Abstract
Elephant endotheliotropic herpesviruses (EEHV) can cause lethal hemorrhagic disease in both African and Asian elephants. At least seven EEHV types have been described, and sensitive real-time PCR tests have been developed for EEHV1A and 1B, which are associated with the majority of characterized Asian elephant deaths. Despite growing knowledge of the different EEHV types, the prevalence of each type within African and Asian elephants remains to be determined and there is considerable need for diagnostic tests to detect and discriminate between each EEHV species for clinical management of African and Asian elephants that develop illness from one or more of these viruses. To begin to address these issues, we developed real-time PCR assays for EEHV2, 3, 4, 5, and 6. Overall, each assay had robust PCR efficiency, a dynamic linear range over 5 log10 concentrations, a limit of detection of 10 copies/test reaction with 100% sensitivity, and low intra- and inter-assay variability. Each assay proved to be specific for the EEHV targets for which it was designed, with the exception of EEHV3 and EEHV4, which was expected because of greater DNA sequence similarity between these two EEHV species than the others. These new tools will be useful for conducting surveys of EEHV prevalence within captive and range country elephants, for diagnostic testing of elephants with suspected EEHV-associated disease, and for managing the treatment of elephants with EEHV-induced illness.
Keywords: elephant, herpesvirus, Proboscivirus, real-time PCR
1. Introduction
Elephantid herpesviruses, commonly known as Elephant endotheliotropic herpesviruses (EEHVs), are most closely related to the Betaherpesvirinae sub-family and are assigned to the Proboscivirus genus (Davison et al., 2009; Richman and Hayward, 2011). EEHVs are associated with lethal hemorrhagic disease in Asian (Elephas maximus) and African (Loxodonta africana) elephants with more than 70 cases of EEHV-associated elephant deaths being reported worldwide (Ehlers et al., 2001; Fickel et al., 2001; Garner et al., 2009; Ossent et al., 1990; Richman and Hayward, 2011; Richman et al., 1999; Richman, Montali, and Hayward, 2000; Zachariah, 2008). EEHV-associated hemorrhagic disease is most common in juvenile captive-born Asian elephants and deaths due to EEHV infection account for approximately 60% of the overall mortality rate of captive-born Asian elephants in North America (Richman and Hayward, 2011). Although the investigation and understanding of the impact of the EEHVs is in it’s infancy, preliminary evidence suggest that overall, they represent a significant source of morbidity and mortality in elephants worldwide.
There are currently seven distinct EEHV types that have been identified based on viral DNA sequence analysis. EEHV2, EEHV3, EEHV4, EEHV5, and EEHV6 differ from EEHV1 by between 15 to 35% at the nucleotide level in several well conserved genes and from each other by 8 to 35% (Latimer et al., 2011; Richman and Hayward, 2011). Therefore, they all rate as distinct species, whereas EEHV1A and EEHV1B are complex partially related chimeric sub-species. EEHV1A and 1B have been responsible for the vast majority of EEHV-associated deaths in elephants (Garner et al., 2009; Latimer et al., 2011; Richman et al., 1999; Zong et al., 2007). EEHV2 has been associated with the deaths of two African elephants while EEHV3 and 4 have been associated with the death of one Asian elephant calf each (Garner et al., 2009; Latimer et al., 2011; Richman et al., 1999). EEHV5 was originally detected in blood samples from an apparently healthy wild-born adult Asian elephant and EEHV4 was detected in blood samples from a clinically ill African elephant calf that survived the infection (Latimer et al., 2011). Although the existence of at least these 7 types of Probosciviruses is known, the prevalence of EEHV infection and distribution of EEHV species amongst in situ or ex situ elephant populations remains to be determined.
