Abstract
Although a large proportion of molecules expressed in the nervous system are conserved from invertebrate to vertebrate, functional properties of such molecules are less characterized. Here, we show that highly conserved hydrolase AHO-3 acts as a novel regulator of starvation-induced thermotactic plasticity in Caenorhabditis elegans. As wild-type animals, aho-3 mutants migrated to the cultivation temperature on a linear thermal gradient after cultivation at a particular temperature with food. Whereas wild-type animals cultivated under food-deprived condition showed dispersed distribution on the gradient, aho-3 mutants exhibited tendency to migrate toward higher temperature. Such an abnormal behavior was completely rescued by the expression of human homologue of AHO-3, indicating that the molecular function of AHO-3 is highly conserved between nematode and human. The behavioral regulation by AHO-3 requires the N-terminal cysteine cluster, which ensures the proper subcellular localization of AHO-3 to sensory endings. Double-mutant analysis suggested that AHO-3 acts in the same pathway with ODR-3, a heterotrimeric G protein alpha subunit. Our results unveiled a novel neural protein in C. elegans, confirming its conserved role in behavioral regulation.
Introduction
The nervous system is one of the most distinctive organs in the animal kingdom. By virtue of that, animals can sense environmental stimuli, memorize the information and modify their behavior. A large fraction of proteins expressed in the nervous system are presumed to be conserved throughout the animal kingdom (Hunt-Newbury et al. 2007; Von Stetina et al. 2007) (http://www.ebi.ac.uk/gxa/). Comparative analyses elucidated that at least 38% of the 20 250 total Caenorhabditis elegans genes share homology with human genes (Lai et al. 2000; Shaye & Greenwald 2011) and that more than half of the 2500 C. elegans transcripts expressed in the nervous system have mammalian homologues (Von Stetina et al. 2007). Characterizations of the conserved neural components through animal species have provided important general insights into the mechanisms of the neural function. For example, G protein signaling pathway is essential for olfactory transduction (Buck 1996; Krieger & Breer 1999; Bargmann 2006) and phototransduction (Fu & Yau 2007; Wang & Montell 2007; Liu et al. 2010), cAMP response element–binding (CREB) protein pathway is a key mechanism for memory formation (Kandel 2001; Josselyn & Nguyen 2005; Kauffman et al. 2010; Nishida et al. 2011), and dopamine signaling is involved in reward learning and responses to food (Schwaerzel et al. 2003; Barron et al. 2010). Nevertheless, the neural functions of many other conserved molecules in the nervous systems still remain unknown.
Caenorhabditis elegans thermotaxis provides a behavioral plasticity paradigm, in which temperature preferences are modified by their cultivation temperature and feeding state (Hedgecock & Russell 1975; Mohri et al. 2005). After cultivation at a certain temperature with food, animals migrate to the cultivation temperature on a thermal gradient without food (Hedgecock & Russell 1975; Mohri et al. 2005; Ito et al. 2006). In contrast, animals cultivated without food disperse from the cultivation temperature (Hedgecock & Russell 1975; Mohri et al. 2005). This behavioral change has been called different names such as ‘thermotactic plasticity induced by starvation’ (Mohri et al. 2005), ‘temperature-food associative learning’ (Kuhara & Mori 2006) or ‘integrative behavior for temperature and feeding state’ (Kodama et al. 2006). We designated this behavior ‘thermotactic plasticity’ in this article.
To investigate the molecular and neural mechanisms underlying thermotactic plasticity, we previously performed a genetic screen to isolate the mutants defective in thermotactic plasticity, which were designated aho (abnormal hunger orientation) mutants (Mohri et al. 2005). Of these, aho-2(nj32) mutants migrated to the cultivation temperature in both well-fed and starved conditions (Mohri et al. 2005; Kodama et al. 2006). We found that aho-2(nj32) mutants exhibited a deletion in the ins-1 gene encoding insulin homologue and showed that insulin-like signaling pathway modulates the neuronal activity of interneurons required for the execution of thermotaxis (Kodama et al. 2006).
In this study, we identified and analyzed the gene responsible for the aho-3(nj15) mutant that has distinct abnormality in thermotactic plasticity. Whereas well-fed aho-3(nj15) mutants migrated to the cultivation temperature on a temperature gradient, starved aho-3(nj15) mutants showed tendency to migrate toward higher temperature. This abnormal phenotype is different from that of aho-2(nj32) mutants. We showed that the aho-3 gene encodes a novel and highly conserved hydrolase. The abnormality in thermotactic plasticity of aho-3 mutants was completely rescued by expressing human homologue of AHO-3, FAM108B1 protein, indicating that the molecular property is highly conserved between nematode and human. It was previously reported that rodent homologues of AHO-3, FAM108 proteins, are found in the membrane fraction of the brain proteome (Blankman et al. 2007; Kang et al. 2008). In addition, other study showed that the conserved N-terminal cysteine cluster of human FAM108 proteins is necessary for its plasma membrane localization (Martin & Cravatt 2009). We show here that AHO-3 acts in sensory neurons and localizes to sensory endings. Furthermore, the N-terminal cysteine cluster of AHO-3 is necessary for its subcellular localization and for its function in thermotactic plasticity. Double-mutant analysis suggested that AHO-3 acts in the same pathway with ODR-3, a heterotrimeric G protein alpha subunit, which is localized to sensory endings (Roayaie et al. 1998; Bargmann 2006). Our results suggest that evolutionarily conserved AHO-3 has important functions in the nervous system for behavioral plasticity.
Results
aho-3 mutants show abnormality in thermotactic plasticity associated with feeding states
We have previously reported that C. elegans exhibits thermotactic plasticity depending on their feeding states using the individual thermotaxis assay with a nonlinear thermal gradient; most of well-fed wild-type animals migrate to their cultivation temperature, whereas few starved animals migrate to their cultivation temperature (Mohri et al. 2005). In this study, we performed the population thermotaxis assay with a linear thermal gradient, which is suitable for quantitatively assessing the migration ability toward a certain temperature (Ito et al. 2006). We used the linear thermal gradient ranging from 17 to 23 °C with 20 °C at the center (Fig. 1A). After cultivation at 17, 20 or 23 °C under well-fed condition, wild-type N2 animals migrated to their cultivation temperature (Fig. 1B,E,H). By contrast, after cultivation at 17, 20 or 23 °C under food-deprived (starved) condition for 3, 2 or 1 h, respectively, most of the starved wild-type animals dispersed and did not migrate to their cultivation temperature (Fig. 1C,F,I; see ‘Experimental procedures’ for details on the starvation conditioning).
Figure 1.
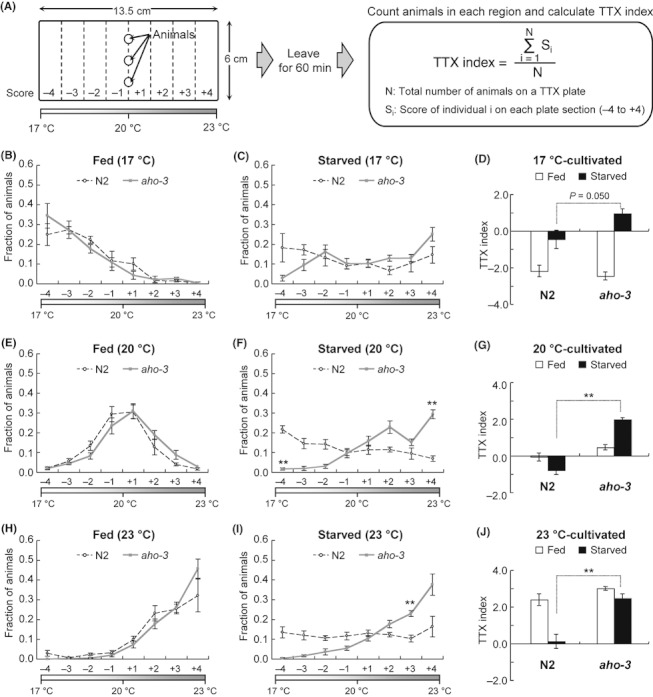
aho-3 mutants show abnormalities in thermotactic plasticity. (A) Procedures for population thermotaxis assay using a linear thermal gradient (Ito et al. 2006); 40–300 animals cultivated at 17, 20 or 23 °C were placed on the centerline of a TTX (thermotaxis) plate and allowed to move freely for 60 min. The animals in each region (from −4 to +4) were counted, and the TTX index was calculated as described. A 17–23 °C thermal gradient was always used in this study, except for Figure S1 in Supporting Information (see Experimental procedures for detail). (B–J) Thermotaxis of well-fed or starved wild-type N2 animals and aho-3(nj15) mutants that were cultivated at 17 °C (B–D), 20 °C (E–G) or 23 °C (H–J). n ≥ 4 assays. Error bars represent SEM. In (B, C, E, F and H, I), statistical significance of values in each region was tested by unpaired t-test with the Dunn–Sidak correction for multiple comparisons; *P < 0.05; **P < 0.01. In (D, G and J), statistical significance of TTX indices was tested by unpaired t-test in comparisons of well-fed N2 animals vs well-fed aho-3 mutants or starved N2 animals vs starved aho-3 mutants; *P < 0.05; **P < 0.01.
To investigate the molecular components in thermotactic plasticity, we conducted a forward genetic screen and isolated aho-3(nj15) mutants that display abnormality in thermotactic plasticity (Mohri et al. 2005). Similar to the well-fed wild-type animals, most of the well-fed aho-3(nj15) mutants cultivated at 17, 20 or 23 °C migrated to their cultivation temperature (Fig. 1B,D,E,G,H,J; P > 0.05; in unpaired t-test compared TTX indices), although distributions of aho-3(nj15) animals seemed to slightly shift toward higher temperature than those of wild-type animals after cultivation at 20 and 23 °C.
Starved aho-3(nj15) mutants conditioned at 17 °C seemed to migrate slightly higher temperature than the starved wild-type animals (Fig. 1C,D), but it was not statistically significant in unpaired t-test (P = 0.050; compared TTX indices). Unlike starved wild-type animals, starved aho-3(nj15) mutants conditioned at 20 °C migrated to the higher-temperature regions (+2, +3 and +4) than their cultivation temperature (Fig. 1F,G; P < 0.01; compared TTX indices), and starved aho-3(nj15) mutants conditioned at 23 °C also migrated to the high-temperature regions (+3 and +4) (Fig. 1I,J; P < 0.01; compared TTX indices). These results indicate that starved aho-3(nj15) mutants show abnormal thermophilic phenotype after 20 or 23 °C cultivation; they migrate to the high-temperature regions on thermal gradient ranging from 17 to 23 °C.
