Abstract
Etoposide (ETP) treatment of ataxia telangiectasia mutated (ATM) and Rad3-related protein (ATR)-, topoisomerase-binding protein-1 (TopBP1) and human MutY homolog (hMYH)-depleted cells results in a significant reduction in apoptotic signaling. The association between ATR or TopBP1 and hMYH increased following ETP treatment. In hMYH knockdown cells, the interaction between ATR and TopBP1 decreased following ETP treatment. We suggest that hMYH functions as a sensor of ETP-induced apoptosis. The results suggest that in the absence of hMYH, cells are unable to recognize the damage signal and the ATR pathway is not activated.
Keywords: human MutY homolog, ataxia telangiectasia mutated, Rad3-related protein, topoisomerase-binding protein-1, apoptosis, etoposide
Introduction
DNA damage response is essential for the maintenance of genome integrity (1–5). As a complex, the DNA damage response involves the recognition of DNA damage, activation of DNA damage-responsive protein kinases, signal amplification by downstream protein kinases and activation of the effector proteins that trigger various cellular processes. At low DNA damage levels, activation of the DNA damage response results in cell cycle arrest and DNA repair. However, at higher levels or under severe conditions, DNA damage response signaling frequently results in cell death by apoptosis (1–5).
Phosphoinositide 3-kinase-related protein kinases, ataxia telangiectasia mutated proteins (ATM) and Rad3-related proteins (ATR), are the key regulators of the DNA damage response (1–8). Once activated, ATM and ATR regulate an array of substrates, including Chk1 and Chk2, which culminate in DNA repair, cell cycle arrest and/or apoptosis. In the canonical model, ATR activation involves the recruitment of the ATR-ATR interacting protein (ATR-ATRIP) and Rad9-Hus1-Rad1 (9-1-1) protein complexes to the DNA damage site via replication protein A (RPA). As a result, the 9-1-1 complex brings topoisomerase-binding protein-1 (TopBP1) (ATR activator) close to ATR for ATR activation (1,4,8). Mammalian TopBP1 functions at the DNA replication checkpoint (9,10) and has multiple BRCA1 C-terminal (BRCT) repeats, which usually function in tandem to bind phosphoproteins (11,12). TopBP1 colocalizes with ATR-ATRIP at the sites of DNA replication stress (9,10). The N-terminus of TopBP1 is required for its recruitment and the resulting activation of ATR via interaction with Rad9 in mammalian cells (13).
Human MutY homolog (hMYH) is a base excision repair DNA glycosylase that excises misincorporated adenine opposite 7,8-dihydro-8-oxoguanine (8-oxoG), a product of oxidative DNA damage. Furthermore, in hMYH-disrupted cells, the phosphorylation of ATR and Chk1 is decreased by hydroxyurea (HU) or ultraviolet (UV) treatment (14). The hMYH is known to interact with 9-1-1 (15), and a recent study revealed that it also interacts with ATR and MutS α via the human MutS homolog (hMSH) 6 subunit (14,16).
The mismatch repair protein hMSH2 may interact with ATR and participate in ATR activation during DNA damage, leading to apoptosis. Therefore, hMSH2-deficient cells are more resistant to apoptosis (17). The mismatch repair and MYH repair pathways share many common features. First, both pathways function immediately following DNA replication to distinguish newly synthesized DNA strands from their parental counterparts (18,19). Second, hMYH and hMSH6 interact with the replication proteins proliferating cell nuclear antigen and RPA and colocalize at the same replication foci (20,21). Finally, both pathways are involved in mutation avoidance following DNA oxidation. Therefore, we suggest that the interaction between hMYH and hATR is the same as hMSH2 and hATR.
Etoposide (ETP), a topoisomerase II inhibitor, is known to induce apoptosis and activate ATR via TopBP1 (22,23); however, it is unclear how hMYH is involved during the ETP response. In this study, MYH, ATR and TopBP1 knockdown cells were treated with ETP. In the absence of these proteins, the cells were more resistant to ETP-induced apoptosis. We determined for the first time that hMYH interacts with hTopBP1 or hATR in HEK293 cells, and that this interaction is increased following ETP treatment. However, when hMYH is disrupted, the interaction between hATR and hTopBP1 is decreased following ETP treatment. Since hATR is inactive, the apoptosis signal cannot transduce to the downstream proteins, specifically p-Chk2 and p-p53.