Previously, a quantitative real-time polymerase chain reaction (qPCR) assay was developed that detects both forms of the EEHV1 species (1A and 1B) and used this assay to screen multiple fluid samples from healthy and clinically ill Asian elephants (Stanton et al., 2010). We demonstrated that EEHV1 is detected commonly in trunk secretions from apparently healthy captive Asian elephants and used viral genotypic analysis on EEHV1 viral DNA to establish the first epidemiologic connection between healthy Asian elephants shedding a virus that had previously been associated with the death of a calf in the same herd. In addition, it was found that knowledge of EEHV viral loads in blood samples and trunk washes is useful for the management of animals infected with EEHV (Stanton et al., 2012). The purpose of the current study was to develop and validate four more similar specific and sensitive qPCR assays that independently detect EEHV2, 3/4, 5, and 6. This work will provide novel assays to include in diagnostic testing for clinically ill elephants and for future studies regarding the overall prevalence of EEHV infection amongst in situ and ex situ Asian elephants.
2. Materials and Methods
2.1. Sample collection and preparation
All samples were collected from Asian elephants as part of their routine medical care or during necropsy. Preparation of DNA samples from North American Proboscivirus (NAP) cases 19, and 31 and 34 (EEHV1A) plus 22 (EEHV4), 27 (EEHV3) and negative control Asian elephants have been previously described (Stanton et al., 2010). DNA was prepared from NAP12 (EEHV2) and NAP35 (EEHV6) (Latimer et al., 2011; Richman et al., 1999) using a Gentra Capture Column Kit (Gentra Systems, Minneapolis, MN) and the manufacturers recommended protocol. DNA was prepared from NAP45 (EEHV1B, Genbank accession JN633891.1) and NAP50 (EEHV5, Genbank accession JN983108.1) using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) and the manufacturers recommended protocol. Genomic elephant DNA used for diluting standards was prepared as described previously (Stanton et al., 2010).
2.2. Real-time qPCR assays
All EEHV qPCR assays were performed using reagents and thermocycling conditions as previously described for the EEHV1 assay (Stanton et al., 2010). DNA primers and 5′-hydrolysis probes were selected using Primer Express software version 3.0 (Applied Biosystems/Life Technologies Inc., Carlsbad, CA). Oligonucleotide primers were ordered from Integrated DNA Technologies, Inc. (Coralville, IA). Hydrolysis probes were ordered from Applied Biosystems/Life Technologies Inc. (Carlsbad, CA). Assays were performed using a StepOne thermocycler and StepOne software package version 2.1 (Applied Biosystems/Life Technologies Inc., Carlsbad, CA) as previously described (Stanton et al., 2010). No template control (NTC) reactions in which water was added instead of test DNA were included with every qPCR assay. Assays were only considered valid and included in this study if NTC reactions produce Cq greater than or equal to 40.
The EEHV2 qPCR amplicon was designed using the DNA polymerase/U38 nucleotide sequence (EEHV2 NAP12 GenBank accession number HM568558). EEHV2 specific oligonucleotides consist of a hydrolysis probe 5′-6FAM-CGACCACGAAAGAATA-MGB/NFQ-3′, forward primer 5′-CCCAGGGACGCCAGTTACTT-3′, and reverse primer 5′-CCAATCGTTAAATCTCTCGCA-3′. A qPCR amplicon to detect both EEHV3 and EEHV4 was designed using nucleotide sequences that are 98% identical over the entire amplicon of both EEHV3 and EEHV4 terminase genes (EEHV3 NAP27 and EEHV4 NAP22, GenBank accession numbers EU658937.1 and JN788931.1 respectively). EEHV3/4 specific oligonucleotides consist of a hydrolysis probe 5′-6FAM-CACGTGATCGCGTCC-MGB/NFQ-3′, a forward primer 5′-TGGGCTTATGTAATCGGTAGC-3′, and a reverse primer 5′-CGTGTGCGAGGAGCACTTATAT-3′. The EEHV5 qPCR amplicon was designed using the DNA polymerase/U38 nucleotide sequence (EEHV5 NAP50 GenBank accession number JN983114.1). EEHV5 specific oligonucleotides consist of the hydrolysis probe 5′-6FAM-AGCCGTGAGAGAAA-MGB/NFQ-3′, a forward primer 5′-CCTGGTTGGCGGAAAGAA-3′, and a reverse primer 5′-GCATCAAAGGGTCACTACACTGTT-3′. The EEHV6 qPCR amplicon was designed using the DNA polymerase/U38 nucleotide sequence (EEHV6 NAP35 GenBank accession number JF692762). EEHV6 specific oligonucleotides consist of the hydrolysis probe 5′-6FAM-CGTGTTTACCGATAGCC-MGB/NFQ-3′, a forward primer 5′-AGGCGTCTCAAAGGGTATGTT-3′, and a reverse primer 5′-TCCCTGAGCGGTGACAGATT-3′. Inspection of all available sequences within Genbank indicate that the hydrolysis probes and primers for all assays bind within areas of sequence that are 100% conserved between known isolates.
