Abstract
Background
Myocardial ischemia can be developed into more serious diseases. Early Detection of the ischemic syndrome in electrocardiogram (ECG) more accurately and automatically can prevent it from developing into a catastrophic disease. To this end, we propose a new method, which employs wavelets and simple feature selection.
Methods
For training and testing, the European ST-T database is used, which is comprised of 367 ischemic ST episodes in 90 records. We first remove baseline wandering, and detect time positions of QRS complexes by a method based on the discrete wavelet transform. Next, for each heart beat, we extract three features which can be used for differentiating ST episodes from normal: 1) the area between QRS offset and T-peak points, 2) the normalized and signed sum from QRS offset to effective zero voltage point, and 3) the slope from QRS onset to offset point. We average the feature values for successive five beats to reduce effects of outliers. Finally we apply classifiers to those features.
Results
We evaluated the algorithm by kernel density estimation (KDE) and support vector machine (SVM) methods. Sensitivity and specificity for KDE were 0.939 and 0.912, respectively. The KDE classifier detects 349 ischemic ST episodes out of total 367 ST episodes. Sensitivity and specificity of SVM were 0.941 and 0.923, respectively. The SVM classifier detects 355 ischemic ST episodes.
Conclusions
We proposed a new method for detecting ischemia in ECG. It contains signal processing techniques of removing baseline wandering and detecting time positions of QRS complexes by discrete wavelet transform, and feature extraction from morphology of ECG waveforms explicitly. It was shown that the number of selected features were sufficient to discriminate ischemic ST episodes from the normal ones. We also showed how the proposed KDE classifier can automatically select kernel bandwidths, meaning that the algorithm does not require any numerical values of the parameters to be supplied in advance. In the case of the SVM classifier, one has to select a single parameter.
Keywords: Myocardial ischemia, Discrete wavelet transform, Kernel density estimation, Support vector machine, QRS complex detection, ECG baseline wandering removal
Background
Coronary artery disease is one of the leading causes of death in modern world. This disease mainly results from atherosclerosis and thrombosis, and it manifests itself as coronary ischemic syndrome [1].
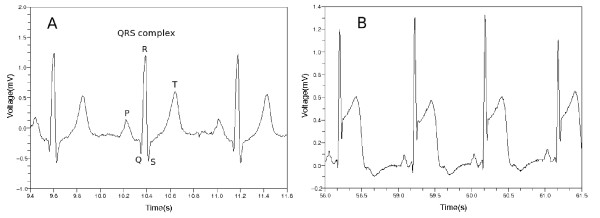
When a patient experiences coronary ischemic syndrome, his or her electrocardiogram (ECG) shows some peculiar appearances. Each segment of ECG can be divided into P, Q, R, S and T waves as shown in Figure 1 where QRS complex and T wave represent ventricular depolarization and repolarization, respectively. In most cases of normal ECG, the ST segment has the same electric potential as the PR segment. When myocardial ischemia is present, however, the electric potential of the ST segment is elevated or depressed with respect to the potential of the PR segment [1,2]. When ischemia occurs, the PR segment is altered, or the ST segment deviates from normal level. If the PR segment moved instead of the ST segment, this looks as if the ST segment itself were modified. This is because the PR segment provides a kind of reference voltage level [1].
Figure 1.
Normal ECG and ST segment elevation. (a) Normal ECG is divided into P, Q, R, S and T parts. The Q, R and S parts are called QRS complex in total. (b) This ECG waveform shows ST segment elevation.