Materials and methods
Cell lines
Human embryonic kidney HEK293 cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Invitrogen Life Technologies) and 1% penicillin-streptomycin solution (Sigma, St. Louis, MO, USA) at 37°C in a 5% CO2 incubator. Prior to the experiments, HEK293 cells were seeded into 6-well plates at a density of 1×106 cells per well and incubated for 24 h.
siRNA construction and transfection into cells
The optimum siRNA sequences for the knockdown of endogenous hMYH were designed and purchased from the Stealth™ RNAi program of Invitrogen Life Technologies. siRNA corresponding to nucleotides 415–439 of the green fluorescence protein (GFP) was used as a negative control. The siMYH and siGFP sequences and transfection method were conducted as previously described (14). ATR siRNA (sc-29763) and TopBP1 siRNA were purchased from Santa Cruz Biotechnology, Inc. (sc-41068; Santa Cruz, CA, USA). ATR and TopBP1 knockdown were conducted according to the manufacturer’s instructions.
Protein extraction and western blot analysis
HEK293 cells were harvested, washed with phosphate-buffered saline (PBS) and lysed with lysis buffer containing 50 mM Tris-HCl phenylmethanesulfonylfluoride, protease and a phosphatase inhibitor cocktail (Sigma), for 1 h at 4°C with occasional vortexing. Protein extracts were collected following centrifugation at 16,000 × g for 20 min, and protein concentration was determined using a Bio-Rad DC protein assay kit (Bio-Rad, Hercules, CA, USA). Protein extracts that were resolved on 8 or 12% sodium dodecyl sulfate (SDS)-polyacrylamide gels were transferred onto PVDF membranes (GE Healthcare Worldwide, Princeton, NJ, USA). The membranes were blocked with 5% non-fat dried milk in Tris-buffered saline with 0.05% Tween-20 and then incubated with antibodies against ATR, Chk1, Chk2, phospho-Chk1 (Ser-345), phospho-Chk2 (Thr-68), β-actin (all from Santa Cruz Biotechnology, Inc.), phospho-ATR (Ser-428), caspase 9, caspase 7, p-p53 (Ser-15; all from Cell Signaling Technology, Inc., Beverly, MA, USA), TopBP1 (Abcam, Cambridge, UK) and hMYH (Abnova, Teipei, Taiwan). Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.). Protein bands were detected using enhanced chemiluminescence (ECL) Pico western blotting detection reagents (Pierce Biotechnology, Inc., Rockford, IL, USA).
Immunofluorescence microscopy
To determine the subcellular location of ATR, TopBP1 and hMYH, HEK293 cells were seeded onto polylysine-coated coverslips and treated with 25 μM ETP for 48 h. At room temperature, cells were fixed with 4% paraformaldehyde in PBS for 30 min and permeabilized with 0.25% Triton X-100 in PBS for 30 min. After blocking with 1% bovine serum albumin in PBS containing 0.5% Tween-20 (PBS-T) for 30 min, cells were incubated with ATR (1:100; Santa Cruz Biotechnology, Inc.), TopBP1 (1:500; Abcam) and MYH monoclonal antibodies (1:100; Abnova) for 2 h. The cells were washed 3 times for 15 min each in PBS and incubated with Alexa 488-conjugated anti-mouse IgG (1:100; Sigma), fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (1:100; Sigma) for 2 h. Cells were rinsed 3 times with 1 ml PBS and analyzed using a confocal fluorescence microscope (Olympus FV-1000; software, Olympus Fluoview; Olympus, Center Valley, PA, USA).