There are two possible explanations for the defect of aho-3(nj15) mutants: starved aho-3(nj15) mutants always seek out 23 °C regardless of their cultivation temperature or just higher temperatures. To examine these possibilities, we used a linear thermal gradient ranging from 20 to 26 °C with 23 °C at the center (Fig. S1A–I in Supporting Information). The starved wild-type animals conditioned at 17 or 23 °C dispersed (Fig. S1B,H in Supporting Information), whereas the starved wild-type animals conditioned at 20 °C migrated to lower-temperature region (Fig. S1E in Supporting Information). aho-3(nj15) mutants also showed abnormality in thermotactic plasticity on this thermal gradient. Few starved aho-3(nj15) mutants conditioned at 17 or 23 °C migrated to lower-temperature region (Fig. S1B,H in Supporting Information), and starved aho-3(nj15) mutants conditioned at 20 °C showed the weaker cryophilic phenotype than wild-type animals (Fig. S1E in Supporting Information). The TTX indices of starved aho-3 mutants were significantly higher than those of starved wild-type animals (Fig. S1C,F,I in Supporting Information). These results suggest that aho-3(nj15) mutants do not always seek out 23 °C but rather show a tendency to accumulate higher temperature than wild-type animals after starvation.
To investigate whether aho-3(nj15) mutants have abnormalities in other behavioral modification, we performed a salt chemotaxis learning assay (Saeki et al. 2001; Tomioka et al. 2006) and an integration test for two opposite chemosensory stimuli, repellent Cu2+ ion and attractant diacetyl (Ishihara et al. 2002). aho-3(nj15) mutants showed defects in both the salt learning behavior and integration behavior (Fig. S2A–F in Supporting Information), whereas they show normal chemotaxis to salt and diacetyl and avoidance of Cu2+ ion (Mohri et al. 2005) (Fig. S2G–H in Supporting Information). Altogether, these results suggest that aho-3(nj15) mutants have abnormalities not only in thermotactic plasticity but also in multiple complex behaviors.
The aho-3 gene encodes a novel and highly conserved hydrolase
To identify the gene responsible for the abnormal thermotactic plasticity of aho-3(nj15) mutants, we performed genetic mapping with the snip-single-nucleotide polymorphisms (SNPs) method (Wicks et al. 2001), and rescue experiments with cosmids or PCR fragments. The aho-3(nj15) mutation was mapped to a 50-kbp region on the chromosome I (Fig. 2A). We found that the abnormal thermotactic plasticity of aho-3(nj15) mutants was rescued by the introduction of the K04G2 cosmid and PCR fragments containing the K04G2.2 gene region (Fig. 2A and Figs S3A–D and S4A–D in Supporting Information). Through DNA sequencing, we identified a C-to-T substitution in the K04G2.2 gene of aho-3(nj15) animals, causing a non-sense mutation (Fig. 2A). The mutation truncated in K04G2.2 product lacking catalytic domain and thus presumably causes in a strong loss of K04G2.2 function. These results suggest that K04G2.2 is the gene responsible for aho-3(nj15) mutants.
Figure 2.
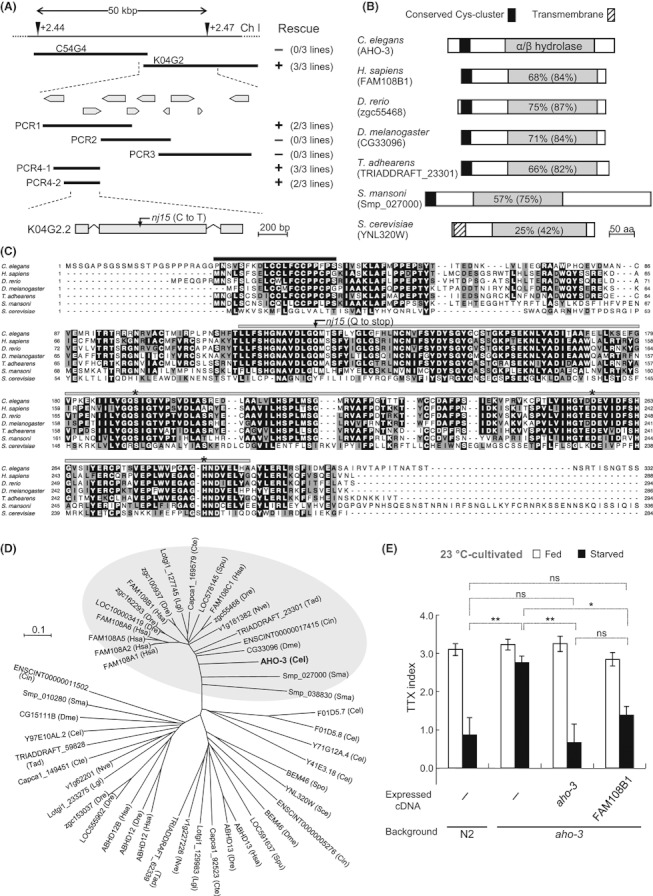
The aho-3 gene encodes a novel protein, which highly conserved among animal species. (A) Position of the aho-3 gene on chromosome I. Arrowheads show the locations of single-nucleotide polymorphisms (SNPs). Results of rescue experiments for abnormal thermotactic plasticity of aho-3(nj15) mutants are indicated as + (rescued) or − (not rescued) on the right side; the rescue experiments were performed with individual thermotaxis assay. Numbers in parentheses indicate the fraction of rescued lines. The gene structure for K04G2.2 is depicted (bottom). The nj15 mutation is shown. (B) Predicted structure of AHO-3 homologues and similar protein. Percentages indicate amino acid identity and similarity (given in parentheses) in the alpha/beta-hydrolase domain between Caenorhabditis elegans AHO-3 and other proteins. (C) Alignment of AHO-3 homologues and similar protein; these proteins are the same as (B). Black bar represents cysteine cluster motif, and gray bar represents alpha/beta-hydrolase domain. Asterisks represent predicted catalytic residues. The nj15 mutation results in Q to STOP at position 127. The C-terminal sequence of Schistosoma mansoni protein is omitted here. (D) Unrooted dendrogram of AHO-3 homologues and similar proteins in 11 animals and two yeasts. Gray ellipse shows AHO-3 homologue group, in which proteins share >70% amino acid sequence similarity with C. elegans AHO-3 in the alpha/beta-hydrolase domains and have the N-terminal cysteine cluster. Cel, Caenorhabditis elegans; Hsa, Homo sapiens; Dre, Danio rerio; Cin, Ciona intestinalis; Spu, Strongylocentrotus purpuratus; Dme, Drosophila melanogaster; Cte, Capitella teleta; Lgi, Lottia gigantea; Sma, Schistosoma mansoni; Nve, Nematostella vectensis; Tad, Trichoplax adhaerens; Sce, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe. (E) Rescue experiment for the abnormality of aho-3(nj15) mutants with human AHO-3 homologue. Thermotaxis of control animals and transgenic aho-3(nj15) mutants expressing C. elegans AHO-3 or human FAM108B1 pan-neuronally. Test animals were cultivated at 23 °C. n ≥ 4 assays. Error bars represent SEM. Tukey’s test was used for multiple comparisons among TTX indices of starved animals; *P < 0.05; **P < 0.01; ns, not significant (P > 0.05).
We also conducted rescue experiments for the salt learning behavior and integration behavior. The defect of aho-3(nj15) mutants in salt chemotactic plasticity was rescued by the PCR fragment containing the K04G2.2 gene (Fig. S2B in Supporting Information), suggesting that K04G2.2 is the gene responsible for the abnormal salt learning behavior of aho-3(nj15) mutants. However, the abnormal integration behavior was not rescued by the PCR fragment (Fig. S2D in Supporting Information). Expression of K04G2.2 cDNA under the control of the pan-neuronal promoter seemed subtly rescued the defects, although it was not significant (Fig. S2D in Supporting Information). These results imply that K04G2.2 is not a gene responsible for the abnormal integration behavior of aho-3(nj15) mutants. However, it is possible that rescue of this defect by K04G2.2 requires strict dose dependency. We designated K04G2.2 as aho-3.
The aho-3 gene encodes a novel protein of 332 amino acid residues that possesses an alpha/beta-hydrolase domain at its C-terminus (Fig. 2B,C). blast searches showed that AHO-3 protein is highly conserved throughout animal species; AHO-3 protein shares 47%–66% amino acid sequence identity and 61%–80% amino acid sequence similarity with FAM108 proteins in Homo sapiens (Chordata), zgc55468 in Danio rerio (Chordata), CG33096 in Drosophila melanogaster (Arthropoda), TRIADDRAFT_23301 in Trichoplax adhaerens (Placozoa), Smp_027000 in Schistosoma mansoni (Platyhelminthes), etc. (calculated utilizing the blastp 2.2.24+ algorithm; Fig. 2B–D and Figs S5 and S6 and Table S1 in Supporting Information). Functions of these AHO-3 homologues in vivo have remained completely unknown, although a few molecular properties of the AHO-3 homologues have been investigated (Blankman et al. 2007; Kang et al. 2008; Martin & Cravatt 2009; Bachovchin et al. 2010; Marrs et al. 2010). Martin and Cravatt reported that the N-terminal cysteine cluster of the human FAM108 proteins is modified with a 16-carbon fatty acid palmitate (Martin & Cravatt 2009). The N-terminal cysteine cluster is conserved in C. elegans AHO-3 and the predicted homologues of AHO-3 in other animal species (Fig. 2B–D and Figs S5 and S6 in Supporting Information). Although we also found similar proteins to AHO-3 in nonanimal species, Arabidopsis thaliana (Streptophyta), Saccharomyces cerevisiae (Ascomycota), Cyanidioschyzon merolae (Rhodophyta), Dictyostelium discoideum (Amoebozoa), etc., the palmitoylation motif is not conserved in those proteins (Fig. 2B–D and Figs S5 and S6 in Supporting Information).
To examine whether AHO-3 is conserved functionally among animal species, we generated transgenic animals that express C. elegans AHO-3 cDNA or human FAM108B1 cDNA in most of the neurons of aho-3(nj15) mutants, and evaluated thermotactic plasticity of those transgenic animals. Both C. elegans AHO-3 expression and human FAM108B1 expression fully rescued the abnormality of aho-3(nj15) mutants (Fig. 2E), suggesting that the molecular function of AHO-3 protein is conserved between nematode and human.
AHO-3 functions in thermotactic plasticity in sensory neurons including AWC
In order to identify the cells in which AHO-3 functions, we analyzed the expression pattern of aho-3. We introduced into wild-type animals fluorescent reporter genes driven by aho-3 promoter. Fluorescence was observed in several tissues (Fig. S7A,B in Supporting Information) and a subset of sensory and interneurons, including AFD (weak fluorescence), AWC (weak) and AIY (strong) neurons that are required for thermotaxis (Mori & Ohshima 1995; Biron et al. 2008; Kuhara et al. 2008), AWB neuron (weak) mediating the avoidance of repellent odors (Troemel et al. 1997) and serotonergic neurons HSN, ADF (strong) and NSM (weak) (Rand & Nonet 1997) (Fig. 3A,B and Fig. S7A–R in Supporting Information).