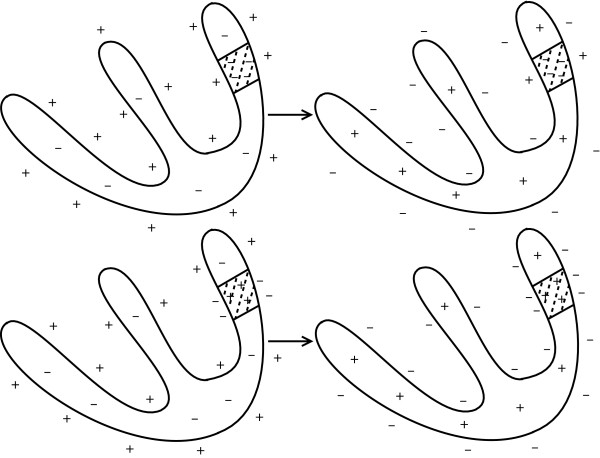
The ST segment deviation is mainly due to injury current in myocardial cells [1]. If the coronary artery becomes blocked by blood clot, some myocytes are affected to be unresponsive to depolarization, or to repolarize earlier than adjacent myocytes. In this case, voltage gradient can occur in the myocytes, and this comes to appear as ST-segment deviation in ECG [1]. Figure 2 shows two cases when the voltage level of the ST segment deviates from its normal position. The left column of the figure shows the distribution of electric charges around myocytes when the heart is in resting state. This is related to the PR segment in ECG. The right column shows the distribution of electric charges right after the ventricles contracted. This is related to the QRS complex and the ST segment in ECG. The shaded region represents the area being affected by myocardial ischemia. In the case of the upper row in Figure 2, there is no voltage gradient at first. After the ventricles contracted, however, the voltage gradient comes to appear because the injured area did not respond to electric depolarization. In the second case of the bottom row, there is no voltage gradient right after the ventricles contracted. In the left figure, however, there was initial voltage gradient, and this makes the PR segment to be modified. The PR segment acts as a reference voltage level when we judge whether the ST segment deviated from normal position. The modified PR segment makes us conclude that there was a ST segment deviation [1].
Figure 2.
Cause of ST segment deviation [[1]]. Left column shows distribution of electric charges before the ventricles contracts. The right column shows the charge distribution after the ventricles contracted. Shaded area represents that the area was affected by ischemia.
There are several approaches to detect ischemic ST deviations. Some researchers used the entropy. Rabbani et al. used the fact that signal perturbation of normal people is lower than the perturbation of ischemic patients. They computed entropy measure of wavelet subband of ECG signal, and classified the ECG by examining which signal exhibited a more chaotic perturbation [3]. Lemire et al. calculated signal entropy at various frequency levels. They computed the entropy in each wavelet scale [4]. Some used adaptive neuro-fuzzy inference system. Pang et al. used Karhunen-Loève transform to extract several feature values. They classified ECG signal by an adaptive neuro-fuzzy inference system [5]. Tonekabonipour et al. used multi-layer perceptron and radial basis function to detect ischemic episode. They classified ECG signals by adaptive neuro-fuzzy network [6]. There are many papers which used artificial neural network. Stamkopoulos et al. used nonlinear principal component analysis to analyze complex data. They classified ECG signal by radial basis function neural network [7]. Maglaveras et al. used neural network optimized with a backpropagation algorithm [8]. Afsar et al. used Karhunen-Loève transform to find feature values, and classified an input ECG by using a neural network [9]. Papaloukas et al. used artificial neural network which was trained by Bayesian regularization method [10]. There are papers studied some other approaches. Bulusu et al. determined morphological features of ECG, and classified the ECG data by support vector machine. Andreao et al. used hidden Markov models to analyze ECG segments. They detected ischemia episode by using median filter and linear interpolation [11]. Faganeli and Jager tried to distinguish ischemic ST episode and non-ischemic ST episode caused by heart rate change. To this end, they computed heart rate values, Mahanalobis distance of Karhunen-Loève transform coefficients and Legendre orthonormal polynomial coefficients [12]. Exarchos et al. used decision tree. They formed decision rules comprising specific thresholds, and developed a fuzzy model to classify ischemic ECG signals [13]. Garcia et al. considered root mean square of difference between the input signal and the average signal composed of first 100 beats. They adopted an adaptive amplitude threshold to classify ECG signal [14]. Murugan and Radhakrishnan used ant-miner algorithm to detect ischemic ECG beats. They calculated several feature values such as ST segment deviation from input ECG signal [15]. Bakhshipour et al. analyzed coefficients resulted from wavelet transform. They examined the relative quotient of the coefficients at each decomposition level of the wavelet transform [16].
We approached this problem by extracting feature values from a ECG waveform. We first found time positions of QRS complexes, and then determined values of the three features. We calculated the feature values for each heart beat, and averaged their values in five successive beats. After that, we classified them by the methods of kernel density estimation and support vector machine.