Co-immunoprecipitation
Co-immunoprecipitation (IP) of endogenous proteins using an ATR antibody was conducted using the ImmunoCruz™ IP/WB Optima B System (Santa Cruz Biotechnology, Inc.) according to the manufacturer’s instructions. IP was conducted using rabbit anti-ATR, and immunoblot (IB) analysis was conducted using rabbit anti-ATR, rabbit anti-TopBP1 and mouse anti-MYH. To determine the effect of ETP on MYH-ATR and MYH-TopBP1 interaction, a similar co-IP procedure was conducted using mouse anti-MYH for co-IP, and IB analysis using rabbit anti-ATR, rabbit anti-TopBP1 and mouse anti-MYH.
Statistical analysis
Experiments were conducted in triplicate and statistical analyses were conducted using the Student’s t-test. Data are expressed as the mean, and P<0.05 was considered to indicate a statistically significant difference.
Results
Effect of ETP on HEK293 cell proliferation
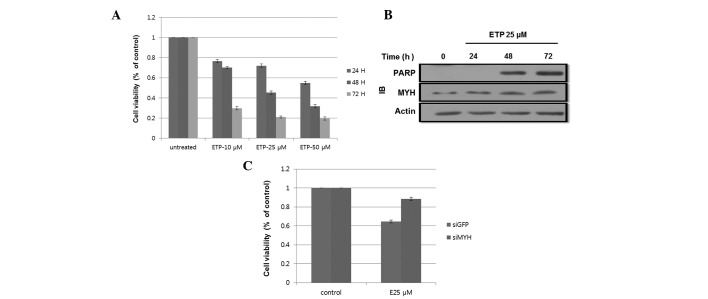
We examined the effect of ETP on cell viability by treating cells with 3 different concentrations of ETP (10, 25 and 50 μM) for various periods of time. After 24, 48 and 72 h of treatment, cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Fig. 1A shows that treatment with ETP greatly reduced cell viability compared with the control cells. ETP induced apoptosis in HEK293 cells in a time- and dose-dependent manner. We determined that treatment with 25 μM ETP was the best condition for our subsequent experiments.
Figure 1.
ETP-induced apoptosis in HEK293 cells. (A) ETP treatment reduced cell viability. Cells were treated with 0, 10, 25 and 50 μM ETP. After 24, 48 and 72 h, 20 μl MTT was added to each well and incubated for an additional 4 h. The results shown are the mean ± SD of 3 independent experiments. (B) ETP-induced cleavage of PARP following 48 h of incubation. Cells were incubated with 25 μM ETP for 24, 48 and 72 h. Cell lysates were subjected to SDS-PAGE. PARP and MYH expression levels were determined by western blotting using the indicated antibodies. (C) hMYH knockdown reduced cell viability following ETP treatment. Cells were transfected with siGFP or siMYH and incubated for 24 h. Cells were then treated with 25 μM ETP for 48 h. Cell viability was determined by the MTT assay. The results shown are the mean ± SD of 3 independent experiments. ETP, etoposide; IB, immunoblot; PARP, poly-ADP ribose polymerase; MYH, MutY homolog; GFP, green fluorescence protein. MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; hMYH, human MYH.
To assess the molecular mechanism of ETP-induced apoptosis, we examined the expression of the 85-kDa cleaved form of poly-ADP ribose polymerase (PARP), an apoptosis marker, and hMYH, an apoptosis-related protein (25). Cells were treated with 25 μM ETP for 24, 48 or 72 h. As shown in Fig. 1B, cleavage of PARP was observed after 48 and 72 h. The hMYH protein level increased in a time-dependent manner, indicating the involvement of hMYH in apoptosis. Taken together, treatment with 25 μM ETP for 48 h was the best condition for the subsequent experiment.
To further determine the effect of hMYH on cell viability, cells transfected with MYH-siRNA or control-siRNA were treated with 25 μM ETP for 48 h. Fig. 1C shows that cell viability, as determined by the MTT assay, decreased by ∼40% in the control cells, while only a 17% decrease was observed in MYH-siRNA cells. These results suggested the involvement of hMYH in apoptotic signaling during ETP treatment.
