nodes and the proximity to open flowers on the day of anthesis. Developing ovules were excised from bolls collected 3 d before anthesis (−3 dpa), −1 dpa, on the day of anthesis (0 dpa), and at +1, +3, +5, +10, +15, +20, +25, +30, and +35 dpa for the isolation of protein or RNA.
Isolation of Cotton DNA and RNA
Genomic DNA was isolated from young, expanding leaves of cotton as described (Wilkins et al., 1994). For RNA, developing fibers were collected from bolls by freezing the excised ovules in liquid N2, and then brushing off the protruding fiber cells. The isolated fiber cells from +5, +10, +15, +20, +25, and +30 dpa were used for RNA extraction (Wan and Wilkins, 1994b; Wilkins and Smart, 1996). Purified total RNA was divided into aliquots, precipitated with ethanol, and stored at −80°C under 70% (v/v) ethanol. For RPAs, the RNA pellets were collected by centrifugation and the pellet was dried under vacuum, and then resuspended to a final concentration of 0.2 mg mL−1 in diethyl pyrocarbonate-treated water.
PCR Amplification and Cloning
PCR was used to amplify partial cDNAs encoding the PM H+-ATPase, PEPCase, MIP, and α-tubulin from approximately 1 × 109 plaque-forming units of a cotton +10-dpa fiber cDNA library constructed in the phagemid vector λZAPII (Stratagene). First-strand cDNA from reverse-transcribed RNA isolated from +10-dpa cotton ovule and fiber tissue was used as the template for PCR amplification of the PPase. Oligonucleotide PCR primer sequences, expected product size, and source targets are listed in Table I. PCR mixture was 1× PCR buffer (10 mm Tris-HCl [pH 8.3] and 50 mm KCl), 1.5 mm MgCl2, 0.8 mm dNTPs, 1.0 μm of each oligonucleotide primer, and 2.5 units of Taq DNA polymerase. Reactions (50 μL) were performed in either a Perkin-Elmer model 480 or an Ericomp thermal cycler (San Diego, CA) using the annealing temperatures listed in Table I. A portion of the cDNA CVA69.24 (Wilkins, 1993), which encodes the 69-kD subunit of the V-ATPase from cotton, was PCR amplified from plasmid DNA containing the cloned cDNA using the above conditions. A portion of the 18S rRNA gene was amplified from cotton genomic DNA using the above conditions. Amplification products from all of the reactions described above were subcloned into the plasmid vector pCRII (Invitrogen, San Diego, CA). Cloned products encoding PEPCase and 18S rRNA were further subcloned into the plasmid vector pBluescript II (Stratagene) to facilitate synthesis of in vitro-transcribed probes for RPAs. The DNA sequences of the cloned fragments to be used as probes were determined from both strands using the Sequenase version II dideoxy sequencing kit according to the manufacturer (Amersham) or using an automated sequencing system (Applied Biosystems). Sequence analysis was performed using either MacDNASIS (Hitachi Software, Palo Alto, CA) or PCGene (IntelliGenetics, Campbell, CA).
Table I.
Oligonucleotide PCR primers used to amplify gene fragments for probes
| Target Gene | Relative Position | Oligonucleotide Primer Sequences (5′–3′)a | Primer Name | Source Template | Product Size | Annealing Temperature |
|---|---|---|---|---|---|---|
| bp | °C | |||||
| PM H+-ATPase | 5′ | TTYCCIGARCAYAARTAYGARAT | LBS106 | Fiber cDNA library | 254 | 50 |
| 3′ | GCRTADATIGTRTARTTYTTCAT | LBS107 | ||||
| V-ATPase (69-kD subunit)b | 5′ | GAAACTGCTAAACTTTTAAG | COT19 | cDNA clone CVA69.24 | 336 | 52 |
| 3′ | CACCTAGTTTCATCCTCC | COT6 | ||||
| PPase | 5′ | CNGAYGCNTGYGAYGCNGCNGG | COT72 | Ovule first-strand cDNA | 615 | 55 |
| 3′ | CYTTNARNGGRTCNCCDATNGTRTC | COT74 | ||||
| PEPCase | 5′ | CCATGGATCTTTGCCTGGAC | COT189 | Fiber cDNA library | 573 | 52 |
| 3′ | GCATTCCAGCAGCAATACC | COT185 | ||||
| α-Tubulin | 5′ | CATGGCTTGYTGTTTGATGTAYCG | COT205 | Fiber cDNA library | 335 | 55 |
| 3′ | CCTCACGAGCCTCAGAGAAYTCTCC | COT206 | ||||
| MIP | 5′ | AAYCCIGCIGTIACITTYGG | LBS103 | Fiber cDNA library | 353 | 50 |
| 3′ | CKIGCIGGRTTCATISA | LBS105 | ||||
| 18S rRNAc | 5′ | CGGAATTCTTGGAGGGCAAGTCTGGT | LBS108 | Cotton DNA | 91 | 55 |
| 3′ | GCCTCGAGTCCAACTACGAGCTTTTTAACT | LBS109 |
RPAs
RPAs were performed using the HybSpeed RPA kit essentially as described by the manufacturer (Ambion, Austin, TX). RNA probes were synthesized by in vitro transcription from linearized plasmid DNA in the presence of 50 μCi of α-[32P]UTP (3000 Ci mmol−1) (Dupont/NEN) using either T7 or SP6 RNA polymerase and were labeled to a specific activity of approximately 1 × 109 cpm μg−1. The 18S rRNA probe was synthesized using 1 μCi of α-[32P]UTP together with the T7 Megashortscript kit as described by the manufacturer (Ambion) and was labeled to a specific activity of approximately 5 × 104 cpm μg−1. Full-length transcripts for probes were purified after electrophoresis in a 5% (w/v) denaturing polyacrylamide vertical slab gel. Trial reactions using 0.2, 1.0, and 2.5 μg of RNA were performed, and a linear increase was observed when comparing the signal from the 1.0-μg sample to that of the 2.5-μg sample, indicating that the reactions were not saturated, even for the 18S rRNA probe (data not shown). One microgram of total cotton fiber RNA was hybridized with 2 × 104 cpm of radiolabeled probe for each reaction, except for the 18S rRNA probe, in which 3 × 103 cpm were added. RPA reaction products were resolved by electrophoresis in a 5% denaturing polyacrylamide gel poured using an analytical comb and electrophoresed using an S2 gel electrophoresis apparatus for optimum resolution (Gibco-BRL). These gels were treated with a solution of 10% (v/v) ethanol, 10% (v/v) acetic acid, transferred to chromatography paper, and then vacuum dried. Protected products were detected by autoradiography at −80°C using reflection film and intensifying screens (DuPont/ NEN), and a radioactive signal was quantified by phosphor imagery (Fujix BAS1000, Fuji Film Co., Tokyo, Japan). If the RPA yielded a doublet of bands, then the combined intensity of both bands was quantified. The sizes of protected products were compared with the migration of radiolabeled products generated from the Century RNA Size Marker kit (Ambion). Assays were repeated at least three times.