Figure 3.
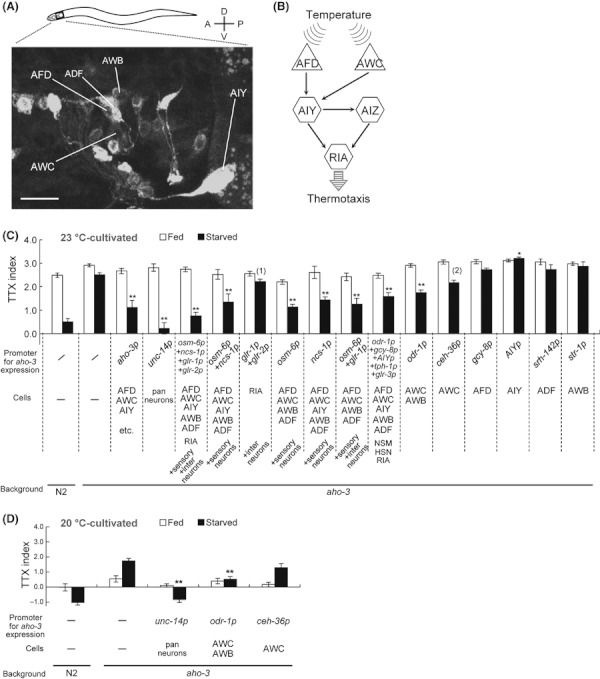
Cell-specific rescue experiments for the abnormal thermotactic plasticity of aho-3 mutants. (A) The expression of aho-3p::cytochrome b5::yfp in the head of an adult wild-type animal. The general ER marker, cytochrome b5::yfp (Rolls et al. 2002), was used to show the expression in cell bodies clearer. Anterior is to the left and dorsal is up. Bars represent 10 μm. A confocal projection (z-stack = 6.4 μm) including only a part of ADF cell body is shown because the strong fluorescence in ADF in other section masks the fluorescence in the AFD cell body. (B) The simplest neural circuit model for thermotaxis (Mori & Ohshima 1995; Biron et al. 2008; Kuhara et al. 2008). AFD and AWC thermosensory neurons (triangle) and downstream AIY, AIZ and RIA interneurons (hexagons) are shown. Arrows represent synaptic connections (White et al. 1986). (C, D) Cell-specific rescue experiments for the abnormal thermotactic plasticity of aho-3(nj15) mutants. Thermotaxis of well-fed or starved animals cultivated at 23 °C (C) or at 20 °C (D). n ≥ 3 assays. Error bars represent SEM. Asterisks represent the comparison of starved transgenic animals with starved aho-3 mutants by Dunnett test; *P < 0.05; **P < 0.01. (1) and (2) represent the comparison of starved transgenic animals with starved aho-3 mutants by unpaired t-test; (1), P = 0.059; (2), P = 0.030.
We conducted cell-specific rescue experiments for thermotactic plasticity of aho-3(nj15) mutants cultivated at 23 °C with a linear thermal gradient from 17 to 23 °C. Expression of aho-3 cDNA under the control of its own promoter or pan-neuronal promoter completely rescued the abnormal thermotactic plasticity of aho-3(nj15) mutants (Fig. 3C). Similarly, AHO-3 expression in ∼40 pairs of sensory and interneurons also completely rescued the abnormality (Fig. 3C and Table S2 in Supporting Information). The abnormality was partially rescued by expressing AHO-3 in ∼20 pairs of sensory neurons, but not rescued by expressing in ∼20 pairs of interneurons (Fig. 3C). AHO-3 expression only in AWC and AWB sensory neurons driven by odr-1 promoter partially rescued the abnormality, but expression in AFD, AIY, ADF or AWB neurons did not (Fig. 3C). Furthermore, the rescue efficiency of the transgenic animals expressing AHO-3 driven by odr-1 promoter was not increased by the additional expression of AHO-3 in AFD, AIY and ADF neurons (Fig. 3C). The AHO-3 expression under the control of the ceh-36prom3, an AWC-specific promoter, also slightly rescued the abnormality of aho-3(nj15) (Fig. 3C). Altogether, these results suggest that AHO-3 functions in a subset of sensory neurons including AWC for thermotactic plasticity.
We also conducted cell-specific rescue experiments for thermotactic plasticity of aho-3(nj15) mutants cultivated at 20 °C using the pan-neuronal promoter, odr-1 promoter and ceh-36prom3 promoter (Fig. 3D). The abnormal thermotactic plasticity was rescued by the expression of AHO-3 under the pan-neuronal promoter (Fig. 3D and Fig. S8A in Supporting Information). The abnormality was partially rescued by the expression of AHO-3 in AWC and AWB under odr-1 promoter (Fig. 3D and Fig. S8B in Supporting Information), but not significantly rescued by the expression only in AWC under ceh-36prom3 promoter (Fig. 3D and Fig. S8C in Supporting Information). Although the site of AHO-3 function might be different depending on the cultivation temperature, it is possible that AWC-specific expression from the extrachromosome array is not stable, resulting in these different results.
We examined whether over-expression of AHO-3 in AWC can affect the thermotactic plasticity after conditioning at 23 °C and at 20 °C (Fig. 4A–F and Fig. S9 in Supporting Information). We expressed AHO-3 using two promoters, odr-1 promoter for the expression in AWC and AWB and str-1 promoter for AWB. After conditioned at 23 °C, starved animals carrying odr-1p::aho-3 showed a tendency to migrate toward higher-temperature region (Fig. 4A,C). Starved animals carrying odr-1p::aho-3 conditioned at 20 °C showed a tendency to accumulate to near their cultivation temperature (Fig. 4D,F). The animals carrying str-1p::aho-3, however, showed no significant difference from wild-type animals both after conditioned at 23 °C and at 20 °C (Fig. 4B,C,E,F). These results suggest that the excess AHO-3 in AWC can affect the thermotaxis plasticity both after cultivation at 23 °C and at 20 °C.
Figure 4.
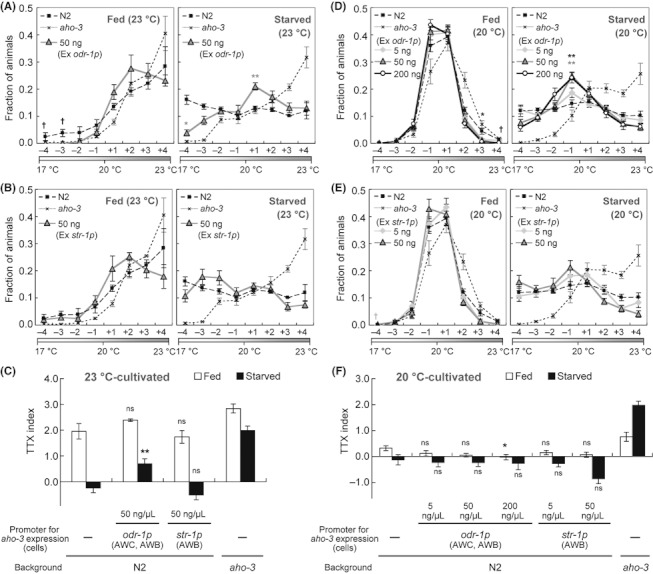
Thermotactic plasticity of animals over-expressing AHO-3. (A–F) Thermotaxis of wild-type animals, transgenic animals and aho-3(nj15) mutants cultivated at 23 °C (A–C) or at 20 °C (D–F) with or without food. The transgenic animals expressing excess AHO-3 in AWC and AWB under the odr-1 promoter (A, D) or only in AWB under the str-1 promoter (B, E) were used. n ≥ 3 assays. Error bars represent SEM. In (A, B and D, E), asterisks represent the comparison of values in individual eight regions by unpaired t-test with the Dunn–Sidak correction for multiple comparison; *P < 0.05; **P < 0.01; colors of asterisks, gray and black, represent the comparisons of N2 animals to each transgenic animals with 50 ng/μL or 200 ng/μL of aho-3cDNA, respectively. Only when all ‘fraction’ values in one dataset were ‘0.00,’ statistical analysis was not performed; in this case, we show a cross with colors, gray and black, representing transgenic animals with 5 ng/μL or 50 ng/μL, respectively. In (C, F), asterisks represent the comparison of transgenic animals with N2 animals by Dunnett test; *P < 0.05; **P < 0.01; ns, not significant (P > 0.05).
To test whether AWC is required for the behavioral plasticity, we examined the thermotactic plasticity of ceh-36(ks86) and ceh-36(ky640) mutants that have a reduction in AWC function because of a mutation in an Otx-type homeobox gene required to specify the AWC cell fate (Lanjuin et al. 2003; Koga & Ohshima 2004). Well-fed ceh-36 mutants showed a tendency to accumulate to the cultivation temperature, and starved ceh-36 mutants showed cryophilic phenotype after cultivation at 20 °C (Fig. 5A,B). These results suggest that the lack of AWC function causes the cryophilic phenotype in thermotactic plasticity. Thus, AWC may induce thermophilic drive or suppress cryophilic drive after starvation. The double-mutant analyses showed that the ceh-36(ky640) mutation at least partially suppressed the abnormal thermophilic phenotype of aho-3(nj15) mutants after starvation (Fig. S10A,B in Supporting Information). Altogether, our results suggest that AHO-3 in AWC plays a role in thermotactic plasticity.
Figure 5.
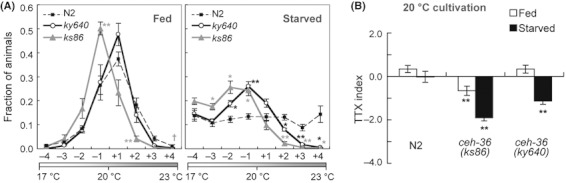
ceh-36 mutants showed the abnormal thermotactic plasticity. (A, B) Thermotaxis of wild-type, ceh-36(ks86) and ceh-36(ky640) mutant animals cultivated at 20 °C with or without food. n ≥ 3 assays. Error bars represent SEM. In (A), asterisks represent the comparison of values in individual eight regions by unpaired t-test with the Dunn–Sidak correction for multiple comparisons; *P < 0.05; **P < 0.01; colors of asterisks, gray and black, represent the comparisons of N2 animals with each mutant, ceh-36(ks86) and ceh-36(ky640), respectively. Only when all ‘fraction’ values in one dataset were ‘0.00,’ statistical analysis was not performed; in this case, we show a cross for ceh-36(ks86). In (B), statistical significance of TTX indices was tested by unpaired t-test in comparisons of N2 animals with mutants; *P < 0.05; **P < 0.01.
aho-3 acts in the same genetic pathway with odr-3 for thermotactic plasticity
Genetic studies have identified several molecular components required for thermotactic plasticity. ins-1 mutants, deficient in the insulin homologue, exhibit a defect in thermotactic plasticity (Kodama et al. 2006). INS-1 antagonizes DAF-2 insulin-like signaling, and this signaling pathway functions in thermotaxis interneurons (Kodama et al. 2006). Similarly, TAX-6 calcineurin was proposed to act downstream of DAF-2 in the thermotaxis interneurons (Kodama et al. 2006; Kuhara & Mori 2006). However, no one has reported genes that function in sensory neurons for thermotactic plasticity after starvation, although there are several other genes whose functional sites were not determined, for example, gcy-28 gene coding receptor-type guanylyl cyclase whose mutations cause a defect in thermotactic plasticity (Tsunozaki et al. 2008).