hMYH, hTopBP1 and hATR deficient cells are resistant to ETP-induced apoptosis and DNA-damage signaling
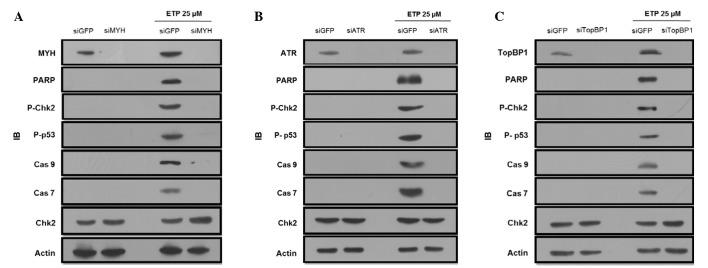
Recently, it has been reported that ATR phosphorylation upon HU or UV treatment was decreased in hMYH-disrupted HEK293 and HaCaT cells (14). ATR is known to phosphorylate Chk1 and Chk2 (14,15), and here, we observed a phosphorylation of Chk2 only in control-transfected cells, but not in siMYH cells following ETP treatment (Fig. 2A). This result suggests that hMYH is required for ATR-mediated Chk2 activation under these conditions. The phosphorylation of Chk1 observed was similar to that of Chk2 (data not shown). Since transactivation of proapoptotic proteins through the p53-dependent signaling pathway is an important apoptotic mechanism, we focused on Chk2 because of its involvement in p53 regulation and ETP-induced apoptosis (22,24). As shown in Fig. 2A, ETP-induced p53 phosphorylation was abrogated in siMYH-transfected cells.
Figure 2.
MYH, ATR and TopBP1 knockdown decreases ETP-induced apoptosis in HEK293 cells. (A) MYH knockdown reduces the activation of apoptosis-related proteins. Control or siMYH transfected cells were treated with 25 μM ETP for 48 h. Total cell lysates were used for IB analysis of p-Chk2 (threonine-68), p-p53 (threonine-15), caspase 9, caspase 7 and MYH. β-actin was used as a loading control. (B and C) The same experiment as in A, however, the cells were transfected with siRNA for either ATR or TopBP1. IB, immunoblot; MYH, MutY homolog; PARP, poly-ADP ribose polymerase; Cas, caspase; GFP, green fluorescence protein; ETP, etoposide; ATR, Rad3-related protein; TopBP1, topoisomerase-binding protein-1.
Caspase is a well-known key molecule in DNA damage-induced apoptosis (22). The activation of caspase 9 and caspase 7 was examined via western blot analysis in hMYH knockdown and control cells. ETP treatment induced the cleavage of inactive 47-kDa procaspase 9 into smaller, detectable, active 37-kDa fragments, and the cleavage of inactive 32-kDa procaspase 7 into the 20-kDa active form in control cells (Fig. 2A). However, the active forms of caspase 9 and 7 were not detected in hMYH knockdown cells following ETP treatment.
To determine the role of ATR relative to Chk2 or p53 in ETP-induced apoptosis, cells were treated with either control or ATR siRNA for 24 h, and then treated with ETP for 48 h. We observed increased phosphorylation or activation of Chk2 and p53 in control siRNA-transfected cells (Fig. 2B). However, the active forms of caspase 9 and 7 were not observed in hATR knockdown cells.
Since hTopBP1 is known to activate hATR, we observed the apoptosis pattern in hTopBP1 knockdown cells. Fig. 2C shows that apoptosis was decreased in hTopBP1 knockdown cells in comparison to the control cells, and similar results were observed in hMYH and hATR knockdown cells.