Preparation of Microsomal Membranes and Tonoplast
Microsomal membranes were prepared essentially as described (Bennett et al., 1984) with some modification to enhance the microsomal preparation, particularly at +10 and +15 dpa. Cotton ovules were isolated and blended in homogenization medium (0.35 mm Suc, 70 mm Tris-HCl [pH 8.0], 10% [v/v] glycerol, 3 mm Na2EDTA, 0.15% [w/v] BSA, 1.5% [w/v] PVP-40, 4 mm DTT, and 1.5 mm PMSF) with a chilled Waring blender for a few seconds. Ovules of +15 dpa and older stages were cut with scissors into small pieces before homogenization. The tissue homogenate was set aside at 0°C for 10 to 15 min to allow foam to settle and was then filtered through three layers of Miracloth (Calbiochem). The filtrate was centrifuged for 15 min at 15,000g at 4°C, and the resulting pellet was discarded. A 2-mL portion of the supernatant was saved as the enzyme extract for PEPCase activity determination. The remainder of the filtrate was centrifuged at 100,000g at 4°C for 40 min, and the microsomal pellet was resuspended in RM (0.35 m Suc, 10 mm Tris-Mes [pH 7.0], 2 mm DTT, and 1.5 mm PMSF). The microsomal suspensions were frozen in liquid nitrogen and saved at −80°C until the next day for enzyme assays or for further membrane purification. Resuspended microsomal membranes were centrifuged in an Eppendorf centrifuge at 16,000g at 4°C for 5 min, and the resulting supernatant was used as the soluble protein fraction for enzyme assays.
To purify vacuolar membrane vesicles (tonoplast), the microsomal suspension was layered onto a 16/27% (w/w) discontinuous Suc gradient made in 10 mm Tris-Mes (pH 7.0), 2 mm DTT, and 1.5 mm PMSF; and centrifuged at 100,000g at 4°C for 35 min in an SW28 rotor (Beckman). The tonoplast fraction, which is at the 16/27% Suc interface (Bennett and Spanswick, 1984), was collected with a Pasteur pipet and diluted in an equal volume of 10 mm Tris-Mes (pH 7.0), 2 mm DTT, and 1.5 mm PMSF. The tonoplast was subsequently centrifuged again at 100,000g at 4°C for 30 min; and the pellet was resuspended in RM, the protein content was assayed, and then the solution was diluted to a concentration of 1 to 5 mg protein mL−1. The tonoplast suspension was further diluted to approximately 200 μg protein mL−1 for enzyme assays.
Purification of Cotton Ovule PM
The PM of cotton ovules were purified by two-phase partitioning (Kjellbom and Larsson, 1984; Larsson, 1985). The microsomal pellet, as prepared above, was resuspended in 0.33 m Suc, 5 mm KCl, and 5 mm potassium phosphate (pH 7.8). One gram of the suspension containing about 6 mg of protein was added to the 7-g phase mixture to give an 8-g phase system with a final composition of 6.5% (w/w) PEG (Mr = 3350), 6.5% (w/w) dextran T-500, 0.33 m Suc, 5 mm KCl, and 5 mm potassium phosphate (pH 7.8). The phase system was thoroughly mixed by 20 to 30 inversions of the tube, then phase separation was facilitated by centrifugation in a swinging bucket rotor at 2000g, at 4°C for 5 min. The PM partitioned into the PEG-rich upper phase, whereas the intracellular membranes partitioned at the interface and in the dextran-rich lower phase (Larsson and Anderson, 1979). The upper phase was removed and repartitioned two additional times with fresh lower phase. The final upper phases were diluted at least 2-fold with RM, and the PM was collected by ultracentrifugation at 100,000g at 4°C for 20 min. The pellet was washed twice with RM, and the pure PM pellet was resuspended in RM for use in enzyme assays.
Protein Assays
Protein concentration was measured by the dye-binding method (Bradford, 1976) using BSA as a protein standard. Membrane protein in RM was first solubilized in 0.05% (v/v) Triton X-100 before the addition of the dye reagent (Bio-Rad). The final concentration of Triton X-100 in the protein assay mixture was 0.005% (v/v).
Enzyme Assays
ATPase activity was assayed as the liberation of Pi from either ATP or PPi and was detected colorimetrically (Ames, 1966; Bennett et al., 1988). ATP and PPi were purchased as disodium salts (Sigma). To deplete the ATP and PPi of sodium ions, they were first treated with HCl, and then desalted with Dowex 50W resin. Finally, the pH was adjusted to 6.5 with BTP (bis-Tris propane [1,3-bis(Tris-[hydroxymethyl]methylamino) propane]). The reaction was initiated by the addition of 8 to 12 μg of membrane protein to a 0.4-mL reaction volume, and the reaction was incubated at 30°C for 30 min. The reactions were terminated and measured spectrophotometrically at 820 nm, as described (Ames, 1966).
V-ATPase activity (Cl− stimulated, vanadate insensitive, and NO3− inhibited) was measured in the presence of 25 mm BTP-Mes (pH 8.0), 3 mm magnesium sulfate, 50 mm ammonium molybdate, 50 mm sodium vanadate, 50 mm sodium azide, 0.1% (w/v) LPC, 3 mm ATP-BTP, and 50 mm KCl or 50 mm KNO3. V-ATPase activity, expressed as micromoles per minute per milligram of protein, was calculated as the difference in Pi released assayed in the presence of Cl− or NO3− ions.
PM H+-ATPase activity (vanadate sensitive) was measured in the presence of 25 mm BTP-Mes (pH 6.5), 3 mm magnesium sulfate, 50 mm ammonium molybdate, 50 μm sodium azide, 0.1% LPC (w/v), 3 mm ATP-BTP, and 50 mm KCl, with or without 50 μm sodium vanadate. PM H+-ATPase activity was calculated as the difference in Pi released assayed in the presence or absence of vanadate (Bennett et al., 1984; Gibrat et al., 1989). Azide and molybdate were added to the reaction mixture to inhibit the mitochondrial and phosphatase activities, respectively (Gallagher and Leonard, 1982).
PPase activity was measured in the presence of 25 mm BTP-Mes (pH 8.0), 3 mm magnesium sulfate, 50 mm ammonium molybdate, 0.1% (w/v) LPC, 3 mm PPi-BTP, and 50 mm KCl. The enzyme activity was expressed as K+-stimulated (difference in the activity assayed in the presence and absence of KCl) and total PPase activity (Rea and Poole, 1985). PPase activity was calculated as one-half of the rate of Pi liberation because the hydrolysis of 1 mol of PPi yields 2 mol of Pi.
