Abstract
Low-grade glioma is predisposed to progress to anaplastic astrocytoma and eventually secondary glioblastoma. The malignant transformation may involve the accumulation of multiple genetic alterations. The purpose of this study was to explore the role of miR-544 in glioma progression and discuss whether it may be a novel biomarker of malignant transformation. The expression of miR-544 was measured in a series of 198 glioma samples (63 low-grade glioma, 44 anaplastic astrocytoma and 91 glioblastoma tumors) using microarrays. Quantitative real-time reverse transcription PCR (qRT-PCR) was used to validate the expression levels of miR-544 in tissue and serum samples in an independent validated cohort (25 low-grade glioma, 21 anaplastic astrocytoma and 20 glioblastoma tumors). A Pearson correlation analysis was performed to examine the correlation between miR-544 levels of tissue and serum samples. Microarrays revealed that the expression levels of miR-544 decreased significantly in anaplastic gliomas (P<0.01) or glioblastoma (P<0.01) compared with low-grade gliomas. In an independent cohort of glioma patients, miR-544 exhibited a progression-associated downregulation in glioma tumors. The levels of miR-544 in serum samples tended to be lower in anaplastic and glioblastoma patients compared with low-grade gliomas, but with no significant difference. The Pearson correlation analysis revealed a weakly positive correlation between tissue and serum levels of miR-544. These data support a significant role for miR-544 aberration in the malignant transformation of glioma. The downregulation of miR-544 in tissue may be used as a novel biomarker.
Keywords: miR-544, glioma, malignant transformation
Introduction
Gliomas are the most common primary tumors of the central nervous system (CNS) and include a variety of histological tumor types and World Health Organization (WHO) malignancy grades. Histologically, astrocytic, oligodendroglial and mixed oligoastrocytic tumors are the most common gliomas (1). Diffuse astrocytoma (WHO grade II) is inherently predisposed to recur locally and spontaneously progress to anaplastic astrocytoma (WHO grade III) and eventually secondary glioblastoma (WHO grade IV) (2). Over the last decades, the molecular mechanisms of tumorigenesis and malignant transformation have been investigated and revealed to be associated with genetic mutations. However, a limited number of molecular alterations have attracted much attention, mainly including TP53 mutations, loss of heterzygosity (LOH) on 17p, isocitrate dehydrogenase 1 gene (IDH1) or IDH2 gene mutations and deletions on chromosomes 6, 9p, 11p, 19q and 22q (3). Therefore, it is crucial to identify novel biomarkers that are involved in the malignant transformation in glioma.
microRNAs (miRNAs) are a class of non-coding short RNAs from 17 to 25 nucleotides in length that play significant roles in post-transcriptional gene regulation (4). The dysregulation of miRNAs has been linked to a series of diseases, including various types of cancer. They take part in complex regulatory networks that influence almost every cellular process and may be involved in oncogenesis (5). The role of miRNAs in the tumorigenesis of glioma is of great interest. It has been demonstrated that miRNAs act as oncogenes or tumor suppressor genes and have modulatory effects on tumor cell proliferation, differentiation, metastasis and invasion (6). The current study explored the role of a single miRNA, miR-544, in the malignant progression of human gliomas by comparing expression levels in primary low-grade gliomas and secondary high-grade gliomas. The expression of miR-544 in tissue and serum samples of glioma patients was analyzed to determine whether it may be used as a biomarker of malignant transformation in glioma.
Patients and methods
Patients and tissue samples
The study was approved by the Ethics Committee of Beijing Tiantan Hospital, Capital Medical University (Beijing, China), and written informed consent was obtained from all patients. In this study, a total of 198 patients with glioma (63 low-grade glioma, 44 anaplastic astrocytoma and 91 glioblastoma tumors) were included in the microarray analysis to identify the expression of miR-544 in tumor samples. Quantitative real-time reverse transcription PCR (qRT-PCR) was used to validate the expression levels of miR-544 in paired tissue and serum samples in an independent validated cohort (25 low-grade glioma, 21 anaplastic astrocytoma and 20 glioblastoma tumors).
Tissue samples were obtained from patients during surgery at the Beijing Tiantan Hospital between January 2006 and December 2009. Histological diagnoses were made according to the 2007 WHO classification. Samples were snap-frozen immediately following surgery and stored at −80°C. Only specimens with a histologically estimated tumor cell content of ≥80% were used for the analyses.