hMYH associates with ATR or TopBP1 in HEK293 cells during ETP treatment
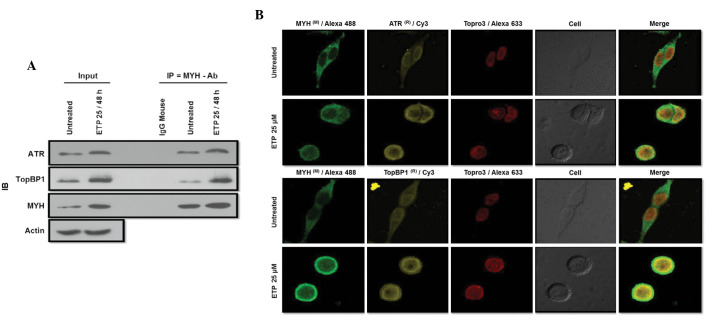
ETP treatment activated hATR, hTopBP1 and hMYH (A-T-M), which triggered a signaling cascade leading to apoptosis. However, it is unclear how A-T-M is activated and regulates the apoptosis pathway. To determine whether hMYH regulates hATR or hTopBP1 in DNA damage models, we conducted a co-IP assay to confirm that hMYH interacts with hATR or hTopBP1. Cells were treated with 25 μM ETP for 48 h, and whole cell extracts were used. As shown in Fig. 3A, ETP treatment induced the association of hMYH-hATR and hMYH-hTopBP1. Our results suggest that hMYH interacts with hATR or hTopBP1 and participates in A-T-M activation during ETP treatment, which leads to the DNA damage response and signals apoptosis.
Figure 3.
Interaction of MYH with ATR or TopBP1 increased following ETP treatment. (A) ATR or TopBP1 interact with MYH increases following treatment with ETP. Cells were treated with 25 μM ETP for 48 h. Cells were lysed and total cell lysates were used for co-IP with MYH antibody. IP samples were analyzed by western blotting with ATR or TopBP1 and MYH antibodies. β-actin was used as a loading control. (B) ATR or TopBP1 and MYH co-localized following treatment with ETP. Cells were cultured overnight on coverglass bottom dishes, then treated with 25 μM ETP for 48 h, and fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Cells were then stained with antibodies against ATR or TopBP1 (FITC; yellow), MYH (Alexa®488/Green) and To-pro®3 (Red). IB, immunoblot; ATR, Rad3-related protein; TopBP1, topoisomerase-binding protein-1; MYH, MutY homolog; IP, immunoprecipitation; Ab, antibody. ETP, etoposide; PBS, phosphate-buffered saline; FITC, fluorescein isothiocyanate.
A critical function of hATR or hTopBP1 activation during genotoxic stress is the accumulation of hATR or hTopBP1 in the nucleus, where signaling proteins form multiple nuclei and interact in response to DNA damage (6,11). To examine the changes in hATR and hTopBP1 localization, we conducted immunofluorescence (IF) experiments. We treated cells with 25 μM ETP for 48 h, and conducted IF assays to determine whether hMYH and hATR or hTopBP1 occupied the same subcellular locations during genotoxic stress. In untreated control cells, light hMYH staining was observed in the cytoplasm (Fig. 3B). However, following ETP treatment, we observed a higher intensity hMYH staining that translocated to the nucleus, indicating an increased expression. Staining of hATR and hTopBP1, which are known to localize to the nucleus, also increased in intensity following ETP treatment. Superimposition of hMYH-dependent green fluorescence with hATR or hTopBP1-dependent yellow fluorescence resulted in yellow-green images in cells. Topro3 Red (red fluorescence) is a nuclear marker. These results are consistent with the interaction of hMYH with hTopBP1 or hATR and their colocalization in cells.
ATR and TopBP1 interaction decreased in hMYH-deficient cells during ETP treatment
The decrease in hMYH expression results in decreased apoptotic signaling following ETP treatment (Fig. 2A). Furthermore, hMYH interacts with hATR or hTopBP1 and this interaction increased following treatment with ETP (Fig. 3A). To observe the implication of hATR, hTopBP1 and hMYH interaction in this pathway, we conducted co-IP assays using ATR antibody. Cells were transfected with siGFP, as a control, or siMYH. Then, cells were treated with or without ETP. Total cell lysates were used in co-IP, and the presence of A-T-M was observed using western blot analysis.
A decreased hMYH expression was observed in MYH knockdown cells (Fig. 4A), while changes in ATR and TopBP1 expression levels were not observed in MYH knockdown cells. Following treatment with ETP, the expression of A-T-M increased. Furthermore, the co-IP result suggests that the interaction between hATR and hTopBP1 was decreased in MYH knockdown cells following treatment with ETP.
Figure 4.