PEPCase (EC 4.1.1.31) activity was assayed spectrophotometrically at 340 nm at 24°C. The reaction was enzymatically coupled to malate dehydrogenase (EC 1.1.1.37), and the rate of NADH oxidation was monitored in the presence of 25 mm BTP-Mes (pH 8.0), 10 mm magnesium chloride, 10 mm NaHCO3, 0.2 mm NADH, 5 mm DTT, 3 mm PEP, and 10 units of malate dehydrogenase (Meyer et al., 1988; Marczewski, 1989; Schuller et al., 1990). The reaction was started by the addition of the enzyme extract, which was prepared as described above.
Denaturing Gel Electrophoresis and Immunoblotting
SDS-PAGE was performed using 10% (w/v) polyacrylamide gels with Laemmli buffers (Laemmli, 1970) and the Mini Protean II electrophoresis system (Bio-Rad). Antibodies raised against V-ATPase, PM H+-ATPase, PPase, PEPCase, MIP, or α-tubulin (Table II) were used for estimating the relative abundance of each protein during development of the cotton ovules. Protein resolved by SDS-PAGE was immediately transferred to PVDF membrane (Millipore) in a Trans-Blot Cell (Bio-Rad) (Towbin et al., 1979). The blots were blocked with 1% (w/v) nonfat dry milk and 0.4% (v/v) Tween 20 in PBS for 2 h at 23°C, and probed with the appropriate antisera (Table II) for 16 h at 23°C. Nonbinding primary antibody was removed by rinsing with PBS. Positive antibody reaction was detected with protein A-alkaline phosphatase conjugate reacted with 5-bromo-4-chloro-3 indolyl phosphate and nitroblue tetrazolium. Relative intensities of the colorimetric reaction products on the immunoblots blots were determined by densitometric scanning using an imaging densitometer (model GS-670, Bio-Rad).
Table II.
Sources of antibodies and dilutions used for immunoblots of ovule protein through development
| Protein Reactivity | Dilution Used | Type of Antiserum | Animal Source | Antigen | Reference |
|---|---|---|---|---|---|
| V-ATPase (69-kD subunit)a | 1:250 | Polyclonal | Rabbit | 69-kD Subunit from mung bean | Matsuura-Endo et al. (1992) |
| V-ATPase (57-kD subunit)a | 1:1000 | Polyclonal | Rabbit | 57-kD Subunit from mung bean | Matsuura-Endo et al. (1992) |
| PM H+-ATPaseb | 1:300 | Polyclonal | Rabbit | AHA1 (A. thaliana PM H+-ATPase gene) fusion protein | Parets-Soler et al. (1990) |
| PPasea | 1:500 | Polyclonal | Rabbit | PPi-binding domain of PPase | Maeshima and Yoshida (1989) |
| PEPCasec | 1:500 | Polyclonal | Rabbit | Maize PEPCase | Albert et al. (1992) |
| MIPa | 1:400 | Polyclonal | Rabbit | VM23 from radish tap root | Maeshima (1992) |
| α-Tubulin | 1:200 | Monoclonal | Mouse | α-Tubulin from chicken embryo brain | Sigma |
Antibodies were raised in the laboratory of Dr. M. Maeshima (Nagoya University, Nagoya, Japan).
The antibodies against PM H+-ATPase were a gift from Dr. R. Serrano (Universidad Politécnica, Valencia, Spain).
Antiserum against PEPCase was raised in the laboratory of Dr. S.S.M. Sun (University of Hawaii, Honolulu) and was provided courtesy of Dr. C. Bassett (U.S. Department of Agriculture-Agricultural Research Service, Kearneysville, WV).
RESULTS
PCR Amplification and Cloning
To obtain probes to assay transcript accumulation throughout development of fiber cells, PCR was used to amplify portions of genes encoding the PM H+-ATPase, the 69-kD subunit of the V-ATPase, PPase, PEPCase, α-tubulin, and MIP from developing cotton fibers. All of the amplified products were ligated into the plasmid vector pCRII (Invitrogen) and transformants were screened for the presence of the expected size insert (Table I). A portion of those plasmids with the expected size insert were used to obtain partial nucleotide sequence to verify the identity of those clones. Individual clones chosen to be used as probes were sequenced completely, and the sequences not previously reported, excluding the primer sequences, were deposited in GenBank and given the following accession numbers: PEPCase, AF008940; α-tubulin, AF009565; PM H+-ATPase, AF009566; MIP, AF009567; and PPase, AF009568. Determination that a clone encodes a particular protein was based on sequence comparison with published gene sequences.
Nucleotide sequences of the five previously uncharacterized clones, encoding the PM H+-ATPase, PPase, PEPCase, MIP, and α-tubulin, were aligned and compared over the corresponding regions, excluding the primer sequences, with individual nucleotide sequences downloaded from the GenBank database. The fragment of the PM H+-ATPase is 85 and 81% identical in nucleotide sequence to the corresponding regions of the genes AHA10 from Arabidopsis thaliana (accession no. S74033) and PHA2 from potato (Solanum tuberosum L., accession no. X76535), respectively. The PPase fragment cloned from cotton ovule RNA is 84% identical over the same region to the tobacco (Nicotiana tabacum) TVP9 PPase clone and 82% identical to the red beet (Beta vulgaris) PPase clone P1 (accession nos. X83730 and L32792, respectively). The cotton α-tubulin clone is highly identical (87% over the same region) to the TUA6 gene from A. thaliana (accession no. M84699) and to the TubA1 gene (85% over the same region) from pea (Pisum sativum, accession no. U12589). The clone of a MIP family gene from cotton is most similar to a root-specific aquaporin from tobacco (78% identity, accession no. X54855) and to δ-tonoplast intrinsic protein from A. thaliana (75% identity, accession no. U39485), which has been demonstrated to be localized to the tonoplast (Daniels et al., 1996). Finally, the fragment of PEPCase amplified from cotton fibers is 78% identical to genes from both soybean (Glycine max, accession no. D10717) and alfalfa (Medicago sativa, accession no. M83086).
The PCR strategy utilized to amplify portions of genes encoding the PM H+-ATPase, the PPase, α-tubulin, MIP, and PEPCase included the potential for the amplification of members of multigene families. Partial sequence data from multiple clones of the PM H+-ATPase, PEPCase, and MIP indicated that the clones analyzed apparently represent the same genes in all three cases (data not shown). However, sequences from different α-tubulin clones indicated that they encoded four unique members of a multigene family (data not shown).