Serum sample collection
Whole blood samples of glioma patients were collected. The samples were allowed to stand at room temperature for 30 min and then centrifuged at 2,500 x g for 15 min at 4°C. To remove cellular contaminants, serum samples were subjected to additional centrifugation at 15,000 x g for 10 min. The supernatants to be used for RNA extraction were snap-frozen and then stored at −80°C.
RNA extraction
Total RNA was extracted from tissues using an mirVana PARIS kit (Ambion, Carlsbad, CA, USA) according to the manufacturer’s instructions. The concentrations of all RNA samples were quantified by NanoDrop 1000 (Nanodrop, Wilmington, Delaware, USA).
RNA extraction from serum was performed using the same kit mentioned previously. In initial experiments, a poor correlation was observed between the RNA concentration, as measured with a NanoDrop spectrophotometer, and the ability to amplify miRNA. To overcome this problem, the RNA volume used in the experiment was unified instead of the RNA concentration, as described by McDonald et al (7). The volume of serum that was extracted and the volume of extracted eluent used for qRT-PCR were consistent throughout the study.
miRNA microarray
The miR-544 expression values of 198 glioma tissues were obtained from Chinese Glioma Genome Atlas (CGGA), a database that focuses on glioma. Briefly, 200 ng total RNA was polyadenylated and then reverse transcribed into cDNA using a biotin-labeled Oligo dT primer with a universal PCR sequence. Following cDNA synthesis, miRNAs were individually interrogated using specific oligonucleotides. A single miRNA-specific Oligo (MSO) was designed against each mature miRNA sequence and miRNA-specific primers were extended using DNA polymerase. Universal primers were used to amplify the cDNA templates and the primer complimentary to the array was fluorescently labeled. Finally, the labeled, single-stranded PCR products were hybridized to the Human v2.0 miRNA Expression BeadChip (Illumina, Inc., San Diego, CA, USA) with 1,146 human miRNAs (97% coverage of the miRBase 12.0 database).
Reverse transcription and qPCR analyses
The TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA, USA) and miRNA-specific stem-loop primers (Applied Biosystems) were used for miRNA reverse transcription. qPCR was performed using TaqMan Gene Expression Master Mix (Applied Biosystems) and individual miRNA primers and hydrolysis probes (Applied Biosystems) according to the manufacturer’s instructions. Relative quantification of miRNA expression was calculated using the 2−ΔΔCT method. RNU6B was used as endogenous control for tissue detection. For serum qRT-PCR analysis, there was no consensus on the use of housekeeping miRNA. Therefore, a healthy donor sample was used as control and processed together with the testing samples as described previously (8).