Our results on the cell-specific rescue, over-expression and double-mutant analyses suggest that AHO-3 acts in sensory neurons including AWC for thermotactic plasticity (Figs 3C,D, 4A–F and 5A,B and Fig. S10A,B in Supporting Information). AWC is capable of sensing distinct stimuli, odor and temperature (Bargmann 2006; Biron et al. 2008; Kuhara et al. 2008). Several molecules have been reported to be important for AWC function. Guanylyl cyclase ODR-1 and G-alpha subunit ODR-3 act in chemotaxis and thermotaxis in AWC (Roayaie et al. 1998; L’Etoile & Bargmann 2000; Kuhara et al. 2008), cGMP-dependent protein kinase EGL-4 plays an important role in olfactory plasticity in AWC (L’Etoile et al. 2002; O’Halloran et al. 2009), and the GCY-28 also functions in AWC for the regulation of odor preferences (Tsunozaki et al. 2008).
To identify candidates that are relevant to AHO-3 function in thermotactic plasticity, we examined the thermotaxis of mutants deficient in AWC function. Fed and starved odr-1(n1933) mutants showed almost normal migration and dispersion after conditioning at both 23 and 17 °C (Fig. 6A,B). In contrast, although odr-3(n1605) mutants showed almost normal thermotactic plasticity after conditioning at 17 °C, they showed abnormal thermotactic plasticity after conditioning at 23 °C; starved odr-3(n1605) mutants conditioned at 23 °C migrated to higher-temperature region, similar to aho-3(nj15) mutants (Fig. 6A,B). egl-4(n479) mutants showed an abnormal thermotactic plasticity after conditioning at both 23 and 17 °C; starved egl-4(n479) mutants conditioned at 23 or at 17 °C migrated to higher- or lower-temperature regions, respectively, which roughly coincide with the cultivation temperature (Fig. 6A,B). These results suggest that the ODR-3 and EGL-4 are necessary for thermotactic plasticity. Abnormality of odr-3(n1605) and egl-4(n479) mutants each was partially rescued by expressing respective cDNA under the control of odr-1 promoter (Fig. S11A,B in Supporting Information), implying that the ODR-3 and EGL-4 function in the thermotactic plasticity either or in both AWC and AWB.
Figure 6.
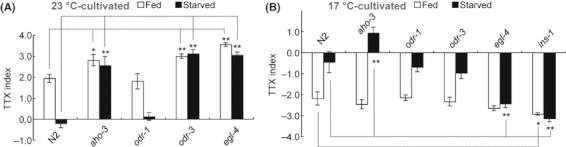
Thermotactic plasticity of mutants that have defects in AWC properties. (A, B) Thermotaxis of well-fed or starved wild-type N2 and mutant animals cultivated at 23 °C (A) or at 17 °C (B). n ≥ 3 assays. Statistical significance of TTX indices was tested by Dunnett test in comparisons of N2 animals with mutants; *P < 0.05; **P < 0.01. In (B), the results of N2 animals and aho-3(nj15) mutants were repeated from Figure 1B–D. ins-1(nr2091) mutants were assayed as control mutants that have defect in thermotactic plasticity after conditioned at 17 °C (Kodama et al. 2006).
To investigate the genetic relationships between aho-3 and other genes such as odr-3, egl-4, gcy-28 and ins-1 that are required for thermotactic plasticity, we constructed double mutants and examined their thermotactic plasticity after cultivation at 20 °C (Fig. 7A–F and Fig. S12A–H in Supporting Information). Similar to aho-3(nj15) mutants, both odr-3(n1605) putative null mutants (Roayaie et al. 1998) and gcy-28(tm2411) strong loss-of-function mutants (Tsunozaki et al. 2008) cultivated under starved condition migrated to higher-temperature region (Fig. 7A and Fig. S12H in Supporting Information). We found that the thermotaxis of starved aho-3;odr-3 double mutants was similar to that of each starved single mutants (Fig. 7C and Fig. S12H in Supporting Information). We also found that starved gcy-28 aho-3 double mutants exhibited slightly enhanced thermophilic phenotype as compared with each single mutants (Fig. 7D and Fig. S12H in Supporting Information), and starved gcy-28;odr-3 double mutants showed similar thermotaxis to each single mutants (Fig. S12A,H in Supporting Information). Behavioral phenotypes of these single and double mutants suggest that aho-3, odr-3 and possibly gcy-28 might act in the same genetic pathway to regulate the thermotactic plasticity.
Figure 7.
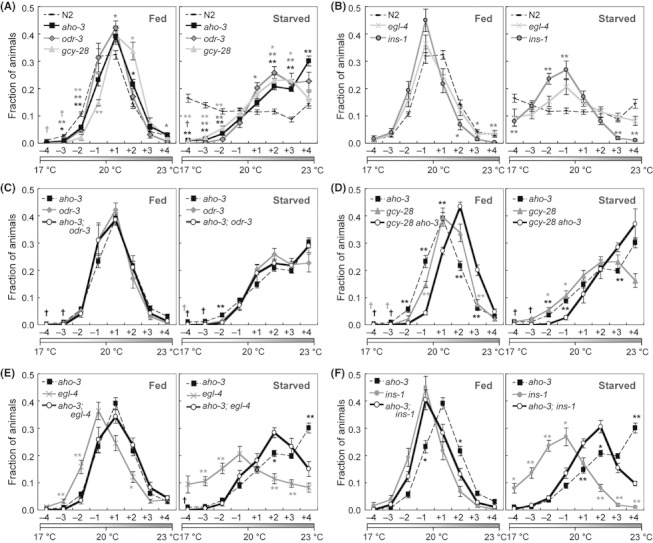
Genetic relationship analysis of aho-3 with genes whose defects cause abnormal thermotactic plasticity. (A, B) Thermotaxis of wild-type N2 and single-mutant animals cultivated at 20 °C with or without food. Phenotypes of aho-3(nj15), odr-3(n1605) and gcy-28(tm2411) are shown in (A), and those of egl-4(n479) and ins-1(nr2091) are shown in (B). (C–F) Thermotaxis of double and single mutants cultivated at 20 °C with or without food. Results of single mutants repeated from (A, B). In all graphs, asterisks represent the comparison of values in individual eight regions by unpaired t-test with the Dunn–Sidak correction for multiple comparisons; *P < 0.05; **P < 0.01; N2 animals vs (A, B) or double mutants vs (C–F) each single mutant. Only when all ‘fraction’ values in one dataset were ‘0.00,’ statistical analysis was not performed; in this case, we show a cross with colors, gray and black, representing single mutants and double mutants, respectively. n ≥ 3 assays. Error bars represent SEM.
Different from aho-3(nj15) mutants, a substantial proportion of starved egl-4(n479) and ins-1(nr2091) putative null mutants (Pierce et al. 2001; Fujiwara et al. 2002) migrated to their cultivation temperature (Fig. 7B). We found that starved aho-3;egl-4 and aho-3;ins-1 double mutants each showed thermotactic phenotype roughly intermediate between that of starved aho-3(nj15) single mutants and of starved egl-4(n479) or ins-1(nr2091) single mutants, respectively (Fig. 7E,F and Fig. S12H in Supporting Information). Similarly, starved double mutants, egl-4;odr-3, ins-1;odr-3, gcy-28;egl-4 and gcy-28;ins-1, each showed thermotactic phenotype roughly intermediate between that of respective single mutants (Fig. S12C–F,H in Supporting Information). These results suggest that aho-3, odr-3 and gcy-28 at least partially act in parallel with egl-4 and ins-1.
We also examined the thermotactic plasticity of three aho-3;odr-3, gcy-28 aho-3 and aho-3;egl-4 double mutants cultivated at 23 °C (Fig. S13A–G in Supporting Information). Phenotype of aho-3;odr-3 mutants was similar to that of each single mutants (Fig. S13C,G in Supporting Information). Correspondingly, gcy-28 aho-3 double mutants showed similar phenotype to gcy-28 single mutants (Fig. S13D,G in Supporting Information). aho-3;egl-4 double mutants, however, exhibited remarkably enhanced thermophilic phenotype in thermotactic plasticity (Fig. S13E,G in Supporting Information). Together with analyses of mutants cultivated at 20 °C, these results suggest that aho-3 functions in the same genetic pathway with odr-3 and gcy-28, whereas aho-3 and egl-4 act in parallel.
It was previously shown that odr-3 encoding G-alpha functions downstream of eat-16 encoding a regulator of G protein signaling protein (RGS) (Kuhara et al. 2008). The loss of EAT-16 causes a hyperactivation of AWC, leading to a cryophilic phenotype (Kuhara et al. 2008). In order to analyze the genetic relationship between aho-3 and odr-3 further, we examined the thermotactic plasticity of aho-3 eat-16 and eat-16;odr-3 double mutants after cultivation at 20 °C (Fig. S14A–E in Supporting Information). Both well-fed and starved eat-16(nj8) nearly null mutants (Kuhara et al. 2008) showed a cryophilic phenotype (Fig. S14A,D,E in Supporting Information). odr-3(n1605) mutation partially suppressed this phenotype (Fig. S14C–E in Supporting Information). Similarly, aho-3(nj15) mutation also showed partial suppression of the eat-16(nj8) phenotype (Fig. S14B,D,E in Supporting Information). Altogether, these results imply that, like odr-3, aho-3 acts downstream of eat-16, although it is also implied that eat-16 at least partially acts in parallel with aho-3 and odr-3.
The predicted catalytic triad and the N-terminal cysteine cluster are essential for AHO-3 function
The AHO-3 protein is highly conserved among animal species and possesses an alpha/beta-hydrolase domain at its C-terminus and a cysteine cluster in N-terminus. Although a few molecular properties of AHO-3 homologues have been previously characterized (Blankman et al. 2007; Kang et al. 2008; Martin & Cravatt 2009; Bachovchin et al. 2010; Marrs et al. 2010), biological functions in vivo have not been well understood. We analyzed the functional significance of domains of the AHO-3 protein for thermotactic plasticity in C. elegans, using recombinant AHO-3 proteins containing respective mutations.