Assays of Transcript Accumulation
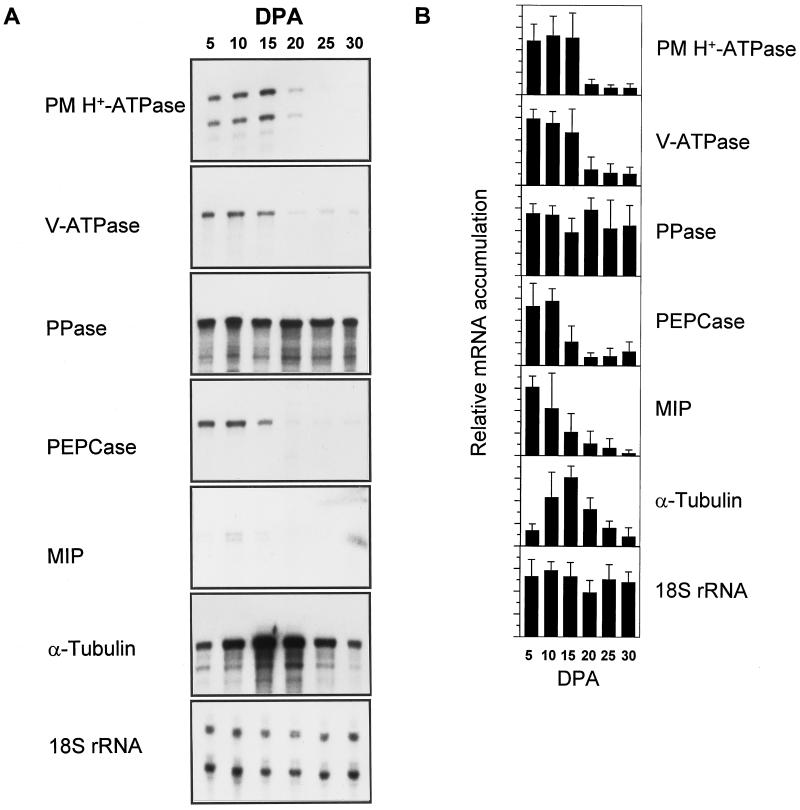
RPAs were utilized to measure the relative transcript abundance of the aforementioned genes during the expansion of developing cotton fibers (Fig. 1). Accumulation of transcripts encoding the PM H+-ATPase was highest during the phase of rapid elongation, from +5 dpa through +15 dpa. In fibers +20 dpa and older, PM H+-ATPase gene transcript abundance dropped to one-fourth of the level of that in fibers earlier in development. A similar pattern was observed for transcripts encoding the 69-kD subunit of the V-ATPase, confirming results of another study (T.A. Wilkins, C.-Y. Wan, W. Kim, F. Vojdani, and M.-P. Hasenfratz, unpublished data). Message levels for α-tubulin followed a similar pattern as for the ATPases, with peak accumulation at +10 and +15 dpa, although the rise and decline of message levels appeared to occur at a slower rate. This peak period of mRNA accumulation corresponds to the period of most rapid cell expansion (Basra and Malik, 1984; T.A. Wilkins, C.-Y. Wan, W. Kim, F. Vojdani, and M.-P. Hasenfratz, unpublished data). The accumulation of mRNA encoding PEPCase and MIP was highest in the earlier stages (+5 and +10 dpa), but then declined after +15 dpa. The detection of MIP mRNA accumulation required much longer exposures, even though all of the mRNA probes were labeled to approximately the same specific activity. Exposure in the +30 dpa lane in Figure 1A was not due to hybridization of the MIP probe, but rather was a signal from a radiolabeled size ladder in the adjacent lane (not shown). In contrast to the two ATPases, the accumulation of transcript encoding the PPase was constitutive through all stages of fiber development.
Figure 1.
A, Autoradiographs of representative results of RPAs of cotton fiber RNA isolated throughout development. Developmental stage in dpa is noted above each lane (DPA). Probes used for RPAs are described in the text. Exposure times varied for each probe. Each assay contained 1 μg of cotton fiber total RNA. B, Quantification of RPAs by phosphor imagery. Bars represent the averages of at least three assays quantified by a Fuji phosphor imager and expressed relative to the highest value. Lines extending above the bars represent the sd. Developmental stage in dpa is noted at the bottom of each lane (DPA). Intensities of the 18S rRNA RPA products are included for comparison in A, but were not used to normalize the values for the other probes.
Use of the 18S rRNA gene probe for RPAs gave consistently equal levels of hybridization signal between the samples in the developmental series, except for a slightly lower signal in samples from +20-dpa fibers. The consistency of signal from this probe indicates that the differences in accumulation observed using the other probes is not due simply to experimental error or to improper RNA quantification. RPAs using probes encoding the PM H+-ATPase, MIP, and 18S rRNA resulted in doublet bands, one of which migrates at the predicted size of the correct protected fragment. These doublets, which are not unusual for this procedure, may result from secondary structure in the mRNA-probe duplex or from base-pairing mismatches that are digested in a fraction of duplexes, either due to PCR errors in the probes used or by cross-hybridization to transcripts from very similar gene family members.
Immunoblot Analysis of Protein Accumulation
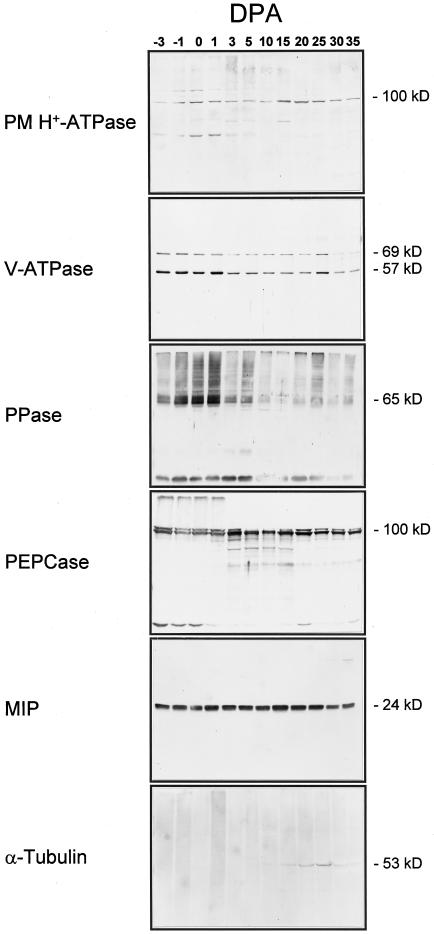
To assay protein accumulation throughout development, immunoblotting was performed using protein isolated from developing ovules and fibers harvested starting at −3 dpa through +35 dpa (Figs. 2 and 3). Initially, protein was also extracted from only fibers at the later developmental stages and used for comparison with protein from ovules and fibers. Immunoblots probed for V-ATPase protein indicated comparable patterns of accumulation throughout development, except for a slightly higher level of V-ATPase protein in the ovule plus fiber extract compared with fiber protein between +15 and +20 dpa (data not shown), which is the period when the embryo enlarges (Wilkins and Jernstedt, 1997).
Figure 2.