We showed techniques of removing baseline wandering and detecting time positions of QRS complexes by discrete wavelet transform. With these explicit methods of dealing with ECG, we could discriminate ischemic ST episode from normal ECG. We did not adopt implicit methods such as artificial neural networks or decision trees, because we considered it was important to utilize explicit features for processes of decision making. The artificial neural network has a kind of black box nature in its hidden layers [17], and a decision tree is apt to include several numerical thresholds [13].
Methods
Materials
We used the European ST-T database from Physionet. European ST-T database has 90 records which are two-channel and each two hours in duration [18,19]. Each record in this database has a different number of ST episodes. Overall there are 367 ischemic ST episodes in the database. Sampling frequency of each ECG data is 250 Hz.
We excluded 5 records because these had some problems. The records e0133, e0155, e0509, and e0611 had no ischemic ST episodes. The record e0163 had so limited ST episode whose length was just 31 seconds.
Removing baseline wandering
The ST segments in ECG can be strongly affected by baseline wandering [20]. Main causes of the baseline wandering are respiration and electrode impedance change due to perspiration [20,21]. The frequency content of the baseline wandering is usually in a range below 0.5 Hz [20,21].
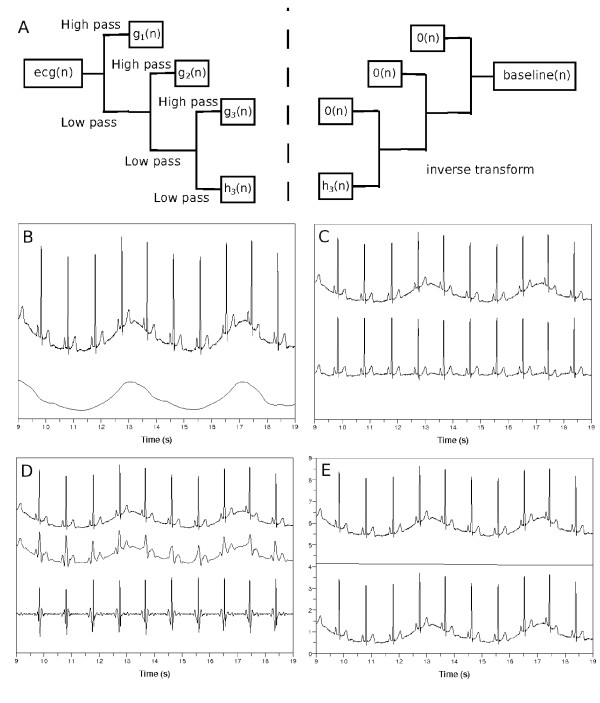
We use discrete wavelet transform to remove baseline wandering in ECG. We transform signal vector into two sequences of coefficients, approximation and detail coefficients sequences [22]. We do this in each step in an iterative fashion, until we get an input signal whose length is smaller than the length of the filter which characterizes the wavelet. In our case, we used Daubechies8 wavelet with filter length of 8. The resulting approximation coefficient sequence becomes the input signal to the next discrete wavelet transform as shown in Figure 3(a) [22].
Figure 3.
Removing baseline wandering in ECG. (a) Discrete wavelet transform of ecg(n) to find coefficient sequences hk(n),gk(n),gk−1(n),· · ·,g1(n). The means zero sequence. (b) Top: input ECG, ecg(n), bottom: wandering baseline in ECG, baseline(n). (c) Top: ecg(n), bottom: ecg(n)-baseline(n). When k is (d) too small or (e) too large, top: ecg(n), middle: baseline(n), bottom: ecg(n)-baseline(n).
In each step, the coefficient sequence implies a band of frequencies. If the sampling frequency of a discrete ECG signal ecg(n) is x, we can determine a continuous and band-limited signal within frequency limits of by Nyquist sampling theorem [23]. Therefore if we have transformed the input signal ecg(n) into the approximation coefficient sequence h1(n) and detail coefficient sequence g1(n), then the frequency content of g1(n) is from to , and the frequency content of h1(n) is below . In this regard, if we have transformed the ecg(n) into the approximation coefficient sequence hk(n), and the detail coefficient sequences gk(n),gk−1(n),· · ·,g1(n), the frequency contents of gk(n),gk−1(n),· · ·,g1(n) become respectively [24,25].