2.3. Calibration curve preparation
Using available DNA sequence data for EEHV3 (EU658937.1) and EEHV5 (JN983114.1), target DNA sequences for these two viruses were amplified by standard PCR, as described previously (Stanton et al., 2010). A 299bp product for the EEHV3 terminase gene was amplified using forward primer 5′-TACGTCATTCCTCGCCCATGTGAA-3′ and reverse primer 5′-TGCTGTAGCGGATCATGTCGAACT-3′. A 500bp product for the EEHV5 POL gene was amplified using forward primer 5′-CCTATGTTATTGTACCTTAGTT-3′ and reverse primer 5′-CAGAACTTTCCACTATCGACA-3′. Both PCR products were cloned into the commercially available pCR Blunt cloning vector (Invitrogen/Life Technologies Inc., Carlsbad, CA) to generate plasmids pRSP724 (EEHV3 terminase) and pRSP796B (EEHV5 DNA polymerase). The recombinant plasmids pRSP724 and pRSP796B were linearized using BamHI or HindIII respectively, purified using a commercially available kit (QIAGEN Inc., Hilden, Germany), and quantified using a commercially available DNA fluorometer kit (Qubit, Invitrogen/Life Technologies Inc., Carlsbad, CA). Oligonucleotides corresponding to the positive DNA strand of the target sequences were ordered from Integrated DNA Technologies, Inc. (Coralville, IA) for construction of the EEHV2 and EEHV6 calibration curves. Lyophilized oligonucleotides were diluted to a concentration of 100 μM (6.02 × 1013 copies/μL) in distilled and deionized sterile water. For plasmids, the following formula was used to calculate copies target sequence per μL: [(6.02 × 1023 copies) × (plasmid concentration in g/μL)]/[(No. of bases) × (660 g/base)]. Linearized plasmids or oligonucleotides were then diluted to known copy number/μL into a solution of 20 ng/μL Asian elephant DNA in AE elution buffer (QIAGEN Inc., Hilden, Germany). The range over which target sequences were tested was 10 – 1 × 105 copies/test reaction.
3. Results
3.1. Assay performance
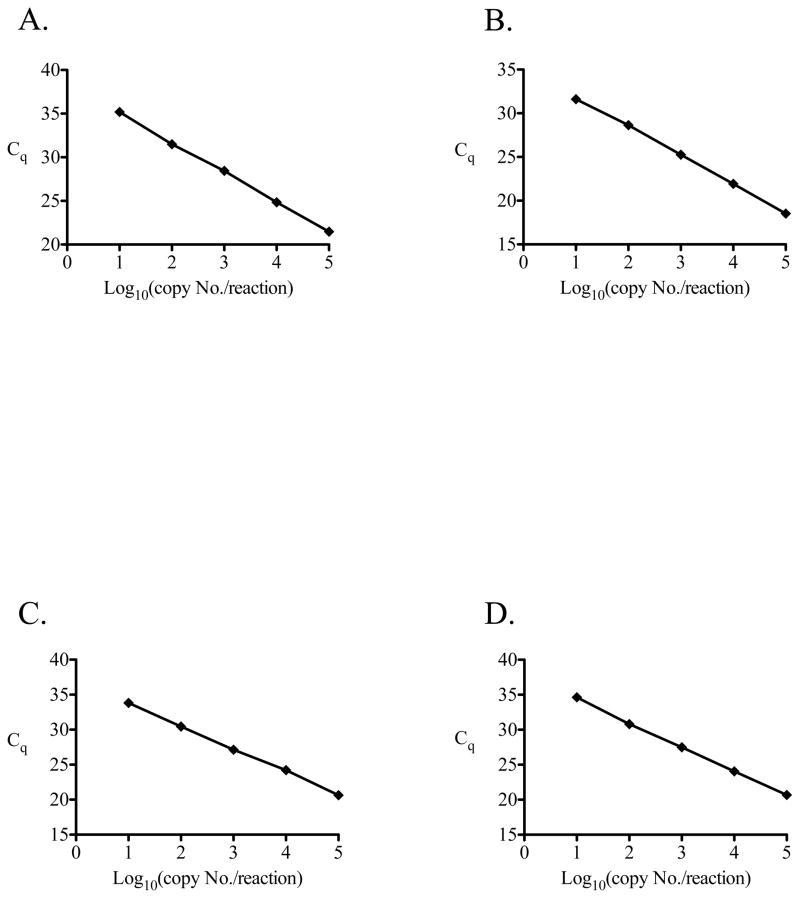
In order to determine the individual assay performance characteristics, a calibration curve containing known quantities of target DNA sequence over a range of 1 – 1 × 105 copies per test reaction was diluted into a solution containing 20 ng/μL of Asian elephant DNA known to be negative for the presence of detectable Elephantid herpesvirus DNA. This allowed the determination of the performance of each assay in a complex solution of DNA that simulates clinical samples as closely as possible. At least 5 independent calibration curves