Mouse AHO-3 homologues were identified as metabolic serine hydrolases (Blankman et al. 2007; Bachovchin et al. 2010). The metabolic serine hydrolases include esterases, lipases, peptidases and amidases (Holmquist 2000; Simon & Cravatt 2010). The majority of these enzymes use an alpha/beta-hydrolase fold and use a Ser–His–Asp catalytic triad (Holmquist 2000; Simon & Cravatt 2010). AHO-3 homologues also possess an alpha/beta-hydrolase domain including catalytic triad predicted by sequence comparison method (Fig. 2C and Fig. S5 in Supporting Information; UniProtKB, http://www.uniprot.org/uniprot/Q5VST6; merops database, http://merops.sanger.ac.uk) (Rawlings et al. 2010).
To determine whether predicted catalytic triad of AHO-3, Ser 191, Asp 256 and His 285, is required for thermotactic plasticity, we constructed mutant aho-3 genes containing mutations that disrupt the probable Ser–His–Asp catalytic triad and expressed it pan-neuronally in aho-3(nj15) mutants (Fig. S15A–E in Supporting Information). The abnormal thermotactic plasticity of aho-3(nj15) mutants was rescued by expressing wild-type AHO-3, but not by mutant AHO-3 proteins (S191A, D256N and/or H285A) (Fig. 8A). These results suggest that the predicted catalytic residues are essential for AHO-3 function in thermotactic plasticity and that AHO-3 acts as an enzyme in this behavioral modification.
Figure 8.
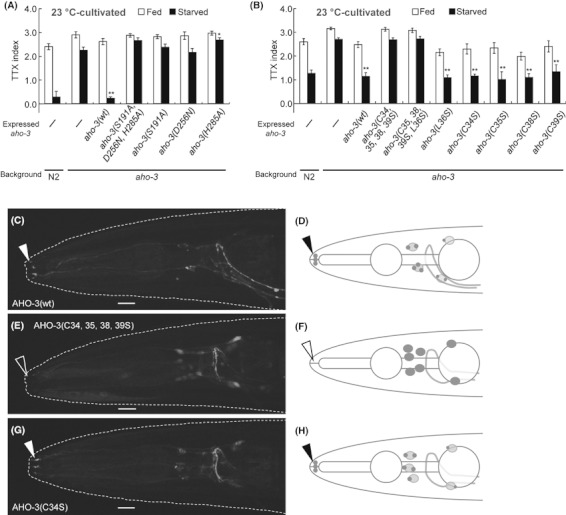
Predicted catalytic residues and the N-terminal cysteine cluster in AHO-3 are essential for thermotactic plasticity. (A, B) Rescue experiment for the abnormal thermotactic plasticity of aho-3(nj15) mutants with recombinant AHO-3::EGFP containing a mutation in the predicted catalytic residues (A) or into the N-terminal cysteines (B). Recombinant proteins were expressed pan-neuronally. Thermotaxis of well-fed or starved animals cultivated at 23 °C. n ≥ 3 assays. Error bars represent SEM. Asterisks represent the comparison of starved transgenic animals with starved aho-3 mutants by Dunnett test; *P < 0.05; **P < 0.01.(C–H) Subcellular localization of AHO-3(wild-type)::EGFP (C), AHO-3(C34, 35, 38, 39S)::EGFP (E) and AHO-3(C34S)::EGFP (G) expressed under the control of the aho-3 promoter in adult aho-3(nj15) mutants. Solid arrowheads point to the localization of AHO-3(wild-type)::EGFP and AHO-3(C34S)::EGFP to sensory endings (C and G). AHO-3(C34, 35, 38, 39S)::EGFP was localized diffusely in cell bodies and not to sensory endings (open arrowhead; E). Schematic diagrams of (C), (E) and (G) are shown in (D), (F) and (H), respectively. Images are Z-stack confocal projection. A rough outline of head of animal is shown (C, E and G). Anterior is to the left. Bars represent 10 μm.
Rat AHO-3 homologues were previously identified as a protein modified by a fatty acid palmitate (Kang et al. 2008). Protein palmitoylation is a post-translational modification in which 16-carbon fatty acid palmitate is added to specific cysteine residues (Linder & Deschenes 2007; Fukata & Fukata 2010). Other group discovered that the human AHO-3 homologues are palmitoylated at the N-terminal cysteine cluster (Martin & Cravatt 2009), which is conserved among animal species (Fig. 2C and Fig. S5 in Supporting Information), and that this cluster is necessary for their own localization to plasma membrane (Martin & Cravatt 2009).
We analyzed the subcellular localization of AHO-3 by expressing recombinant AHO-3 proteins fused with fluorescent proteins under the control of the aho-3 promoter. The AHO-3::EGFP expression by the aho-3 promoter rescued the abnormal thermotactic plasticity of aho-3(nj15) mutants (Fig. 3C). These AHO-3::EGFP proteins were observed in sensory endings, cell bodies as punctiform and often the nerve ring (Fig. 8C,D and Fig. S16A–F in Supporting Information). The punctate stainings of the AHO-3 protein in cell bodies were observed very near to the stainings of Golgi markers, mannosidase::YFP and MIG-23::GFP (Rolls et al. 2002; Nishiwaki et al. 2004) (Fig. S16A–L in Supporting Information). We examined whether the subcellular localization is changed depending on the feeding states, but there was no apparent difference between fed and starved animals under the 23 °C cultivation (Table S3 in Supporting Information).
To analyze whether the N-terminal cysteine cluster of AHO-3 is required for its localization, we constructed translational AHO-3::EGFP containing mutations in the N-terminal cysteines (Cys 34, 35, 38 and 39) and generated transgenic animals. The recombinant AHO-3(C34, 35, 38 and 39S)::EGFP was localized diffusely in cell bodies and not to sensory endings (Fig. 8E,F and Table 1). Another mutant AHO-3(C34S)::EGFP containing a mutation in only one cysteine was localized to sensory endings and cell bodies in punctiform, although it seemed to be moderately diffused (Fig. 8G–H and Table 1). These results suggest that the N-terminal cysteine cluster of AHO-3 is necessary for its subcellular localization and that a certain number of cysteines may be essential for its localization to sensory endings.
Table 1.
Subcellular localization analysis of recombinant AHO-3
| Subcellular location | Recombinant AHO-3 | Category AHO-3 fluorescence | Total n | ||
|---|---|---|---|---|---|
| Strong | Weak | Invisible | |||
| Sensory endings | Wild type | 60 | 17 | 3 | 80 |
| C34S | 65 | 17 | 2 | 84 | |
| C34, 35, 38, 39S** | 18 | 24 | 42 | 84 | |
| Punctuations in cell bodies | Wild type | 62 | 14 | 4 | 80 |
| C34S** | 2 | 37 | 45 | 84 | |
| C34, 35, 38, 39S** | 0 | 0 | 84 | 84 | |
| Diffusion in cell bodies | Wild type | 4 | 56 | 20 | 80 |
| C34S** | 42 | 35 | 7 | 84 | |
| C34, 35, 38, 39S** | 61 | 23 | 0 | 84 | |
P < 0.01.
Localization of AHO-3 was evaluated in adults carrying each types of aho-3p::aho-3cDNA::egfp. Animals were cultivated at 20 °C with food. We categorized fluorescence intensity in sensory endings and cell bodies into strong, weak and invisible. Statistical analysis by a chi-square test using a 2 × 3 contingency table was performed to compare the AHO-3(wild type) with each recombinant AHO-3.
We also examined the requirement of the N-terminal cysteine cluster of AHO-3 for thermotactic plasticity. The abnormal thermotactic plasticity of aho-3(nj15) mutants was not rescued by pan-neuronal expression of the recombinant AHO-3(C34, 35, 38 and 39S)::EGFP or the recombinant AHO-3(C35, 38 and 39S, L36S)::EGFP keeping only one cysteine, whereas the abnormality was fully rescued by expressing the recombinant AHO-3(C34, 35, 38 or 39S)::EGFP keeping three cysteines (Fig. 8B and Fig. S17A–F in Supporting Information). These results suggest that the N-terminal cysteine cluster of AHO-3 is necessary and a certain number of cysteines in the cluster are essential for thermotactic plasticity. Altogether, these results suggest that the proper subcellular localization of AHO-3 to sensory endings is crucial for thermotactic plasticity.
Discussion
We here reported that the highly conserved novel hydrolase AHO-3 is required for C. elegans behavioral plasticities. Rescue experiments suggest that the molecular function of the AHO-3 protein is conserved between nematode and human. We propose that the AHO-3 protein plays conserved important roles in neural function across animal species.
The function of AHO-3 in thermotactic plasticity in Caenorhabditis elegans
It was previously shown that C. elegans exhibits thermotactic plasticity; well-fed animals migrate to their cultivation temperature, and starved animals avoid the cultivation temperature on the nonlinear thermal gradient (Mohri et al. 2005). Our study showed that starved animals disperse on the linear thermal gradient except for the 20 °C-cultivated starved animals assayed in the thermal gradient ranging from 20 to 26 °C: these animals migrated to the colder region. These different results can be caused by the differences in the steepness of the thermal gradients or starting-point temperature, because variations in these factors affect the thermotaxis of well-fed animals (Ramot et al. 2008; Nakazato & Mochizuki 2009; Jurado et al. 2010; Beverly et al. 2011). In all of these assays, however, animals modify their behavior after starvation. We call all forms of these plasticities ‘thermotactic plasticity’ in this article.
aho-3 mutants exhibit abnormality in thermotactic plasticity. Like wild-type animals, well-fed aho-3 mutants migrated to the cultivation temperature on a thermal gradient (Fig. 1B,E,H and Fig. S1A,D,G in Supporting Information). Whereas starved wild-type animals showed dispersed distribution or migration to colder region, starved aho-3 mutants exhibited tendency to migrate toward higher temperature than the starved wild-type animals (Fig. 1C,F,I and Fig. S1B,E,H in Supporting Information). The simplest model for thermotaxis in C. elegans is that the behavior is a result of balanced regulation of two opposing thermophilic and cryophilic drives (Hedgecock & Russell 1975). Based on this model, we could assume that starvation conditioning changes the strength of these opposing drives. Considering the behavioral abnormality of aho-3 mutants, it is likely that AHO-3 protein suppresses the thermophilic drive or promotes the cryophilic drive in response to the starvation signal.
Our cell-specific rescue experiments showed that AHO-3 functions in many sensory neurons including the AWC thermosensory neuron for thermotactic plasticity (Fig. 3C,D). The AHO-3 expression in both AWC and AWB neurons partially rescued the abnormality of aho-3 mutants after cultivation at 20 and 23 °C (Fig. 3C,D). The expression in only AWC partially rescued the abnormality after cultivation at 23 but not at 20 °C (Fig. 3C,D). The over-expression analysis, however, showed that the excess AHO-3 in AWC but not in AWB caused an abnormal thermotactic plasticity both after cultivation at 20 °C and at 23 °C (Fig. 4A–F). In addition, the reduction in AWC function partially suppressed the defect of aho-3 mutants after cultivation at 20 °C (Fig. S10A,B in Supporting Information). Altogether, our results suggest that AHO-3 acts in AWC for thermotactic plasticity, although the AHO-3 activity in other neurons is also important.