Statistical analysis
Comparisons of miR-544 levels among patients with various tumor grades were performed using Student’s t-tests. The Pearson correlation analysis was used to examine the correlation between miR-544 levels of paired tissue and serum samples. All statistical analyses were performed using GraphPad Prism 5. A two-sided P<0.05 was considered to indicate a statistically significant result.
Results
miR-544 expression is decreased in anaplastic glioma and glioblastoma tissue samples
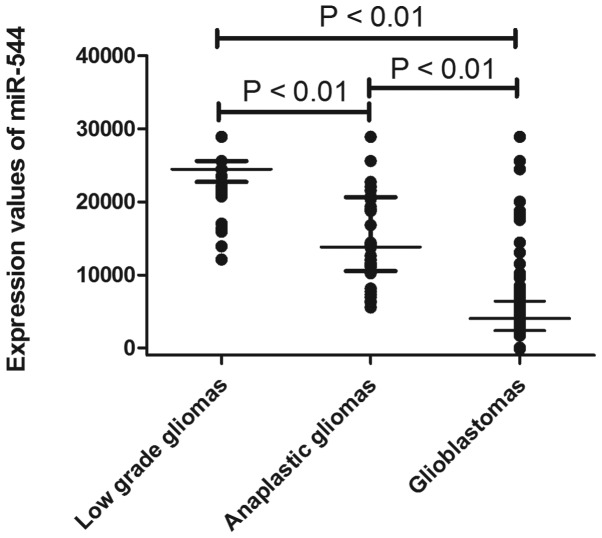
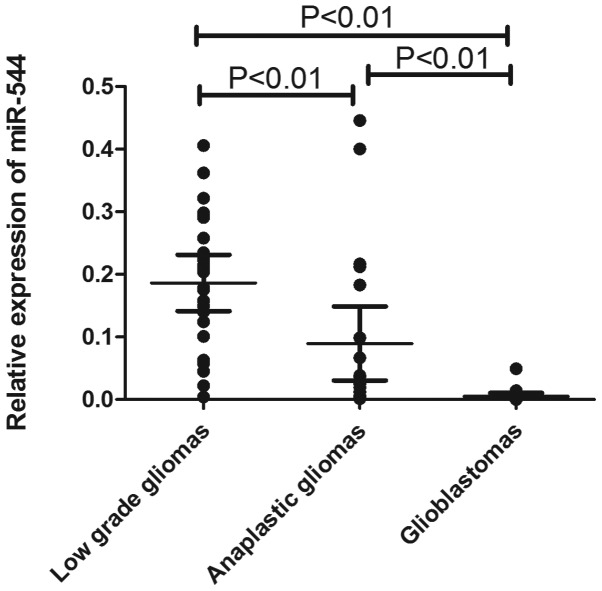
The expression of miR-544 was measured in a series of 198 glioma samples via microarray. As shown in Fig. 1, miR-544 expression decreased significantly in anaplastic gliomas (P<0.01) and glioblastomas (P<0.01) compared with low-grade gliomas. To further confirm this result, qRT-PCR was performed to examine miR-544 expression levels in an independent cohort containing 66 tumor samples (25 low-grade glioma, 21 anaplastic astrocytoma and 20 glioblastoma tumors). As shown in Fig. 2, miR-544 was significantly downregulated in anaplastic gliomas (P<0.01) and glioblastomas (P<0.01) compared with low-grade gliomas.
Figure 1.
Expression levels of miR-544 in tissue samples via microarray. The expression levels of miR-544 decreased significantly in anaplastic gliomas (P<0.01) or glioblastoma (P<0.01) compared with low-grade gliomas.
Figure 2.
Expression levels of miR-544 in tissue samples of the validation cohort. The expression levels of miR-544 decreased significantly in anaplastic gliomas (P<0.01) or glioblastoma (P<0.01) compared with low-grade gliomas.
miR-544 expression is not significantly different in glioma, anaplastic astrocytoma or glioblastoma serum samples
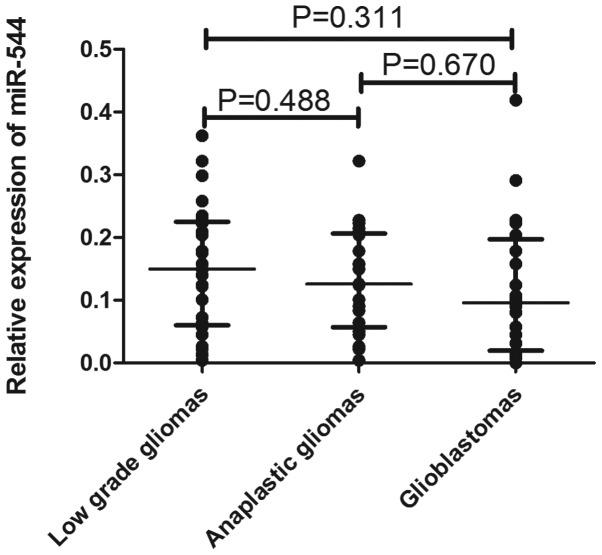
To explore whether miR-544 may be a soluble biomarker of malignant transformation, the expression of miR-544 was detected in the validation cohort containing 66 serum samples (25 low-grade glioma, 21 anaplastic astrocytoma and 20 glioblastoma tumors), which were paired with tissue samples, by qRT-PCR assay. As shown in Fig. 3, there was no significant difference in miR-544 expression among the 3 groups, although the levels tended to be lower in anaplastic and glioblastoma patients.
Figure 3.