were carried out over a range of 10 – 1 × 105 copies target sequence per test reaction for each assay and representative curves are shown in Fig. 1. Using data generated from the calibration curves we determined the PCR efficiency, linear dynamic range, limit of detection (LOD), and the intra assay and inter assay variation for each EEHV qPCR assay (Tables 1–3 and Figure 1). Intra- and inter-assay variability for the published EEHV1 assay is included for comparison (Tables 2 and 3). Each assay had a robust PCR efficiency (90–110%), a dynamic linear range over 5 log10 concentrations (10 – 1 × 105), a limit of detection of 10 copies/test reaction with 100% sensitivity, and low intra-assay and inter-assay variation.
Figure 1.
Representative graphs displaying calibration curves that were used for validation of EEHV qPCR assays: EEHV2 (A), EEHV3/4 (B), EEHV5 (C), and EEHV6 (D). Calibrations curves are shown over a target concentration range of 10 to 100,000 target copies per reaction and assay parameters are shown in Table 1.
Table 1.
EEHV qPCR standard curve assay parameters assessed using calibration curves.
| qPCR assay | Linear Dynamic Range (copies per reaction) | Limit of Detection (100% Sensitivity) | PCR % Efficiency | R2 | Slope | Y-intercept |
|---|---|---|---|---|---|---|
| EEHV2 | 10 – 100,000 | 10 | 96.553 | 0.998 | −3.407 | 38.508 |
| EEHV3/4 | 10 – 100,000 | 10 | 100.483 | 0.999 | −3.31 | 35.145 |
| EEHV5 | 10 – 100,000 | 10 | 102.607 | 0.998 | −3.261 | 37.037 |
| EEHV6 | 10 – 100,000 | 10 | 94.186 | 0.998 | −3.47 | 37.942 |
Table 3.
Reproducibility of real-time PCR for EEHV1, 2, ¾, 5, and 6
| Input copies | EEHV1 | EEHV2 | EEHV3/4 | EEHV5 | EEHV6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean±SDa | CVb | Mean±SDa | CVb | Mean±SDa | CVb | Mean±SDa | CVb | Mean±SDa | CVb | |
| 10 | 34.43±0.14 | 0.40 | 34.73±0.81 | 2.34 | 31.77±0.26 | 0.81 | 30.90±0.19 | 0.60 | 34.71±0.36 | 1.02 |
| 102 | 31.16±0.30 | 0.96 | 31.50±0.16 | 0.51 | 28.66±0.08 | 0.27 | 28.02±0.15 | 0.53 | 31.05±0.39 | 1.26 |
| 103 | 27.67±0.26 | 0.93 | 28.34±0.45 | 1.58 | 25.39±0.15 | 0.61 | 24.67±0.07 | 0.27 | 27.71±0.34 | 1.21 |
| 104 | 24.22±0.21 | 0.88 | 24.74±0.09 | 0.38 | 22.03±0.16 | 0.73 | 21.26±0.04 | 0.18 | 24.28±0.36 | 1.49 |
| 105 | 21.52±0.19 | 0.87 | 21.39±0.09 | 0.40 | 18.68±0.14 | 0.74 | 17.82±0.03 | 0.19 | 20.85±0.16 | 0.80 |
Mean and standard deviation of the threshold cycle (Cq) from 5 independent experiments carried out between 1–3 months.