To remove baseline wandering, we should choose appropriate wavelet scale. We follow argument similar to that presented by Arvinti et al. except that they used stationary wavelet transform instead of its discrete counterpart [26]. We remove signal components whose frequency content is less than 1/2 Hz [20,21]. If we have transformed the ECG signal ecg(n) into coefficient sequences hk(n),gk(n),gk−1(n),· · ·,g1(n), the frequency contents of hk(n) and gk(n) become and respectively, where x is the sampling frequency. If we choose k as , the frequency content of the approximation coefficient sequence hk(n) becomes less than 1/2 Hz. Thus, we assign zero sequence to all the detail coefficient sequences gk(n),gk−1(n),· · ·,g1(n), and calculate inverse transform of to form the baseline(n) in the bottom of Figure 3(b). If we subtract baseline(n) from ecg(n), we obtain the flattened signal like the one shown in Figure 3(c).
If we select a wrong wavelet scale k to find coefficient sequences of ecg(n), we obtain disappointing results. The flattened signal in Figure 3(c) is obtained when k is , where 250 is the sampling frequency expressed in Hz. When select k=4 to use h4(n),g4(n),g3(n),g2(n),g1(n), we obtain a plot in Figure 3(d). The middle waveform, baseline(n), resulted from the inverse discrete wavelet transform of . This middle waveform is too detailed, so the bottom waveform ecg(n)-baseline(n) was negatively affected. When we select k=12, see Figure 3(e), the bottom waveform was not different from the input waveform ecg(n).
We adopt a discrete wavelet transform to retain the details of the ECG waveform because filtering by some cut-off frequency can deteriorate the quality of the ECG waveforms [27].
Detecting QRS complexes
We have to select an appropriate wavelet scale to capture proper time positions of QRS complexes. We will deal with only the flattened ECG waveform ecg(n)-baseline(n) referred in the previous section. We will denote it as fecg(n).
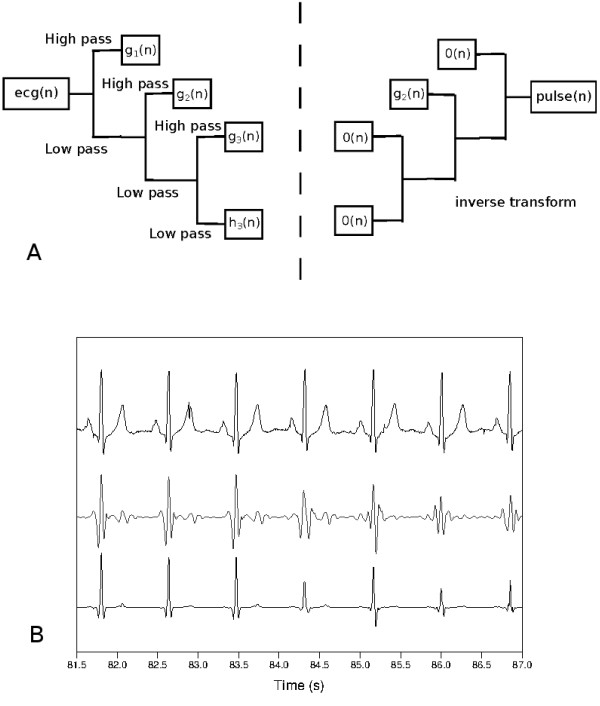
First, we determine the sequences of wavelet coefficients of the fecg(n), obtaining hk(n),gk(n),gk−1(n),· · ·,g1(n) where , x is sampling frequency. We assign zero to all the coefficient sequences except one, gj(n). Then, we calculate inverse transform of (approximation coefficients), (detail coefficients, onward), to obtain pulse(n). To find a protruding segment, that is, a QRS complex, we compute the score for each wavelet scale j,
| (1) |
We select the wavelet scale j which produces the largest drop of scorej−scorej + 1(j≥2). The bottom waveform in Figure 4(b) shows the time positions of QRS complexes when selecting this suitable wavelet scale.
Figure 4.
Selection of wavelet scale to find the time positions of QRS complexes. (a) Discrete wavelet transform and inverse transform. (b) Top: A flattened ECG waveform, fecg(n). Middle: waveform resulted from the inverse transform, pulse(n). Bottom: fecg(n).