It was reported that AWC regulates the activity of downstream AIY interneuron that promotes thermophilic drive (Kuhara et al. 2008; Ohnishi et al. 2011). Our study suggested that AWC is important for thermotactic plasticity (Fig. 5A,B), but the calcium-imaging analysis did not detect any significant difference in the responses of AWC or AIY of fed and starved animals (Fig. S18A–G in Supporting Information). The changes in AWC or AIY activity after starvation may be too subtle to detect.
Double-mutant analyses suggested that aho-3 acts in the same genetic pathway with odr-3 encoding G protein alpha subunit (Fig. 7C and Figs S12H, S13C and S13G in Supporting Information). ODR-3 is required for thermosensation in AWC and is localized to sensory endings (Roayaie et al. 1998; Bargmann 2006; Kuhara et al. 2008). Our results showed that AHO-3 localized to sensory endings like ODR-3 (Fig. 8C). These results imply that AHO-3 acts in the ODR-3-mediated G protein signaling pathway to inhibit the thermophilic drive after starvation in AWC.
Our over-expression experiments, however, showed that the excess AHO-3 in AWC did not simply induce cryophilic phenotype (Fig. 4A,C,D,F). Because AWC transmits both excitatory and inhibitory signals to AIY (Kuhara et al. 2008; Ohnishi et al. 2011), it is possible that AHO-3 modifies these AWC transmissions in complex manner, regulating thermotactic plasticity after starvation.
Molecular function of the highly conserved AHO-3 protein across animal species
The novel hydrolase AHO-3 is highly conserved from flat animals to human (47%–66% amino acid sequence identity and 61%–80% similarity; Fig. 2B–D, Figs S5 and S6 in Supporting Information). Previous studies showed that mammalian AHO-3 homologues are expressed in the brain (Bachovchin et al. 2010) (EMBL-EBI, http://www.ebi.ac.uk/gxa/). Rescue experiment with human homologue of AHO-3 suggests that molecular properties of AHO-3 are conserved between nematode and human (Fig. 2E). Although in vivo functions of AHO-3 homologues have remained unknown, a few molecular properties have been evaluated (Blankman et al. 2007; Kang et al. 2008; Martin & Cravatt 2009; Bachovchin et al. 2010; Marrs et al. 2010).
The activity-based profiling showed that mouse AHO-3 homologues belong to the metabolic serine hydrolase superfamily (Blankman et al. 2007; Bachovchin et al. 2010), whose members mostly possess an alpha/beta-hydrolase domain including a catalytic triad (Holmquist 2000; Simon & Cravatt 2010). All AHO-3 homologues also have an alpha/beta-hydrolase domain including predicted catalytic triad (Fig. 2C, Fig. S5 in Supporting Information), which is required for AHO-3 function in thermotactic plasticity (Fig. 8A). It is yet unknown what kind of substrates AHO-3 homologues react to. So far, only one of the AHO-3 similar proteins, ABHD12 (see Fig. 2D), was suggested to hydrolyze 2-arachidonoylglycerol, an endogenous ligand for cannabinoid receptors, in mouse brain (Blankman et al. 2007; Marrs et al. 2010). It is possible that AHO-3 proteins play a role in the degradation of small neural molecules, for example some sorts of ligand or intracellular messenger.
Human AHO-3 homologues were identified as a protein modified by a fatty acid palmitate (Martin & Cravatt 2009). Palmitoylation is a unique lipid-based post-translational modification in that it is reversible (Linder & Deschenes 2007; Fukata & Fukata 2010). The reversible nature allows palmitoylation to regulate diverse aspects of neuronal protein trafficking, localization and functions related to neurite outgrowth and neural plasticity (Linder & Deschenes 2007; Fukata & Fukata 2010). N-terminal cysteine cluster, the putative palmitoylation motif in human AHO-3 homologues (Martin & Cravatt 2009), is conserved in all AHO-3 homologues (Fig. 2B,C and Fig. S5 in Supporting Information). We showed here that the putative palmitoylation motif of C. elegans AHO-3 is necessary for its localization to sensory endings and for thermotactic plasticity (Fig. 8B–H). Given the property of palmitoylation, our results imply that palmitoylation may regulate the AHO-3 localization that is necessary for its function and consequently modify multiple behaviors such as thermotactic plasticity. Further molecular, biochemical and behavioral analyses on AHO-3 using C. elegans and other model animals should predict its functions in the nervous system and general roles in behavioral modifications through animal species.
Experimental procedures
Strains and maintenance
Caenorhabditis elegans strains were maintained with E. coli OP-50 and handled according to standard procedures (Brenner 1974). We used the following strains: wild-type Bristol strain (N2), wild-type Hawaiian strain (CB4856) for mapping with the snip-SNPs method, IK850 aho-3(nj15) I backcrossed 10 times with N2 (six times by phenotype of abnormal thermotactic plasticity and four times by nj15 genotype), IK849 aho-3(nj15) I backcrossed six times with N2 only for mapping in Figure 2A and Figure S4 in Supporting Information and for behavioral tests in Figure S2E–H in Supporting Information, JC2154 hen-1(tm501) X, MT3644 odr-3(n1605) V, IK852 gcy-28(tm2411) I, IK571 egl-4(n479) IV, IK607 ins-1(nr2091) IV, CX2349 odr-1(n1933) X, ceh-36(ks86) X, ceh-36(ky640) X, eat-16(nj8) I, aho-3(nj15) I; odr-3(n1605) V, gcy-28(tm2411) aho-3(nj15) I, gcy-28(tm2411) I; odr-3(n1605) V, aho-3(nj15) I; egl-4(n479) IV, egl-4(n479) IV; odr-3(n1605) V, gcy-28(tm2411) I; egl-4(n479) IV, aho-3(nj15) I; ins-1(nr2091) IV, ins-1(nr2091) IV; odr-3(n1605) V, gcy-28(tm2411) I; ins-1(nr2091) IV, egl-4 (n479) ins-1 (nr2091) IV, aho-3(nj15) eat-16(nj8) I, eat-16(nj8) I; odr-3(n1605) V, aho-3(nj15) I; ceh-36(ky640) X and transgenic strains derived from them. All transgenic strains were made essentially as described (Mello et al. 1991), with the co-injection marker ges-1p::NLS-GFP (pKDK66) for the rescue experiment strains and over-expression experiment strains, rol-6gf (pRF4) or ges-1p::NLS-TagRFP (pNAS88) for the subcellular localization test strains, and rol-6gf (pRF4; for Fig. S7B,O–Q in Supporting Information), ges-1p::NLS-GFP (pKDK66; for Fig. S7R in Supporting Information) or none (for Fig. 3A, Fig. S7A,C–N in Supporting Information) for the expression pattern test strains. Test plasmids were injected at 2–50 ng/μL. In Figure 3C,D, rescue experiment strains carried aho-3 cDNA fused with each cell-specific promoter, except for aho-3p and ceh-36p fused with aho-3 cDNA::egfp. In Figure S11A in Supporting Information, odr-3(n1605); Ex[odr-1p::odr-3 cDNA] was made by outcrossing with N2; Ex[odr-1p::odr-3 cDNA] (Kuhara et al. 2008). At least two independent lines were tested for each rescue and over-expression experiment, except for unc-14p::aho-3 cDNA and odr-1p::aho-3 cDNA in Figure 3D and Figure S8A–C and for genomic PCR fragment in Figure S2B,D and S3A–D in Supporting Information. We tested the same one line in Figure S2B,D and S3A–D in Supporting Information.
Behavioral analysis
The population thermotaxis assay
The population thermotaxis assay was performed as previously reported (Ito et al. 2006) with some modifications according to Mohri et al. (Mohri et al. 2005) for evaluating thermotactic plasticity associated with feeding states. Equipment for establishing the linear thermal gradient was used as described (Hedgecock & Russell 1975; Ito et al. 2006). A stable, linear thermal gradient was established on a 60-cm-long aluminum platform by using two water baths at 5 and 35 °C. TTX (thermotaxis) plate (13.5 cm × 6 cm, 1.8 cm height) containing 10 mL of TTX medium (2% agar, 0.3% NaCl, 25 mm potassium phosphate, pH 6.0) was placed on the platform. The extra space between the bottom of the TTX plate and the platform was filled with water to increase the thermal conductivity as much as possible. Except for Figure S1 in Supporting Information, the center of the 13.5-cm-long agar surface in TTX plate was adjusted at 20 °C, and linear thermal gradient ranging from 17 to 23 °C was established on the agar surface. In Figure S1 in Supporting Information, the center of TTX plate was adjusted at 23 °C, and linear thermal gradient from 20 to 26 °C was established.
We cultivated animals under uncrowded and well-fed condition at respective temperatures, 17, 20 or 23 °C, on a 6-cm plate containing 14 mL of nematode growth medium (NGM) with 2% agar, on which E. coli OP-50 was seeded. Except for Figures 1B–D and 6B, well-fed naive animals were collected and washed twice with NG buffer (0.3% NaCl, 1 mm CaCl2, 1 mm MgSO4, 25 mm potassium phosphate, pH 6.0) in population thermotaxis assay with well-fed animals. This NG buffer was pre-incubated and kept at 20 °C (except for Fig. S1 in Supporting Information) or at 23 °C (only for Fig. S1 in Supporting Information). Approximately 40–300 animals were placed at the center of the TTX plate, and excess water was removed with tissue paper immediately. The TTX plates were left undisturbed and animals were allowed to move freely for 60 min. After animals were killed by chloroform gas or immobilized with ice, the adult animals in each of the eight regions were counted. The TTX index, defined as shown in Figure 1A, was calculated. In population thermotaxis assay with starved animals, the well-fed naive animals were collected and washed twice with NG buffer and placed on starvation-conditioning plate (2% agar, 1 mm CaCl2, 1 mm MgSO4, 25 mm potassium phosphate, pH 6.0) without food; those NG buffer and starvation-conditioning plates were pre-incubated and kept at each cultivation temperature. Excess water was removed immediately after placement. After cultivation without food for designated time (see below), animals were collected with NG buffer kept at 20 °C (except for Fig. S1 in Supporting Information) or at 23 °C (only for Fig. S1 in Supporting Information), and approximately 40–300 animals were placed at the center of the TTX plate. The subsequent procedure was the same as described for the well-fed animals. In Figures 1B–D and 6B, we performed population thermotaxis assay almost similar to described above, except that animals were picked instead of washed when they were transferred from a plate onto other plate (e.g., from NGM plate onto starvation-conditioning plate) and that 18–55 animals were placed on the TTX plate. Starvation-conditioning plates contained a high-osmolarity ring of 8 m glycerol at the periphery in Figures 1B–D and 6B to prevent the animals from swimming off the agar; the 8 m glycerol ring does not affect thermotaxis (Mohri et al. 2005). In this study, we designated starvation time 3 h at 17 °C, 2 h at 20 °C and 1 h at 23 °C, respectively, because it was reported that behavioral change induced by starvation sufficiently occurs after cultivation at 17 °C for 3 h or after cultivation at 25 °C for 1 h (Mohri et al. 2005).