Immunoblots of protein purified from cotton locules throughout development. Developmental stage in dpa is noted above each lane (DPA). The V-ATPase blot was probed with a mixture of antibodies raised to the 69-kD subunit and antibodies to the 57-kD subunit. Protein for the PM H+-ATPase blot was from microsomes (14 μg of protein per lane). Protein for the V-ATPase, PPase, and MIP blots was from tonoplast-enriched membranes (8 μg of protein per lane). Total soluble protein was used for blots probed with PEPCase and α-tubulin antibodies (50 μg of protein per lane).
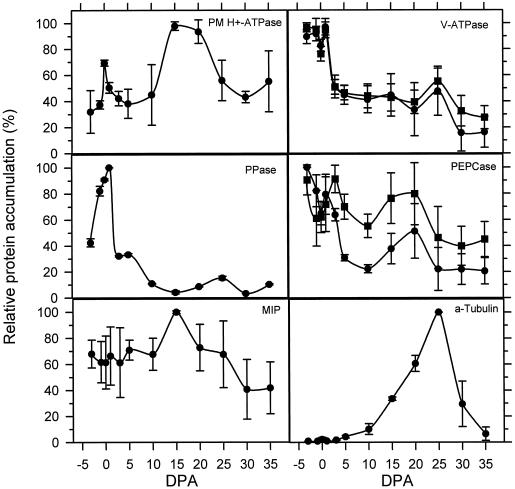
Figure 3.
Quantification of signal from immunoblots by scanning densitometry. The intensity of bands of the correct size on immunoblots was quantified using a scanning densitometer (Bio-Rad) and is expressed as a percentage of the highest value, which was set at 100% for each repetition. Values are the averages of quantifications of at least three blots for each antibody, and bars above and below each point represent one sd. Values in the V-ATPase panel represent quantification of the 69-kD subunit (•) and the 57-kD subunit (▪). In the PEPCase panel, quantifications of the 100-kD band (•) and the 95-kD band (▪) are shown. Developmental stage in dpa is noted on the x axis (DPA).
From these results we concluded that the major contribution of protein in the fiber and ovule extract was from the fiber cells. Polyclonal antibodies raised to the A. thaliana PM H+-ATPase (Parets-Soler et al., 1990) detected a band of the expected size of approximately 100 kD at greatest levels in microsomes from tissues harvested closest to the peak period of expansion, at around +15 dpa. The PM H+-ATPase protein level declined in samples taken at later points in development (+20 dpa and beyond). The 57-kD and 69-kD subunits of the V-ATPase were detected using a mixture of antibodies raised to the comparable proteins purified from mung bean (Matsuura-Endo et al., 1992) and displayed changes in accumulation in the tonoplast that paralleled each other throughout development. The levels of V-ATPase subunit accumulation were highest in early stages (−3 through +1 dpa), and then dropped to less than one-half of those levels through the period of expansion, except for a small peak at +25 dpa. Antibodies raised to the PPase from mung bean (Maeshima and Yoshida, 1989) detected many proteins, but the most prominent bands were of the expected size (approximately 60–65 kD). As with the V-ATPase, the PPase protein accumulated to highest levels in samples from −1 through +1 dpa, after which the levels declined except for a minor peak at +25 dpa. An additional band of approximately 24 kD was also detected, which comigrates with the band detected by the TIP antibody and probably represents reaction of the PPase polyclonal antibodies to cotton TIP protein. This reaction is probably the result of TIP contamination of the PPase protein preparation used to raise the antibodies, since antibodies raised to a PPase peptide did not cross-react to a 24-kD band on comparable immunoblots (data not shown).
Antibodies raised to maize PEPCase gave complex banding patterns at different stages of development. In all samples a doublet of bands was detected migrating at the expected sizes of approximately 100 and 110 kD. The relative intensities of cross-reaction to these bands varies throughout development, with peak accumulation of the lower band in samples from +3 dpa on, whereas the upper band showed greatest accumulation at +20 dpa, decreasing progressively in the samples from the last three time points. Bands of unexpected sizes detected by the PEPCase antibodies varied in accumulation throughout development and may represent nondenatured multimers or breakdown products. Antibodies raised to the VM23 MIP protein of radish (Maeshima and Yoshida, 1989) cross-reacted with a protein of approximately 24 kD in tonoplast-enriched membranes at nearly equal levels throughout development, with a small peak at +15 dpa. Antibodies recognizing α-tubulin (Sigma) detected a protein of approximately 53 kD in total protein from +10 dpa and later, with peak abundance at +25 dpa, then decreasing in samples from +30 and +35 dpa. One should note that the changes in specific protein accumulation detected in purified membrane fractions reflect differences relative to the total amount of membrane in the cell and to the amount of that membrane purified at each developmental stage. The samples were not standardized to allow determination of the absolute amount of any specific protein in a cell throughout development.
Enzyme Activity Assays
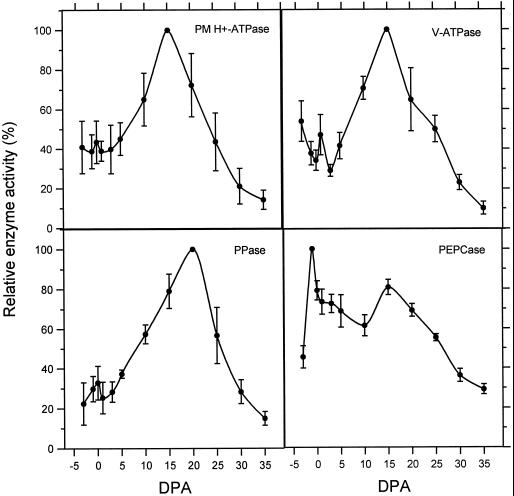
The ATPase activity of the PM H+-ATPase and the V-ATPase were assayed in microsomes (data not shown) and in PM- or tonoplast-enriched fractions (Fig. 4) using a published procedure that relies on the differential sensitivity of the V- and P-type ATPases to inhibitors (Gallagher and Leonard, 1982; Bennett et al., 1984). The pyrophosphatase activity of the PPase was also measured in microsomes (data not shown) and tonoplast-enriched fractions (Fig. 4) as described (Rea and Poole, 1985). Care was taken to measure enzyme activity as soon as possible after protein purification and to use samples that had experienced minimal freeze/thaw cycles, since activity does decrease slightly over time and after a freeze/thaw event (data not shown). The activity of the PM H+-ATPase displays the lowest levels of activity early and late in fiber development (<+10 dpa and >+25 dpa), whereas the highest activity was measured in samples from +15 dpa. The activity of the PM H+-ATPase was highly enriched in PM purified by two-phase partitioning (Fig. 4) as compared with activity in total microsomes (data not shown). The activity of the V-ATPase displayed a very similar pattern through development, with peak activity at +15 dpa. As with both H+-ATPases, the PPase displayed rising levels of activity in samples from +3 through +15 dpa but, in contrast, the peak of PPase activity was at +20 dpa rather than +15 dpa. Likewise, the activities of the vacuolar enzymes were greater in tonoplast-enriched fractions isolated by Suc density-gradient fractionation (Fig. 4) than in total microsomes (data not shown). The patterns of enzyme activity in total microsomes were essentially equivalent to the patterns observed in enriched membrane preparations, indicating that enrichment for the membrane fractions was performed with equivalent efficiency throughout the developmental series. The activity of PEPCase was measured in total protein (Fig. 4). Highest PEPCase activity levels were measured in samples from −1 dpa, which represents a sharp increase from the activity at −3 dpa. Activity declined through +10 dpa, with another peak at +15 dpa, then declined again through fiber development.