Coefficient of variation
Table 2.
Intra-assay accuracy of real-time PCR for EEHV1, 2, ¾, 5, and 6
| Input copies | EEHV1 | EEHV2 | EEHV3/4 | EEHV5 | EEHV6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Mean±SDa | CVb | Mean±SDa | CVb | Mean±SDa | CVb | Mean±SDa | CVb | Mean±SDa | CVb | |
|
| ||||||||||
| 10 | 35.51±0.43 | 1.23 | 34.66±0.66 | 1.92 | 32.02±0.60 | 1.87 | 31.38±0.37 | 1.18 | 34.40±0.62 | 1.79 |
| 102 | 32.37±0.22 | 0.67 | 31.53±0.12 | 0.37 | 28.60±0.13 | 0.46 | 28.35±0.11 | 0.37 | 30.83±0.09 | 0.30 |
| 103 | 28.86±0.06 | 0.21 | 28.25±0.18 | 0.62 | 25.27±0.10 | 0.40 | 24.89±0.07 | 0.29 | 27.55±0.09 | 0.33 |
| 104 | 25.47±0.04 | 0.14 | 24.72±0.03 | 0.14 | 21.86±0.04 | 0.17 | 21.49±0.13 | 0.61 | 24.12±0.09 | 0.37 |
| 105 | 22.10±0.04 | 0.18 | 21.35±0.04 | 0.20 | 18.62±0.07 | 0.37 | 18.09±0.14 | 0.76 | 20.87±0.04 | 0.18 |
Mean and standard deviation of the threshold cycle (Cq) from 5 replicates
Coefficient of variation
3.2. Assay specificity
To validate the specificity of the four newly generated qPCR assays, we utilized archival Asian elephant DNA samples had previously been characterized for the presence of DNA for all known EEHV species (Table 4). A total of 10 archival DNA samples were examined, including samples that had been used previously to validate our EEHV1 qPCR assay (Stanton et al., 2010). We considered a positive assay to be one in which the Cq was less than 40 and those samples with Cq equal to or greater than 40 were considered negative. Any assay in which a NTC reaction produced a positive Cq was considered invalid and not included in the data set. Each assay which was developed proved to be specific for the designed EEHV. The only assay that did exhibit cross-reactivity between different EEHV species was the EEHV3/4 assay and this was an expected result based on the high DNA sequence similarity between the EEHV3 and EEHV4 amplicon sequences and the primer design used.
Table 4.
Specificity validation of multiple real-time qPCR assays detecting known EEHV species.
| EEHV Speciesa | NAP No.b | Samplec | EEHV1 (Cq) | EEHV2 (Cq) | EEHV3/4 (Cq) | EEHV5 (Cq) | EEHV6 (Cq) |
|---|---|---|---|---|---|---|---|
| 1A | 31 | Blood | 23.7d | >40 | >40 | >40 | >40 |
| 1A | 33 | Blood | 28.7d | >40 | >40 | >40 | >40 |
| 1A/B | 19 | Blood | 27.9d | >40 | >40 | >40 | >40 |
| 1B | 45 | TWe | 16.4 | >40 | >40 | >40 | >40 |
| 2 | 12 | Blood | >40 | 29.5 | >40 | >40 | >40 |
| 3 | 27 | Colon | >40d | >40 | 26.4 | >40 | >40 |
| 4 | 22 | Blood | >40d | >40 | 26.6 | >40 | >40 |
| 5 | 50 | Blood | >40 | >40 | >40 | 33.3 | >40 |
| 6 | 35 | Blood | >40 | >40 | >40 | >40 | 36.5 |
| None | Nonef | Blood | >40 | >40 | >40 | >40 | >40 |
EEHV species previously detected in archival elephant DNA sample.