Salt chemotaxis learning assay
We performed salt chemotaxis learning assay according to previous reports (Tomioka et al. 2006) with some modifications. Test animals were cultivated under well-fed condition at 20 °C. We used ‘liquid conditioning method’ and assayed on 6-cm test plates with a salt gradient made by an agar plug containing 50 mm NaCl. The calculation of Chemotaxis index was modified as shown in Figure S2A in Supporting Information. Detailed methods are described in Supplementary materials.
Interaction assay, Chemotaxis assay and Avoidance assay
The procedure for interaction assay between chemotaxis to odorants and avoidance of Cu2+ ion was according to the previous report (Ishihara et al. 2002). 1/100 diacetyl and 100 mm Cu2+ ion were used. Test animals were cultivated under well-fed condition at 17, 20 or 25 °C. The procedures for assaying chemotaxis to volatile odorant and assaying avoidance from Cu2+ ion were according to Bargmann et al. (Bargmann et al. 1993) and Wicks et al. (Wicks et al. 2000), respectively. Test animals were cultivated under well-fed condition at 25 °C.
Molecular biology
aho-3 cDNA was amplified from yk1293d3 and cloned into pPD49.26 to generate pNAS1 using the Acc65I and EcoRI sites. aho-3 cDNA from pNAS1 was inserted into each promoter construct to generate specific promoter::aho-3 cDNA plasmids. Specific promoters are unc-14p for pan-neuronal (Ogura et al. 1997), osm-6p for ∼20 pairs of sensory neurons (Collet et al. 1998; Kodama et al. 2006), ncs-1p for AIY and ∼10 pairs of sensory neurons (Gomez et al. 2001), glr-1p for ∼15 pairs of interneurons (Hart et al. 1995; Maricq et al. 1995), glr-2p for ∼10 pairs of interneurons (Brockie et al. 2001), odr-1p for AWC and AWB (L’Etoile & Bargmann 2000), ceh-36prom3 for AWC (Etchberger et al. 2007), gcy-8p for AFD (Inada et al. 2006), ttx-3p for AIY (Kodama et al. 2006; Kuhara & Mori 2006), tph-1p for ADF, HSN and NSM (Sze et al. 2000), srh-142p for ADF (Sagasti et al. 1999; Chang & Bargmann 2008), glr-3p for RIA (Brockie et al. 2001) and str-1p for AWB (Troemel et al. 1997) (Table S2 in Supporting Information). The aho-3 genomic sequence including 4 kb of the promoter region was amplified from the N2 genome by PCR and cloned into pBluescript II SK+ to generate pFUG2. The 4-kb promoter region of aho-3 was amplified from pFUG2 and cloned into pPD95.75 or into pNAS1 to generate aho-3p::gfp (pNAS14) and aho-3p::aho-3 cDNA (pNAS15) using the BamHI site. We cloned aho-3 cDNA amplified from unc-14p::aho-3cDNA (pNAS3) and EGFP amplified from ttx-7::EGFP (Tanizawa et al. 2006) (generated from pEGFP-N1; Takara Bio) into pNAS3 to generate unc-14p::aho-3 cDNA::EGFP (pNAS42); aho-3 cDNA and EGFP was fused with NsiI site. aho-3p::aho-3 cDNA::EGFP (pNAS50) was generated from pNAS15 and pNAS42 using the Acc65I, EcoRI and PvuI sites. aho-3p::aho-3 cDNA::CFP (pNAS141) and aho-3p::aho-3 cDNA::DsRed-monomer (pNAS140) were constructed from pNAS50; aho-3 cDNA and the marker genes were fused with NsiI sites. FAM108B1 cDNA was amplified from EHS1001-10687 (Open Biosystems, Inc.) and cloned to make unc-14p::FAM108B1 cDNA (pNAS72). We generated recombinant aho-3 cDNAs, used in Figure 8 and Figures S15 and S17 in Supporting Information, by site-directed mutagenesis from pNAS3 or pNAS42 and constructed unc-14p::aho-3 cDNA(recombinant)::EGFP. From those plasmids containing unc-14p fusion recombinant aho-3 cDNAs and from pNAS50, we constructed aho-3p::aho-3 cDNA(recombinant)::EGFP using the Acc65I and NsiI sites. aho-3p::Mans::YFP (pNAS132) was constructed from pNAS50 and glr-3p::Mans::YFP (pUBA23) using the SphI and Acc65I sites. aho-3p::mig-23::GFP (pNAS136) was constructed from pNAS50 and mig-23::GFP using the BamHI sites. aho-3p::cytochrome b5::yfp (pNAS128) and odr-1p::cytochrome b5::yfp (pNAS129) were constructed from pNAS50 or odr-1p::aho-3 cDNA::egfp (pNAS96) and from glr-3p::cytochrome b5::yfp using the SphI and Acc65I sites. odr-1p::cytochrome b5::cfp (pNAS158) was constructed from pNAS129 and from AIYp::Mans::cfp using the XmaI and AatII sites. tph-1p::NLS-tagRFP (pNAS160) was constructed from tph-1p::GFP (pOKU82) and pNAS88 using the SalI and StyI sites. unc-14p::odr-3cDNA (pNAS77) was generated from pNAS3 and odr-3 cDNA. unc-14p::egl-4.a cDNA (pNAS85) and odr-1p::egl-4.a cDNA (pNAS99) were generated from egl-4.a cDNA vector and pNAS3 or odr-1p::aho-3 cDNA (pNAS8), respectively.
Sequence analysis of AHO-3
We used the Pfam program (http://www.sanger.ac.uk/resources/databases/pfam.html) and the sosui program (http://bp.nuap.nagoya-u.ac.jp/sosui/) to analyze the structure of the C. elegans AHO-3 protein, and it was predicted that the AHO-3 possesses an alpha/beta-hydrolase domain and no transmembrane segment. The sosui program was also used to predict the transmembrane segment of S. cerevisiae YNL320W in Figure 2B. Because extents of alpha/beta-hydrolase domains predicted from Pfam program were slightly different from one another in AHO-3 homologues and similar proteins, those extents of domains in Figure 2B are predicted from sequence alignment with the domain sequence in C. elegans AHO-3 protein, which was predicted from Pfam program. To search the AHO-3 homologues and AHO-3 similar proteins in 17 biological species, we use Web blast services provided by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/), Joint Genome Institute (http://www.jgi.doe.gov/) and WormBase (http://www.wormbase.org/). In the human proteome, except for the FAM108 proteins, most similar proteins to C. elegans AHO-3 were ABHD12, ABHD12B and ABHD13, consistent with a previous report (Simon & Cravatt 2010) and a database (Tree families database, http://www.treefam.org/). Figure 2D and Figure S6 in Supporting Information show dendrograms of similar proteins to FAM108 proteins, ABHD12 proteins and ABHD13 protein in 11 animal species and 2 or 6 other species.
Analyses of expression and localization of AHO-3
All of the fluorescence images were taken with a confocal laser scanning microscope Fluoview FV1000 (Olympus) except for the image in Figure S7R in Supporting Information, which was taken with an Axioplan2 light microscope (Zeiss). We identified the neurons expressing a reporter gene under the control of the aho-3 promoter, based on their position and morphology or their co-expression with cell-specific markers, odr-1p::cytochrome b5::cfp, gcy-8p::tagRFP, AIYp::tagRFP and tph-1p::NLS-tagRFP. We observed and evaluated recombinant AHO-3::EGFP localization in head neurons of adult animals with an Axioplan2 light microscope in Table 1 and Table S3 in Supporting Information. Recombinant AHO-3::EGFP proteins were expressed under the control of the aho-3 promoter in aho-3(nj15) mutants. Test animals were cultivated at 20 °C (Table 1) or at 23 °C (Table S3 in Supporting Information). We scored the fluorescence intensity in sensory endings, punctiform staining of cell bodies and entire cell bodies by using a three-point scale (strong, weak and invisible).
In vivo calcium imaging
In vivo calcium imaging was performed essentially according to previous reports (Kuhara et al. 2008; Ohnishi et al. 2011). Detailed methods are described in Supporting Information.
Statistics
All behavioral assays were performed at least three times in separate experiments. In all Figures, error bars represent standard error of mean (SEM). To compare the distributions of animals in thermotaxis assay, statistical significance of ‘fraction of animals’ in each region was tested by the unpaired t-test with the Dunn–Sidak correction for multiple comparisons. Only when all ‘fraction of animals’ values in one dataset were ‘0.00,’ statistical analysis was not performed; in this case, we show a cross in each Figures. To compare other values, statistical significance was tested by the unpaired t-test, Tukey’s test or Dunnett test.
In the localization analysis of AHO-3, more than 25 animals (Table 1) or 10 animals (Table S3 in Supporting Information) were tested at each trial in 3 days. We scored the fluorescence intensity by using a three-point scale, and statistical analysis by a chi-square test was performed to compare the wild-type AHO-3 with each mutated AHO-3 or the AHO-3 in fed animals with the AHO-3 in starved animals.
Acknowledgments
We would like to thank the Caenorhabditis Genetic Center for providing strains and cosmid clones; Y. Kohara for yk cDNA clones; M.M. Rolls for organelle markers; H. Inada for making N2-CB4856 hybrid strains; C.I. Bargmann for odr-1(n1933) and odr-3(n1605) mutants, the srh-142, str-1 and odr-1 promoters, and the odr-3 cDNA clone; S. Mitani at the National Bioresource Project (Japan) for gcy-28(tm2411) mutants; G. Ruvkun for ins-1(nr2091) mutants; T. Ishihara for hen-1(tm501) mutants; P. Swoboda for the osm-6 promoter; P. Nef for the ncs-1 promoter; B. Wedel, D. Garbers and H. Inada for the gcy-8 promoter; J.F. Etchberger and O. Hobert for the ceh-36 AWC-specific promoter and the ttx-3 AIY-specific promoter; J. Sze for the tph-1 promoter; M. Okumura for the unc-14 and glr-1 promoters; Y. Tanizawa for the glr-2 and glr-3 promoters; J. McGhee for the ges-1 promoter; M. Fujiwara for the egl-4 cDNA clone; N. Hisamoto for the mig-23::GFP vector; A. Fire for pPD vectors; the C. elegans Sequence Consortium for updated C. elegans genome information; and S. Nakano, Y. Tsukada, T. Kimata, T. Sugi, H. Inada and Mori laboratory members for stimulating discussions. NN was supported by the Japan Society for the Promotion of Science. IM is a Research Director of CREST (JST). This work was supported by Grant-in-Aid for Scientific Research on Innovative Areas ‘Neural Diversity and Neocortical Organization’ from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to IM. The Caenorhabditis Genetics Center is funded by the NIH National Center for Research Resources (NCRR).