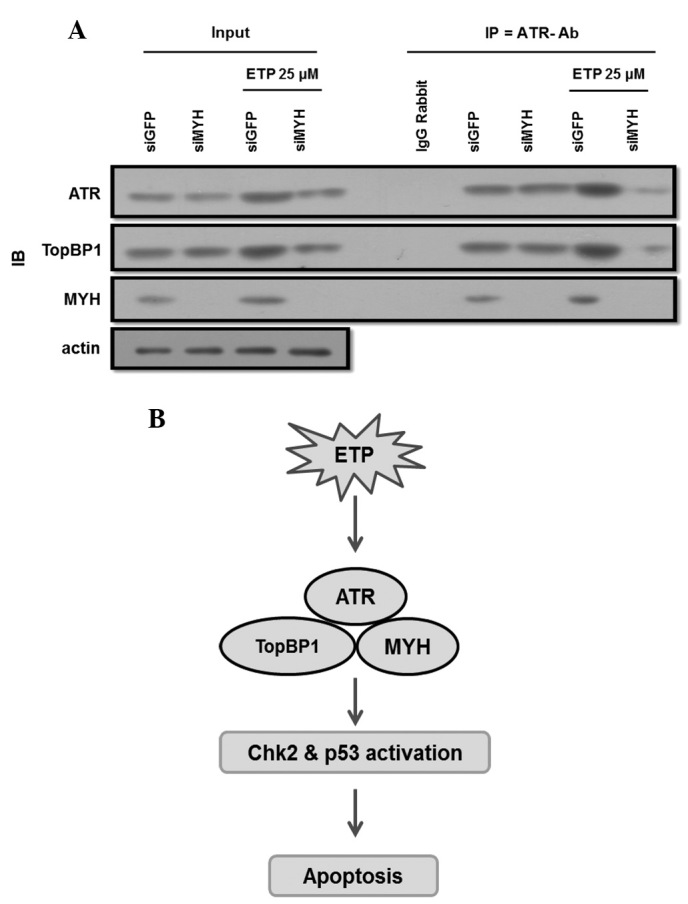
Knockdown of hMYH abrogated the interaction between ATR and TopBP1. (A) Co-IP and western blot analysis were conducted using lysates of cells transfected with siGFP or siMYH for 24 h. Cells were treated with 25 μM ETP for 48 h and co-IP was conducted using an ATR antibody. IP samples were further analyzed by western blotting with ATR, MYH and TopBP1 antibodies. Rabbit IgG was used as a control for co-IP. (B) A model of hMYH-mediated ATR activation in DNA damage signaling. DNA damage signaling involving ATR pathway activation is initiated by hMYH. For ATR activation, hMYH interacts with ATR and TopBP1 at damaged sites, leading to the activation of Chk2 and p53, culminating in cell cycle arrest and apoptosis. IB, immunoblotting; ATR, Rad3-related protein; TopBP1, topoisomerase-binding protein-1; hMYH, human MutY homolog; ETP, etoposide; GFP, green fluorescence protein; IP, immunoprecipitation; Ab, antibody.
Discussion
hMYH interacts not only with hATR, but also with the human MutS homologs (hMSH2/hMSH6) via the hMSH6 subunit (14,16). hMSH2 directly participates in ATR recruitment and activation, leading to DNA damage signaling and subsequent apoptosis. Therefore, hMSH2 deficiency increases the resistance of cells to apoptosis (17). Another study suggests that the nuclear isoforms of hMYH initiate cell death by sensing adenine opposite 8-oxoG during nuclear DNA replication, thus suppressing tumorigenesis. In addition, accumulation of 8-oxoG in mitochondrial DNA and initiation cell death by MYH may also contribute to the tumor suppression (25).
In this study, we implicated the DNA glycosylase, hMYH, in ETP-induced apoptosis, and revealed that hMYH knockdown cells reduce Chk2 (T-68) and p53 (T-15) phosphorylation. We also observed similar results in ATR and TopBP1 knockdown cells (Fig. 2). This result suggests the possibility that hATR, hTopBP1 and hMYH (A-T-M) function in the same pathway. This was accompanied by the suppression of proapoptotic protein expression and decreased apoptosis. The interaction between hMYH with ATR and TopBP1 is dependent on ETP treatment (Fig. 3).