Expression levels of miR-544 in serum samples of the validation cohort. The expression levels of miR-544 tended to be lower in anaplastic and glioblastoma patients compared with low-grade gliomas but with no significant difference.
miR-544 expression in tissue and serum samples has a weakly positive correlation
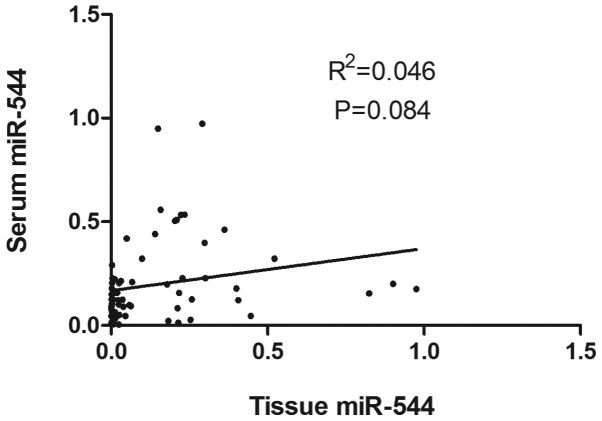
To further explore the correlation between miR-544 expression in paired tissue and serum samples, a Pearson correlation analysis of miR-544 levels between 66 tissue and serum samples of glioma patients was performed. As shown in Fig. 4, there was a weakly positive correlation between tissue and serum levels of miR-544 (R2=0.046, P=0.084).
Figure 4.
Pearson correlation analysis of miR-544 levels between 66 tissue and serum samples. There was a weakly positive correlation between tissue and serum levels of miR-544 but with no significant difference.
Discussion
Low-grade gliomas are well differentiated and grow slowly but show a consistent tendency to diffusely infiltrate the surrounding brain tissue. They almost invariably recur and this is often associated with the progression to a higher malignancy (9). The median survival of low-grade glioma patients is between 5 and 10 years (10). However, the median survival time of patients with anaplastic astrocytomas is less than 3 years, while for glioblastoma this is less than one year (11). In addition, resistance to chemotherapeutic drugs and therapeutic irradiation often occurs in the malignant transformation of gliomas (12). Thus, novel biomarkers for the detection of malignant transformation are required to form predictions and select the best possible treatment regimen for glioma patients. Over the last few decades, several genes associated with the malignant progression of gliomas have been identified. The TP53 mutation appears to be a relatively early event during the development of an astrocytoma, whereas the loss or mutation of PTEN and amplification of EGFR are characteristic of higher grade tumors (13–15). The loss of 9p21 is confined to WHO grade III tumors, whereas mutations at codon 132 of IDH1 may occur following the formation of a low-grade glioma and drive the progression of the tumor to a glioblastoma (16,17). The current study has identified that a non-coding RNA, miR-544, exhibited a progression-associated downregulation in glioma tumors, which may be used as a novel biomarker in malignant transformation.
Previous studies have indicated that a variety of miRNAs play significant roles in glioma. miR-21 was the first miRNA to be linked with glioma malignancy and acted as an oncogene (6). The expression levels of miR-21 were elevated in human glioma cells and tissues and also correlated with tumor grade (18). miR-181a and miR-181b were observed as tumor suppressors that inhibit growth and induce the apoptosis of glioma cells (19). The expression of miR-221 and 222 were upregulated in human glioblastoma and they induced an increase in cell migration and growth by targeting the protein phosphate PTPμ, whereas let-7 reduced the in vitro proliferation and migration of glioma cells (20,21).
Using genome-wide miRNA profiling, a previous study identified a 23-miRNA expression signature that may distinguish glioblastoma from anaplastic astrocytoma with 95% accuracy (22). In the current study, the expression of miR-544 was measured using a microarray in a series of glioma tissues with various tumor grades. It was revealed that miR-544 expression decreased significantly in anaplastic gliomas or glioblastoma compared with low-grade gliomas and the further qRT-PCR assay confirmed the results. A previous study also reported that miR-544 was significantly downregulated in osteosarcoma and may affect its pathogenesis by targeting the cMYC transcript (23). This is in accordance with the finding that the dysregulation of miR-544 may be involved in the process of tumorigenesis and malignant progression.
Circulating miRNAs are significant tumor markers due to their invasiveness and ease of detection. To further explore whether circulating miR-544 may be a non-invasive biomarker for malignant transformation of gliomas, its expression levels were measured in serum samples. The results revealed that the levels tended to be lower in anaplastic and glioblastoma patients compared with low-grade gliomas, but with no significant difference. There was a weakly positive correlation between the tissue and serum levels of miR-544. The majority of previous studies have demonstrated the same trend of alteration between circulating miRNAs and tissue miRNAs, either an increase or decrease, in various types of cancer (24–26). However, a different correlation between circulating miRNAs and tissue miRNAs has also been observed. Wulfken et al observed 109 miRNAs at higher levels in the serum of patients with renal cell carcinoma, but only 36 miRNAs were upregulated in tissue samples (27). Another study reported that approximately 66% of the released miRNAs reflect the cellular miRNA content of malignant mammary epithelial cells (28). These studies provide potential evidence for the hypothesis that circulating miRNAs reflect various aspects of human physiological status and only a subset of circulating miRNAs have a tumor-specific origin (27,29). Conversely, the field of detecting circulating miRNAs for use in cancer diagnosis is new and thus the lack of standard protocol may also contribute to the conflicting results. Therefore, certain issues remain to be resolved, including type of samples, use of appropriate normalization controls, standard protocols for sample preparation and concentration measurement (7). Taken together, it is difficult for circulating miR-544 to be a stable biomarker of the malignant transformation of glioma.