Supporting Information/Supplementary material
The following Supporting Information can be found in the online version of the article:
Appendix S1 Experimental procedures.
Figure S1 Abnormal thermotactic plasticity of aho-3 mutants on the 20–26 °C thermal gradient.
Figure S2 aho-3 mutants show defects in the salt learning behavior and integration behavior.
Figure S3 Rescue experiment of aho-3 mutants with K04G2.2 gene for the abnormal thermotactic plasticity.
Figure S4 Rescue experiments to identify the gene responsible for aho-3 mutants.
Figure S5 Alignment of AHO-3 homologs and similar proteins.
Figure S6 AHO-3 novel protein is highly conserved among animal species.
Figure S7 A reporter gene expression under control of the aho-3 promoter.
Figure S8 Cell-specific rescue experiments for aho-3 mutants conditioned at 20 °C.
Figure S9 Locomotive ability of animals overexpressing AHO-3 in AWC.
Figure S10 The analysis with aho-3;ceh-36 double mutants.
Figure S11 Rescue experiment for the abnormal thermotactic plasticity of odr-3 and egl-4 mutants.
Figure S12 Genetic relationship analysis among genes whose defect cause abnormal thermotactic plasticity; for 20 °C-cultivation.
Figure S13 Genetic relationship analysis of aho-3 with odr-3, gcy-28 and egl-4; for 23 °C-cultivation.
Figure S14 Genetic relationship analysis among aho-3, odr-3 and eat-16.
Figure S15 Expressions and localizations of AHO-3 mutated in the predicted cataritic triad.
Figure S16 Subcellular localization assay of AHO-3.
Figure S17 Expressions and localizations of AHO-3 mutated in the N-terminal cysteines.
Figure S18 In vivo calcium imagingof AWC and AIY according to temperature change in aho-3 mutants.
Table S1 AHO-3 homologs and similar proteins
Table S2 Expression patterns driven by each promoter
Table S3 Subcellular localization analysis of AHO-3 with fed and starved animals
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening. Proc. Natl Acad. Sci. USA. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. 2006. Chemosensation in C. elegans. In: WormBook (ed The C. elegans Research Community, WormBook) doi: 10.1895/wormbook.1.123.1 http://www.wormbook.org.
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Barron AB, Søvik E, Cornish JL. The roles of dopamine and related compounds in reward-seeking behavior across animal phyla. Front. Behav. Neurosci. 2010;4:163. doi: 10.3389/fnbeh.2010.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly M, Anbil S, Sengupta P. Degeneracy and neuromodulation among thermosensory neurons contribute to robust thermosensory behaviors in Caenorhabditis elegans. J. Neurosci. 2011;31:11718–11727. doi: 10.1523/JNEUROSCI.1098-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron D, Wasserman S, Thomas JH, Samuel AD, Sengupta P. An olfactory neuron responds stochastically to temperature and modulates Caenorhabditis elegans thermotactic behavior. Proc. Natl Acad. Sci. USA. 2008;105:11002–11007. doi: 10.1073/pnas.0805004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem. Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck LB. Information coding in the vertebrate olfactory system. Annu. Rev. Neurosci. 1996;19:517–544. doi: 10.1146/annurev.ne.19.030196.002505. [DOI] [PubMed] [Google Scholar]
- Chang AJ, Bargmann CI. Hypoxia and the HIF-1 transcriptional pathway reorganize a neuronal circuit for oxygen-dependent behavior in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2008;105:7321–7326. doi: 10.1073/pnas.0802164105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Yau KW. Phototransduction in mouse rods and cones. Pflugers Arch. 2007;454:805–819. doi: 10.1007/s00424-006-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Sengupta P, McIntire SL. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- Gomez M, De Castro E, Guarin E, Sasakura H, Kuhara A, Mori I, Bartfai T, Bargmann CI, Nef P. Ca2+ signaling via the neuronal calcium sensor-1 regulates associative learning and memory in C. elegans. Neuron. 2001;30:241–248. doi: 10.1016/s0896-6273(01)00276-8. [DOI] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Russell RL. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1975;72:4061–4065. doi: 10.1073/pnas.72.10.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist M. Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000;1:209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada H, Ito H, Satterlee J, Sengupta P, Matsumoto K, Mori I. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics. 2006;172:2239–2252. doi: 10.1534/genetics.105.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Iino Y, Mohri A, Mori I, Gengyo-Ando K, Mitani S, Katsura I. HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell. 2002;109:639–649. doi: 10.1016/s0092-8674(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Ito H, Inada H, Mori I. Quantitative analysis of thermotaxis in the nematode Caenorhabditis elegans. J. Neurosci. Methods. 2006;154:45–52. doi: 10.1016/j.jneumeth.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr. Drug Targets CNS Neurol. Disord. 2005;4:481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- Jurado P, Kodama E, Tanizawa Y, Mori I. Distinct thermal migration behaviors in response to different thermal gradients in Caenorhabditis elegans. Genes Brain Behav. 2010;9:120–127. doi: 10.1111/j.1601-183X.2009.00549.x. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialog between genes and synapses. Biosci. Rep. 2001;21:565–611. doi: 10.1023/a:1014775008533. [DOI] [PubMed] [Google Scholar]
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, Davis NG, El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama E, Kuhara A, Mohri-Shiomi A, Kimura KD, Okumura M, Tomioka M, Iino Y, Mori I. Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes Dev. 2006;20:2955–2960. doi: 10.1101/gad.1479906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Ohshima Y. The C. elegans ceh-36 gene encodes a putative homeodomain transcription factor involved in chemosensory functions of ASE and AWC neurons. J. Mol. Biol. 2004;336:579–587. doi: 10.1016/j.jmb.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Krieger J, Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- Kuhara A, Mori I. Molecular physiology of the neural circuit for calcineurin-dependent associative learning in Caenorhabditis elegans. J. Neurosci. 2006;26:9355–9364. doi: 10.1523/JNEUROSCI.0517-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhara A, Okumura M, Kimata T, Tanizawa Y, Takano R, Kimura KD, Inada H, Matsumoto K, Mori I. Temperature sensing by an olfactory neuron in a circuit controlling behavior of C. elegans. Science. 2008;320:803–807. doi: 10.1126/science.1148922. [DOI] [PubMed] [Google Scholar]
- Lai CH, Chou CY, Ch’ang LY, Liu CS, Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjuin A, VanHoven MK, Bargmann CI, Thompson JK, Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- L’Etoile ND, Bargmann CI. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron. 2000;25:575–586. doi: 10.1016/s0896-6273(00)81061-2. [DOI] [PubMed] [Google Scholar]
- L’Etoile ND, Coburn CM, Eastham J, Kistler A, Gallegos G, Bargmann CI. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- Liu J, Ward A, Gao J, Dong Y, Nishio N, Inada H, Kang L, Yu Y, Ma D, Xu T, Mori I, Xie Z, Xu XZ. C. elegans phototransduction requires a G protein-dependent cGMP pathway and a taste receptor homolog. Nat. Neurosci. 2010;13:715–722. doi: 10.1038/nn.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- Marrs WR, Blankman JL, Horne EA, et al. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat. Neurosci. 2010;13:951–957. doi: 10.1038/nn.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri A, Kodama E, Kimura KD, Koike M, Mizuno T, Mori I. Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics. 2005;169:1437–1450. doi: 10.1534/genetics.104.036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I, Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature. 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- Nakazato K, Mochizuki A. Steepness of thermal gradient is essential to obtain a unified view of thermotaxis in C. elegans. J. Theor. Biol. 2009;260:56–65. doi: 10.1016/j.jtbi.2009.05.027. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Sugi T, Nonomura M, Mori I. Identification of the AFD neuron as the site of action of the CREB protein in Caenorhabditis elegans thermotaxis. EMBO Rep. 2011;12:855–862. doi: 10.1038/embor.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K, Kubota Y, Chigira Y, Roy SK, Suzuki M, Schvarzstein M, Jigami Y, Hisamoto N, Matsumoto K. An NDPase links ADAM protease glycosylation with organ morphogenesis in C. elegans. Nat. Cell Biol. 2004;6:31–37. doi: 10.1038/ncb1079. [DOI] [PubMed] [Google Scholar]
- Ogura K, Shirakawa M, Barnes TM, Hekimi S, Ohshima Y. The UNC-14 protein required for axonal elongation and guidance in Caenorhabditis elegans interacts with the serine/threonine kinase UNC-51. Genes Dev. 1997;11:1801–1811. doi: 10.1101/gad.11.14.1801. [DOI] [PubMed] [Google Scholar]
- O’Halloran DM, Altshuler-Keylin S, Lee JI, L’Etoile ND. Regulators of AWC-mediated olfactory plasticity in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000761. doi: 10.1371/journal.pgen.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N, Kuhara A, Nakamura F, Okochi Y, Mori I. Bidirectional regulation of thermotaxis by glutamate transmissions in Caenorhabditis elegans. EMBO J. 2011;30:1376–1388. doi: 10.1038/emboj.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramot D, MacInnis BL, Lee HC, Goodman MB. Thermotaxis is a robust mechanism for thermoregulation in Caenorhabditis elegans nematodes. J. Neurosci. 2008;28:12546–12557. doi: 10.1523/JNEUROSCI.2857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB, Nonet ML. Synaptic transmission. In: Riddle DL, Blunenthal T, Meyer BJ, Preiss JR, editors. C. elegans II. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 611–643. [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Hall DH, Victor M, Stelzer EH, Rapoport TA. Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki S, Yamamoto M, Iino Y. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J. Exp. Biol. 2001;204:1757–1764. doi: 10.1242/jeb.204.10.1757. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–1806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J. Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J. Biol. Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Tanizawa Y, Kuhara A, Inada H, Kodama E, Mizuno T, Mori I. Inositol monophosphatase regulates localization of synaptic components and behavior in the mature nervous system of C. elegans. Genes Dev. 2006;20:3296–3310. doi: 10.1101/gad.1497806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, Iino Y. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- Tsunozaki M, Chalasani SH, Bargmann CI. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron. 2008;59:959–971. doi: 10.1016/j.neuron.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina SE, Watson JD, Fox RM, Olszewski KL, Spencer WC, Roy PJ, Miller DM. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 2007;8:R135. doi: 10.1186/gb-2007-8-7-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- White J, Southgate E, Thomson J, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