In hMYH knockdown cells, ATR and TopBP1 interaction decreases following ETP treatment (Fig. 4A). We suggest that hMYH functions as a sensor in ETP induced apoptosis. In the absence of hMYH, cells cannot recognize the damage signal, and thus the ATR pathway is not activated, which results in tumor development (14,25).
Based on these observations, we suggest new pathways for A-T-M sensor activation (Fig. 4B). This pathway is initiated by MYH, which recruits and activates ATR-related proteins in ETP-induced apoptosis. In summary, binding of MYH directly participates in ATR and TopBP1 activation in DNA damage signaling, leading to apoptosis. An MYH protein deficiency increases the resistance of cells to apoptosis.
Acknowledgments
This study was supported by the project from the Advanced Technology Center (ATC) Support Program of the Ministry of Knowledge Economy (Republic of Korea; Grant No. 10033024), the Basic Science Research Program through the National Research Foundation of Korea (NRF; Grant No. 2012R1A1A3005889) and the World Class University (WCU; Grant No. R33-2008-000-1071) program through the Korea Science and Engineering Foundation funded by the Ministry of Education, Science and Technology.
References
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Barket J, Bartkova J, Lukas J. DNA damage signaling guards against activated oncogenes and tumor progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 3.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoint in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 6.Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA repair. 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Hurley PJ, Bunz F. ATM and ATR: components of an integrated circuit. Cell cycle. 2007;6:414–417. doi: 10.4161/cc.6.4.3886. [DOI] [PubMed] [Google Scholar]
- 8.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–628. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mäkiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Mäkelä TP, Syväoja JE. BRCT domain-containing protein TopBP1 Functions in DNA replication and damage response. J Biol Chem. 2001;276:30399–30406. doi: 10.1074/jbc.M102245200. [DOI] [PubMed] [Google Scholar]
- 10.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phosphor-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 13.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoints signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahm SH, Park JH, Ko SI, Lee YR, Chung IS, Chung JH, Kang LW, Han YS. Knock-down of human MutY homolog (hMYH) decreases phosphorylation of checkpoint kinase 1 (Chk1) induced by hydroxyurea and UV treatment. BMB Rep. 2011;44:352–357. doi: 10.5483/BMBRep.2011.44.5.352. [DOI] [PubMed] [Google Scholar]
- 15.Luncsford PJ, Chang DY, Shi G, Bernstein K, Madabushi A, Patterson DN, Lu AL, Toth EA. A structural hinge in eukaryotic MutY homologues mediates catalytic activity and Rad9-Rad1-Hus1 checkpoint complex interactions. J Mol Biol. 2010;403:351–370. doi: 10.1016/j.jmb.2010.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/Human MutS homolog 6. J Biol Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 17.Pabla N, Ma Z, Mcllhatton MA, Fishel R, Dong Z. hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J Biol Chem. 2011;286:10411–10428. doi: 10.1074/jbc.M110.210989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 19.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 20.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 21.Flores-Rozas H, Clark D, Kolodner RD. Proliferation cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 22.Karpinich NO, Tafani M, Rothman RJ, Russo MA, Farber JL. The course of Etoposide-induced apoptosis from damage to DNA and p53 activation tomitochondrial release of cytochrome c. J Biol Chem. 2002;277:16547–16552. doi: 10.1074/jbc.M110629200. [DOI] [PubMed] [Google Scholar]
- 23.Rossi R, Lidonnici MR, Soza S, Biamonti G, Montecucco A. The dispersal of replication proteins after Etoposide treatment requires the cooperation of Nbs1 with ataxia telangiectasis Rad3-related/Chk1 pathway. Cancer Res. 2006;66:1675–1683. doi: 10.1158/0008-5472.CAN-05-2741. [DOI] [PubMed] [Google Scholar]
- 24.Xia Y, Ongusaha P, Lee SW, Liou YC. Loss of Wip1 sensitizes cells to stress- and DNA damage-induced apoptosis. J Biol Chem. 2009;284:17428–17437. doi: 10.1074/jbc.M109.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102:677–682. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]