Figure 4.
Enzyme activity in protein extracts from developing cotton locules. Each point represents the average of at least three independent assays, with bars above and below representing one sd. Purified PM was used for the PM H+-ATPase assays, whereas tonoplast-enriched membranes were used for V-ATPase and PPase assays. Total soluble protein was assayed for PEPCase activity. The average specific activity of PM H+-ATPase at 15 dpa was 152.4 ± 26.6 nmol Pi mg−1 min−1, of V-ATPase at 15 dpa was 103.7 ± 23.4 nmol Pi mg−1 min−1, of PPase at 20 dpa was 142.3 ± 27.4 nmol PPi mg−1 min−1, and of PEPCase at −1 dpa was 0.1 ± 0.01 unit mg−1, where 1 unit equals the oxidation of 1 μmol NADH min−1 using a molar extinction coefficient of 6.22 × 103.
DISCUSSION
The regulation of cell expansion is a vital process of plant growth and development and is subject to control by environmental and hormonal factors. Cotton fibers afford us the opportunity to study the development of a single cell type that undergoes very rapid, virtually synchronized cell elongation (Basra and Malik, 1984; Tiwari and Wilkins, 1995) and that can be harvested in pure preparations for RNA and protein purification. Here we have characterized the expression of the proton pumps, a putative water channel, an enzyme that contributes to the production of osmoticum, and a cytoskeletal protein by assaying mRNA accumulation, protein accumulation, and enzyme activity. To measure RNA accumulation, we chose to use the RPA because it is very sensitive, quantitative, and requires only a small amount of RNA. These experiments were performed using tissue isolated from carefully staged greenhouse-grown plants, which, in our hands, showed very predictable developmental timing, even from season to season (Hasenfratz et al., 1995). Accumulation of 18S rRNA transcript was assayed throughout development for comparison with accumulation of the mRNAs tested. Protein samples for immunoblots were standardized to the amount of total protein in each sample. To make comparisons between levels of mRNA and protein accumulation, one must assume equal variation in the accumulation of 18S rRNA and cellular protein throughout development.
PCR-based strategies were used to isolate novel clones to serve as probes for PM H+-ATPase, PPase, PEPCase, MIP, and α-tubulin. Cotton is an allotetraploid, making complete characterization of large gene families, such as the PM H+-ATPase (Ewing and Bennett, 1994; Harper et al., 1994) and MIP (Weig et al., 1997) gene families, intractable. In other plants, the PPase and PEPCase have been found to be encoded by low-copy-number genes or small gene families (Kim et al., 1994; Chollet et al., 1996), whereas α-tubulin is encoded by a large gene family in Arabidopsis (Kopczak et al., 1992). For all but PEPCase, degenerate oligonucleotide primers were used, not in a strategy designed to isolate all of the potential gene family members, but rather with the expectation of isolating perhaps the most predominantly expressed gene family member. Of the PCR clones isolated and characterized by sequence analysis, only clones of α-tubulin appeared to originate from multiple genes (data not shown), suggesting that the recovered PM H+-ATPase and PPase clones may represent the predominantly expressed isoform. The cotton MIP clone isolated is most similar to the δ-TIP gene from A. thaliana (Daniels et al., 1996). Notably, it is 99% identical over the region cloned to another MIP isolated from cotton fibers (Ferguson et al., 1997) (GenBank accession no. U62778), and thus may represent an allele of that gene. MIP proteins are encoded by large multigene families in plants (Chrispeels and Agre, 1994; Weig et al., 1997), so it is difficult to say whether this gene represents the predominant isoform expressed in fibers. The very low accumulation of transcript, compared with relatively abundant protein detected by immunoblotting, suggests that this gene may not be the predominant MIP expressed during cotton fiber development.
This and previous work suggest that there are multiple developmental programs controlling gene expression through cotton fiber development (Wilkins and Jernstedt, 1997). One pattern is represented by those genes expressed at their highest levels slightly before and during the period of peak fiber expansion, followed by a sharp decline in message accumulation by +20 dpa, when the rate of expansion slows sharply (Basra and Malik, 1984). Transcripts arising from the PM H+-ATPase, V-ATPase, PEPCase, α-tubulin, and MIP genes accumulate to their highest levels in the days just prior to the peak period of expansion (+12–15 dpa; Basra and Malik, 1984).
Other genes display this same pattern of expression throughout cotton fiber development. A gene encoding a fiber-specific lipid-transfer protein is most highly expressed at about +15 dpa and is probably involved in cutin deposition on the surface of the fiber cell (Ma et al., 1995). Likewise, genes encoding expansin and endo-1,4-β-glucanase are expressed at highest levels between +9 and +15 dpa (Shimizu et al., 1997). These proteins act by loosening or cleaving bonds in the cell wall to allow cell expansion (Fry, 1995; Cosgrove, 1997). In addition, the E6 gene displays this same pattern of gene expression, verified to be regulated at the level of transcription (John and Crow, 1992; Rinehart et al., 1996). As with the genes we have analyzed, the accumulation of transcripts from these three genes declines by +20 dpa. This program of gene expression is consistent with proteins that are involved in the process of rapid cell expansion and primary cell wall deposition, which occurs during this period of cotton fiber development.
In general, the patterns of transcript and protein accumulation correlate with the patterns of activity for the PM H+-ATPase and PEPCase, although the peaks of mRNA accumulation precede the peaks of protein accumulation and enzyme activity by a few days. In contrast, the V-ATPase protein level is highest early in fiber development, when enzyme activity is relatively low. The peak of V-ATPase activity at +15 dpa, with no corresponding change in protein accumulation, suggests posttranslational activation of this enzyme, perhaps at the level of assembly. Accumulation of α-tubulin message peaks sharply at +15 dpa, which is 10 d prior to the peak of protein accumulation and could reflect a long half-life for α-tubulin turnover. The constant level of MIP protein accumulation throughout development may indicate that the protein is quite stable or that the VM23 antibodies detected protein(s) other than the one assayed by RPAs, the mRNA of which declined dramatically beyond +10 dpa.