After finding the locations of QRS complexes, we choose QRS onset and offset points in each QRS complex. We search QRS onset point in backward direction from a peak point in each QRS complex. We take the QRS onset point if the point is at the place of changing direction of rising and falling of fecg(n) twice. In the same way, we take the QRS offset point in forward direction from the peak point.
Algorithm 1 shows a process of removing baseline wandering and detecting QRS complexes.
Algorithm 1
A procedure to find time positions of QRS onset, peak and offset points. This procedure includes the method of removing baseline wandering in ECG. nBeats stands for the number of QRS peaks. It is the length of the sequences idx_QRS_Onset(n), idx_QRS_Peak(n) and idx_QRS_Offset(n). Input: Sampling_Hz, ecg(n)Output: idx_QRS_Onset(n), idx_QRS_Peak(n), idx_QRS_Offset(n)k Discrete wavelet transform (DWT) of ecg(n) into hk(n),gk(n),gk−1(n),· · ·,g1(n)fori=1 to kdogi(n)
Discrete wavelet transform (DWT) of ecg(n) into hk(n),gk(n),gk−1(n),· · ·,g1(n)fori=1 to kdogi(n) {// means zero sequence.} end for Inverse wavelet transform (IDWT) of hk(n),gk(n),gk−1(n),· · ·,g1(n) into baseline(n) DWT of fecg(n) into hk(n),gk(n),gk−1(n),· · ·,g1(n)hk(n)
{// means zero sequence.} end for Inverse wavelet transform (IDWT) of hk(n),gk(n),gk−1(n),· · ·,g1(n) into baseline(n) DWT of fecg(n) into hk(n),gk(n),gk−1(n),· · ·,g1(n)hk(n) >gk(n)
>gk(n) fori=1 to k−1do
fori=1 to k−1do
(n) gi(n)
gi(n)
gi(n)
end forfori=1 to k−1do
gi(n) (n)
(n)
IDWT of hk(n),gk(n),gk−1(n),· · ·,g1(n) into pulse(n)
gi(n)
end for chosen_scale 
 IDWT of hk(n),gk(n),gk−1(n),· · ·,g1(n) into pulse(n) Make idx_QRS_Peak(n) by searching for local maxima of needle(n)fori=1 to nBeatsdoifthen
IDWT of hk(n),gk(n),gk−1(n),· · ·,g1(n) into pulse(n) Make idx_QRS_Peak(n) by searching for local maxima of needle(n)fori=1 to nBeatsdoifthen
whiledo
end while
whiledo
end while
idx_QRS_Onset(i) idx_QRS_Peak(i)−j
idx_QRS_Peak(i)−j
whiledo
end while
whiledo
end while
idx_QRS_Offset(i) idx_QRS_Peak(i) + j
idx_QRS_Peak(i) + j
else
· · ·{//When QRS complex protrudes downward, code is same with reversing directions of inequality signs.}
end ifend for
Feature formation for classification problems
We deal with the flattened waveform, fecg(n), to obtain the values of the features. We take voltage level of QRS onset point as the reference from which we measure voltage deviation [2,28]. We denote the mean value of electric potentials at QRS onset points as . We consider this value as an effective zero voltage, so we measure voltage deviation from the .
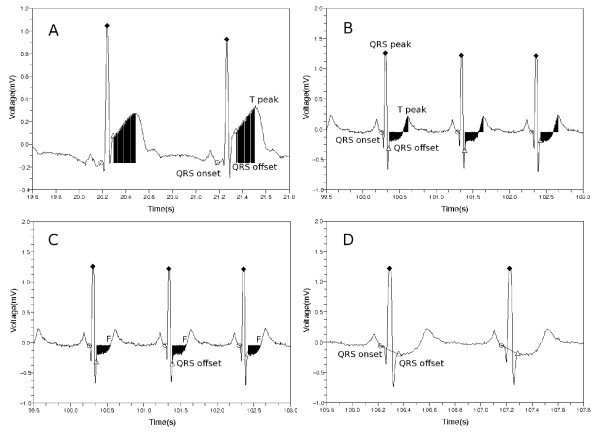
To form the first feature, we sum up all the voltage deviation from QRS offset point to T wave peak point as shown in Figure 5(a) and (b).