North American Proboscivirus (NAP) number assigned to the test sample. If no NAP number was previously assigned to the case, the elephant’s name is given.
Tissue or fluid from which the test DNA sample was obtained.
Previously published in Stanton et al. 2010.
Trunk wash
Negative control Asian elephant with no detectable EEHV in genomic DNA sample
4. Discussion
It was shown that real-time PCR assays designed to recognize specific sequences in either the DNA polymerase gene or the terminase gene of EEHV2-6 had high specificity, sensitivity and reproducibility for the detection of the appropriate virus in a variety of previously characterized elephant DNA samples (Tables 1, 2 and Figure 1).
One hypothesis for the devastating lethal effects of EEHV1 in juvenile Asian elephants is because it is the result of a cross-species transmission between an endogenous African elephant virus to Asian elephants (Richman et al., 1999). To answer this question, more studies are needed to assess the prevalence of the different EEHV types within captive and range country Asian and African elephant populations. Previous studies from our group have shown that the EEHV1 qPCR assay detects reliably virus DNA in a wide variety of selected samples, including necropsy tissue and blood from symptomatic animals as well as trunk washes or blood from asymptomatic animals undergoing primary or reactivated infections and shedding (Stanton et al., 2010). The generation of the assays described in this study will provide the requisite tools needed to embark on studies to determine which EEHVs are endogenous to African and Asian elephants.
A difficult management issue for EEHV disease is the short time-frame between appearance of clinical signs and death. However, several Asian elephants have survived severe EEHV-associated disease (Schmitt et al., 2000; Stanton et al., 2012). The survival of these animals was assisted in part by aggressive supportive therapies and administration of anti-herpesviral medications. These novel qPCR assays will facilitate rapid and early diagnosis of potential disease, which would prompt initiation of treatment procedures known to have been associated with the increased rate of survival of other EEHV infected elephants. In addition, these qPCR assays could be used for regular monitoring of susceptible animals where one might even detect emerging viremia prior to the appearance of clinical signs.
Highlights.
Real-time PCR assays were developed for EEHV 2, 3, 4, 5, and 6
The new EEHV assays are sensitive and specific
The assays are useful for determining EEHV prevalence in in situ and ex situ elephants
The assays will be useful for diagnosing suspected cases of EEHV-associated illness
The assays will be useful for managing treatment of elephants with EEHV-induced illness
Acknowledgments
The authors thank Erin Latimer and the Houston zoo for providing control samples for this study. Support for this study was provided by the Houston Zoo and a grant from the Dan L. Duncan Foundation, as well as by grants from the International Elephant Foundation and the Morris Animal Fund to the NZP Elephant Herpesvirus Laboratory as well as by NIH grant R01 AI24576 to GSH at Johns Hopkins University. Sally Nofs was supported by National Institutes of Health training grant T32-AI-07471.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jeffrey J. Stanton, Email: stantonj@bcm.edu.
Sally Nofs, Email: nofs@bcm.edu.
Rongsheng Peng, Email: rpeng@bcm.edu.
Gary S. Hayward, Email: ghayward@jhmi.edu.
Paul D. Ling, Email: pling@bcm.edu.