Another pattern of gene expression is displayed by the PPase gene, for which message accumulation was constitutive through fiber development. Likewise, the 18S rRNA gene was constitutively expressed. Genes encoding actin, endoxyloglucan transferase, and Suc synthase also display constitutive mRNA accumulation through fiber development (Shimizu et al., 1997).
It is interesting that the PPase enzyme activity changes dramatically during expansion, with a peak at +20 dpa, a few days after the peak rate of fiber expansion, whereas the greatest protein accumulation occurs at +1 dpa, when the levels are severalfold higher than those at +15 or +20 dpa. As with the V-ATPase, the highest levels of PPase activity simultaneous with relatively low levels of protein accumulation are indicative of posttranslational activation of the PPase enzyme. The peak of PPase activity occurred at about +20 dpa, a few days later than the peaks of activity of the two H+-ATPases, which may suggest that the PPase plays more of a role in the process of secondary wall deposition.
An additional pattern of gene expression through fiber development is apparent from analysis of other cotton genes. This pattern is characterized by highest levels of mRNA accumulation later in fiber development, during the phase of secondary cell wall formation, approximately +17 dpa through +35 dpa. The genes encoding cellulase synthase, celA1 and celA2, display greatest accumulation beyond +17 dpa (Pear et al., 1996). Likewise, the H6 gene and FbL2A genes are expressed primarily during the later stages of fiber development (John and Keller, 1995; Rinehart et al., 1996). None of the genes characterized in our study displayed low levels of expression early in development, which is indicative of this class of genes.
The defined patterns of mRNA accumulation presented here suggest that gene expression through fiber development may be controlled by common regulatory elements, and that the patterns are consistent with the presumed functions of these enzymes in the major cellular processes of turgor regulation, cell wall deposition, and cytoskeleton formation. Preliminary research suggests that genes encoding turgor control enzymes and cell wall-related proteins respond differently to hormonal signals, perhaps indicating coordinated responses to multiple signaling cascades (W. Kim and T.A. Wilkins, unpublished data). Discrepancies between protein accumulation and enzyme activity, as in the cases of the V-ATPase and PPase, suggest posttranslational mechanisms of regulation, which are the topics of current research. When considering molecular genetic approaches to manipulation of fiber properties, it will be important to know the contributions of posttranscriptional and posttranslational regulation of enzyme activity, in addition to the tissue-specific and developmental patterns of transcription. It is clear that only after gaining a thorough understanding of the biology of fiber development will predictable manipulation of fiber properties be accomplished.
ACKNOWLEDGMENTS
The authors are grateful to Dr. Alan B. Bennett for allowing some of this work to be performed in his laboratory and to Dr. Ching-Yi Wan for isolating the α-tubulin clone. We acknowledge the generous gifts of antibodies from Dr. Ramon Serrano, Valencia, Spain, and Dr. Carole Bassett, U.S. Department of Agriculture-Agricultural Research Service, Kearneysville, WV.
Abbreviations:
- dpa
days postanthesis
- H+-ATPase
proton-translocating ATPase
- LPC
l-α-lysophosphatidylcholine
- MIP
major intrinsic protein
- PEPCase
phophoenolpyruvate carboxylase
- PM
plasma membrane
- PPase
proton-translocating pyrophosphatase
- RM
resuspension medium
- RPA
RNase protection assay
- V-ATPase
vacuolar H+-ATPase
Footnotes
This work was supported by funding from Cotton Incorporated and the U.S. Department of Energy. L.B.S. was supported by a National Science Foundation postdoctoral fellowship in plant biology (no. BIR-9203665).
LITERATURE CITED
- Albert HA, Martin T, Sun SSM. Structure and expression of a sugarcane gene encoding a housekeeping phosphoenolpyruvate carboxylase. Plant Mol Biol. 1992;20:663–671. doi: 10.1007/BF00046451. [DOI] [PubMed] [Google Scholar]
- Ames BN. Assay of inorganic phosphate, total phosphate, and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- Barkla BJ, Pantoja O. Physiology of ion transport across the tonoplast of higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:159–184. doi: 10.1146/annurev.arplant.47.1.159. [DOI] [PubMed] [Google Scholar]
- Basra AS, Malik CP. Development of the cotton fiber. Int Rev Cytol. 1984;89:65–113. [Google Scholar]
- Bennett AB, Leigh RA, Spanswick RM. H+-ATPase from vacuolar membranes of higher plants. Methods Enzymol. 1988;157:579–590. doi: 10.1016/0076-6879(88)57106-9. [DOI] [PubMed] [Google Scholar]
- Bennett AB, O'Neill SD, Spanswick RM. H+-ATPase activity from storage tissue of Beta vulgaris I. Identification and characterization of an anion-sensitive H+-ATPase. Plant Physiol. 1984;74:538–544. doi: 10.1104/pp.74.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AB, Spanswick RM. H+-ATPase activity from storage tissue of Beta vulgaris II. H+/ATP stoichiometry of an anion-sensitive H+-ATPase. Plant Physiol. 1984;74:545–548. doi: 10.1104/pp.74.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Agre P. Aquaporins: water channel proteins of plant and animal cells. Trends Biochem Sci. 1994;19:421–425. doi: 10.1016/0968-0004(94)90091-4. [DOI] [PubMed] [Google Scholar]
- Cosgrove D. Biophysical control of plant cell growth. Annu Rev Plant Physiol. 1986;37:377–405. doi: 10.1146/annurev.pp.37.060186.002113. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Creeping walls, softening fruit, and penetrating pollen tubes: the growing roles of expansins. Proc Natl Acad Sci USA. 1997;94:5504–5505. doi: 10.1073/pnas.94.11.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA. Organization of cortical microtubules in plant cells. Curr Opin Cell Biol. 1995;7:65–71. doi: 10.1016/0955-0674(95)80046-8. [DOI] [PubMed] [Google Scholar]
- Daniels MJ, Chaumont F, Mirkov TE, Chrispeels MJ. Characterization of a new vacuolar membrane aquaporin sensitive to mercury at a unique site. Plant Cell. 1996;8:587–599. doi: 10.1105/tpc.8.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing NN, Bennett AB. Assessment of the number and expression of P-type H+-ATPase genes in tomato. Plant Physiol. 1994;106:547–557. doi: 10.1104/pp.106.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DL, Turley RB, Kloth RH. Identification of a δ-TIP cDNA clone and determination of related A and D genome subfamilies in Gossypium species. Plant Mol Biol. 1997;34:111–118. doi: 10.1023/a:1005844016688. [DOI] [PubMed] [Google Scholar]
- Fry SC. Polysaccharide-modifying enzymes in the plant cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:497–520. [Google Scholar]