In summary, miR-544 expression decreased significantly in anaplastic gliomas or secondary glioblastomas compared with low-grade gliomas. In an independent cohort of glioma patients in validation studies, miR-544 exhibited a progression-associated downregulation in glioma tumors. These data support a significant role for miR-544 aberration in the malignant transformation of glioma and thus the downregulation of miR-544 in tissue may be used as a novel biomarker. Further studies are required to characterize in detail the molecular mechanisms of miR-544 action.
References
- 1.Martinho O, Granja S, Jaraquemada T, et al. Downregulation of RKIP is associated with poor outcome and malignant progression in gliomas. PLoS One. 2012;7:e30769. doi: 10.1371/journal.pone.0030769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellano L, Giamas G, Jacob J, et al. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci. 2012;17:700–712. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 8.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe K, Sato K, Biernat W, et al. Incidence and timing of p53 mutations during astrocytoma progression in patients with multiple biopsies. Clin Cancer Res. 1997;3:523–530. [PubMed] [Google Scholar]
- 10.Jung TY, Jung S, Moon JH, Kim IY, Moon KS, Jang WY. Early prognostic factors related to progression and malignant transformation of low-grade gliomas. Clin Neurol Neurosurg. 2011;113:752–757. doi: 10.1016/j.clineuro.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Stupp R, Reni M, Gatta G, Mazza E, Vecht C. Anaplastic astrocytoma in adults. Crit Rev Oncol Hematol. 2007;63:72–80. doi: 10.1016/j.critrevonc.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Weller M, Malipiero U, Aguzzi A, Reed JC, Fontana A. Protooncogene bcl-2 gene transfer abrogates Fas/APO-1 antibody-mediated apoptosis of human malignant glioma cells and confers resistance to chemotherapeutic drugs and therapeutic irradiation. J Clin Invest. 1995;95:2633–2643. doi: 10.1172/JCI117965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 15.Weber RG, Sabel M, Reifenberger J, et al. Characterization of genomic alterations associated with glioma progression by comparative genomic hybridization. Oncogene. 1996;13:983–994. [PubMed] [Google Scholar]
- 16.Bigner SH, Matthews MR, Rasheed BK, et al. Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol. 1999;155:375–386. doi: 10.1016/S0002-9440(10)65134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti A, Aguennouz M, La Torre D, et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J Neurooncol. 2009;93:325–332. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- 19.Shi L, Cheng Z, Zhang J, et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 20.Quintavalle C, Garofalo M, Zanca C, et al. miR-221/222 over-expession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPmu. Oncogene. 2012;31:858–868. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee ST, Chu K, Oh HJ, et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J Neurooncol. 2011;102:19–24. doi: 10.1007/s11060-010-0286-6. [DOI] [PubMed] [Google Scholar]
- 22.Rao SA, Santosh V, Somasundaram K. Genome-wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol. 2010;23:1404–1417. doi: 10.1038/modpathol.2010.135. [DOI] [PubMed] [Google Scholar]
- 23.Thayanithy V, Sarver AL, Kartha RV, et al. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brase JC, Johannes M, Schlomm T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 25.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 26.Yang CC, Hung PS, Wang PW, et al. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J Oral Pathol Med. 2011;40:397–404. doi: 10.1111/j.1600-0714.2010.01003.x. [DOI] [PubMed] [Google Scholar]
- 27.Wulfken LM, Moritz R, Ohlmann C, et al. MicroRNAs in renal cell carcinoma: diagnostic implications of serum miR-1233 levels. PLoS One. 2011;6:e25787. doi: 10.1371/journal.pone.0025787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pigati L, Yaddanapudi SC, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zen K, Zhang CY. Circulating MicroRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32:326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]