References
- Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–7. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Burkhardt S, Goltz M, Bergmann V, Ochs A, Weiler H, Hentschke J. Genetic and ultrastructural characterization of a European isolate of the fatal endotheliotropic elephant herpesvirus. J Gen Virol. 2001;82:475–82. doi: 10.1099/0022-1317-82-3-475. [DOI] [PubMed] [Google Scholar]
- Fickel J, Richman LK, Montali R, Schaftenaar W, Goritz F, Hildebrandt TB, Pitra C. A variant of the endotheliotropic herpesvirus in Asian elephants (Elephas maximus) in European zoos. Vet Microbiol. 2001;82:103–9. doi: 10.1016/s0378-1135(01)00363-7. [DOI] [PubMed] [Google Scholar]
- Garner MM, Helmick K, Ochsenreiter J, Richman LK, Latimer E, Wise AG, Maes RK, Kiupel M, Nordhausen RW, Zong JC, Hayward GS. Clinico-pathologic features of fatal disease attributed to new variants of endotheliotropic herpesviruses in two Asian elephants (Elephas maximus) Vet Pathol. 2009;46:97–104. doi: 10.1354/vp.46-1-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer E, Zong JC, Heaggans SY, Richman LK, Hayward GS. Detection and evaluation of novel herpesviruses in routine and pathological samples from Asian and African elephants: identification of two new probosciviruses (EEHV5 and EEHV6) and two new gammaherpesviruses (EGHV3B and EGHV5) Vet Microbiol. 2011;147:28–41. doi: 10.1016/j.vetmic.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossent P, Guscetti F, Metzler AE, Lang EM, Rubel A, Hauser B. Acute and fatal herpesvirus infection in a young Asian elephant (Elephas maximus) Vet Pathol. 1990;27:131–3. doi: 10.1177/030098589002700212. [DOI] [PubMed] [Google Scholar]
- Richman LK, Hayward GS. Elephant Herpesviruses. In: Fowler REMaM., editor. Fowler’s Zoo and Wild Animal Medicine: Current Therapy. Saunders; St. Louis: 2011. pp. 496–502. [Google Scholar]
- Richman LK, Montali RJ, Garber RL, Kennedy MA, Lehnhardt J, Hildebrandt T, Schmitt D, Hardy D, Alcendor DJ, Hayward GS. Novel endotheliotropic herpesviruses fatal for Asian and African elephants. Science. 1999;283:1171–6. doi: 10.1126/science.283.5405.1171. [DOI] [PubMed] [Google Scholar]
- Richman LK, Montali RJ, Hayward GS. Review of a Newly Recognized Disease of Elephants Caused by Endotheliotropic Herpesviruses. Zoo Biol. 2000;19:383–392. doi: 10.7589/0090-3558-36.1.1. [DOI] [PubMed] [Google Scholar]
- Schmitt DL, Hardy DA, Montali RJ, Richman LK, Lindsay WA, Isaza R, West G. Use of famciclovir for the treatment of endotheliotrophic herpesvirus infections in Asian elephants (Elephas maximus) J Zoo Wildl Med. 2000;31:518–22. doi: 10.1638/1042-7260(2000)031[0518:UOFFTT]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Stanton JJ, Zong JC, Eng C, Howard L, Flanagan J, Stevens M, Schmitt D, Wiedner E, Graham D, Junge RE, Weber MA, Fischer M, Mejia A, Tan J, Latimer E, Herron A, Hayward GS, Ling PD. Kinetics of viral loads and genotypic analysis of elephant endotheliotropic herpesvirus-1 infection in captive Asian elephants (Elephas Maximus) Journal of Zoo and Wildlife Medicine. 2012 doi: 10.1638/1042-7260-44.1.42. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton JJ, Zong JC, Latimer E, Tan J, Herron A, Hayward GS, Ling PD. Detection of pathogenic elephant endotheliotropic herpesvirus in routine trunk washes from healthy adult Asian elephants (Elephas maximus) by use of a real-time quantitative polymerase chain reaction assay. Am J Vet Res. 2010;71:925–33. doi: 10.2460/ajvr.71.8.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah A, Richman LK, Latimer E, Hayward GS, Kalaivannan N, Zachariah A, Balan S, Gafoor A, Easwaran EK. Fatal Endotheliotropic Elephant Herpes Virus Mortality In Free Ranging and Captive Asian Elephants in South India. 2008 International Elephant Conservation and Research Symposium; 2008. p. 60. [Google Scholar]
- Zong JC, Latimer E, Heaggans S, Richman LK, Hayward GS. Pathogenesis and Molecular Epidemiology of Fatal Elephant Endotheliotropic Disease Associated with the Expanding Proboscivirus Genus of the Betaherpesvirinae. International Elephant Conservation and Research Symposium; 2007. pp. 23–35. [Google Scholar]