- Gallagher SR, Leonard RT. Effect of vanadate, molybdate, and azide on membrane-associated ATPase and soluble phosphatase activities of corn roots. Plant Physiol. 1982;70:1335–1340. doi: 10.1104/pp.70.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibrat R, Grouzis JP, Rigaud J, Galtier N, Grignon C. Electrostatic analysis of effects of ions on the inhibition of corn root plasma membrane Mg2+-ATPase by the bivalent orthovanadate. Biochim Biochem Acta. 1989;979:46–52. doi: 10.1016/0005-2736(89)90521-x. [DOI] [PubMed] [Google Scholar]
- Goddard RH, Wick SM, Silflow CD, Snustad DP. Microtubule components of the plant cell cytoskeleton. Plant Physiol. 1994;104:1–6. doi: 10.1104/pp.104.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Manney L, Sussman MR. The plasma membrane H+-ATPase gene family in Arabidopsis: genomic sequence of aha10 which is expressed primarily in developing seeds. Mol Gen Genet. 1994;244:572–587. doi: 10.1007/BF00282747. [DOI] [PubMed] [Google Scholar]
- Hasenfratz MP, Tsou CL, Wilkins TA. Expression of two related vacuolar H+-ATPase 16-kilodalton proteolipid genes is differentially regulated in a tissue-specific manner. Plant Physiol. 1995;108:1395–1404. doi: 10.1104/pp.108.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME, Crow LJ. Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc Natl Acad Sci USA. 1992;89:5769–5773. doi: 10.1073/pnas.89.13.5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME, Keller G. Characterization of mRNA for a proline-rich protein of cotton fiber. Plant Physiol. 1995;108:669–676. doi: 10.1104/pp.108.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Kim EJ, Rea PA. Isolation and characterization of cDNAs encoding the vacuolar H+-pyrophosphatase of Beta vulgaris. Plant Physiol. 1994;106:375–382. doi: 10.1104/pp.106.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellbom P, Larsson C. Preparation and polypeptide composition of chlorophyll-free plasma membranes from leaves of light-grown spinach and barley. Physiol Plant. 1984;62:501–509. [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis contains at least 6 expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson C. Plasma membrane. In: Linskens HF, Jakson JF, editors. Cell Components. Berlin: Springer-Verlag; 1985. pp. 85–104. [Google Scholar]
- Larsson C, Anderson B. Two phase method for chloroplasts, chloroplast elements and mitochondria. In: Reid E, editor. Plant Organelles, Vol. 9. Chichester, UK: Ellis Horwood; 1979. pp. 34–46. [Google Scholar]
- Ma DP, Tan H, Si Y, Creech RG, Jenkins JN. Differential expression of a lipid transfer protein gene in cotton fiber. Biochim Biochem Acta. 1995;1257:81–84. doi: 10.1016/0005-2760(95)00077-p. [DOI] [PubMed] [Google Scholar]
- Maeshima M. Characterization of the major integral protein of vacuolar membrane. Plant Physiol. 1992;98:1248–1254. doi: 10.1104/pp.98.4.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M, Yoshida S. Purification and properties of vacuolar membrane proton-translocating inorganic pyrophosphatase from mung bean. J Biol Chem. 1989;264:20068–20073. [PubMed] [Google Scholar]
- Marczewski W. Kinetic properties of phosphoenolpyruvate carboxylase from lupin nodules and roots. Physiol Plant. 1989;76:539–543. [Google Scholar]
- Matsuura-Endo C, Maeshima M, Yoshida S. Mechanism of the decline in vacuolar H+-ATPase activity in mung bean hypocotyls during chilling. Plant Physiol. 1992;100:718–722. doi: 10.1104/pp.100.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Reizer J, Schroeder JI, Chrispeels MJ. The vacuolar membrane protein γ-TIP creates water specific channels in Xenopus oocytes. EMBO J. 1993;12:2241–2247. doi: 10.1002/j.1460-2075.1993.tb05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CR, Rustin P, Wedding RT. A simple and accurate spectrophotometric assay for phosphoenolpyruvate carboxylase activity. Plant Physiol. 1988;86:325–328. doi: 10.1104/pp.86.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parets-Soler A, Pardo JM, Serrano R. Immunocytolocalization of plasma membrane H+-ATPase. Plant Physiol. 1990;93:1654–1658. doi: 10.1104/pp.93.4.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiol. 1992;99:1271–1274. doi: 10.1104/pp.99.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA, Poole RG. Proton-translocating inorganic pyrophosphatase in red beet (Beta vulgaris L.) tonoplast vesicle. Plant Physiol. 1985;77:46–52. doi: 10.1104/pp.77.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart JA, Petersen MW, John ME. Tissue-specific and developmental regulation of cotton gene FbL2A. Plant Physiol. 1996;112:1331–1341. doi: 10.1104/pp.112.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert AM, Benedict CR, Berlin JD, Kohel RJ. Cotton fiber development-kinetics of cell elongation and secondary wall thickening. Crop Sci. 1973;13:704–709. [Google Scholar]
- Schuller KA, Turpin DH, Plaxton WC. Metabolite regulation of partially purified soybean nodule phosphoenolpyruvate carboxylase. Plant Physiol. 1990;94:1429–1435. doi: 10.1104/pp.94.3.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T. Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol. 1997;38:375–378. doi: 10.1093/oxfordjournals.pcp.a029178. [DOI] [PubMed] [Google Scholar]
- Silk WK. Quantitative descriptions of development. Annu Rev Plant Physiol. 1984;35:479–518. [Google Scholar]
- Tiwari SC, Wilkins TA. Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Bot. 1995;73:746–757. [Google Scholar]
- Towbin J, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. Isolation of multiple cDNAs encoding the vacuolar H+-ATPase subunit b from developing cotton (Gossypium hirsutum L.) ovules. Plant Physiol. 1994a;106:393–394. doi: 10.1104/pp.106.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994b;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Weig A, Deswarte C, Chrispeels MJ (1997) The major intrinsic protein family of Arabidopsis has 23 members that form three distinct groups with functional aquaporins in each group. 114: 1347–1357 [DOI] [PMC free article] [PubMed]
- Wilkins TA. Vacuolar H+-ATPase 69-kilodalton catalytic subunit cDNA from developing cotton (Gossypium hirsutum) ovules. Plant Physiol. 1993;102:679–680. doi: 10.1104/pp.102.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins TA, Jernstedt JA (1997) Cell biology and molecular genetics of developing cotton fibers. In AS Basra, ed, Cotton Fibers. Food Products Press, New York (in press)
- Wilkins TA, Smart LB. Isolation of RNA from plant tissue. In: Krieg PA, editor. A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis. Inc., New York: Wiley-Liss; 1996. pp. 21–41. [Google Scholar]
- Wilkins TA, Wan CY, Lu CC. Ancient origin of the vacuolar H+-ATPase 69-kilodalton catalytic subunit superfamily. Theor Appl Genet. 1994;89:514–524. doi: 10.1007/BF00225389. [DOI] [PubMed] [Google Scholar]