Figure 5.
Features used in the classification process. Area between QRS offset and T peak with respect to the reference mean voltage in cases of (a) ST segment elevation and (b) ST segment depression. (c) Normalized and signed sum of voltage deviations from the QRS offset to the first point F at which voltage becomes equal to the reference voltage. (d) Slope from the QRS onset point to the QRS offset point. Markers ○, 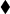 and
and  designate QRS onset, peak and offset points respectively.
designate QRS onset, peak and offset points respectively.
| (2) |
The second feature is similar to the first feature with an exception of the ending position of the sum. We terminate the summation as we reach the first point, F, at which the voltage becomes equal to the reference voltage , see Figure 5. When doing this, we add the signed values of the voltage deviation to find whether the area is lower or higher with respect to the reference voltage. Then we divide the value by the voltage at QRS peak point. The second feature value is given as follows.
| (3) |
The third feature is a slope from the QRS onset point to the QRS offset point.
| (4) |
We calculate these three feature values for each heart beat. Then we average these values in five successive beats, and arrange the three mean values as .
Algorithm 2 shows the pseudo-code of computing feature values.
Algorithm 2
A procedure to compute feature values. nBeatsdenotes the number of QRS peaks. It is the length of the sequences idx_QRS_Onset(n), idx_QRS_Peak(n) and idx_QRS_Offset(n). ncl is equal to nBeats/5. Input:fecg(n), idx_QRS_Onset(n), idx_QRS_Peak(n), idx_QRS_Offset(n)Output: {//cl can be S (ST episode) or N (normal).} mean_idx_diff1 mean_idx_diff2
mean_idx_diff2 {//mean_idx_diff1and mean_idx_diff2are truncated into integers.}
{//mean_idx_diff1and mean_idx_diff2are truncated into integers.}  fori=1 to nBeatsdok
fori=1 to nBeatsdok idx_QRS_Peak(i) + mean_idx_diff2m
idx_QRS_Peak(i) + mean_idx_diff2m idx_QRS_Peak(i)−mean_idx_diff1end forfori=1 to nBeats/5end for
idx_QRS_Peak(i)−mean_idx_diff1end forfori=1 to nBeats/5end for
Classification by kernel density estimation
We approximate probability density at a point by considering the other points. Let us assume we have d-dimensional points . We can estimate the probability density at a point y as where V is a small volume around y, and K is a number of enclosed points in the volume V [29]. We replace the term by d-dimensional Gaussian function as follows [30].
| (5) |
If we assume that the covariance matrix 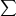 is a diagonal matrix with each diagonal element , the probability density at the point y is given as follows [31].
is a diagonal matrix with each diagonal element , the probability density at the point y is given as follows [31].
| (6) |
We classify a test point by examining posterior probabilities in which the test point belongs to two classes, normal or ischemic ST episode. We assume we have nS points , and nN points . The first and the second set designate training sets of ischemic ST episode and normal part, respectively. Each point is described by three components .
We compute posterior probability in which the test point ybelongs to each class by Bayes’ theorem as follows [29].
| (7) |
The prior probability is given as or . The likelihood and reads as
| (8) |
| (9) |
The quantities and are called kernel bandwidths. We calculate these bandwidths for each class (N or S) and component . These kernel bandwidths impact accuracy of kernel density estimation [32].
We have ncltraining points where cl denotes class, N (normal) or S (ischemic ST episode). For each component of the feature vector, we calculate the mean value of differences as follows.
| (10) |
We choose half of the mean, , as kernel bandwidth for each class cl (N or S), and component i.
Classification with the use of support vector machine
Let us assume we have ncltraining points . Each point is described as in a three-dimensional feature space. We construct support vector machine classifier by solving the following optimization problem [33]
| (11) |
The target label is specified as 1 (normal) or -1 (ischemic ST episode). The parameter C controls the trade-off between the slack variable (ξi) penalty and the margin (wT·w) penalties [29]. The dual form of the above classifier reads as follows
| (12) |
where the matrix H is expressed as [33]. When we classify a new pattern y, we examine decision function, . Whenever the input training set was changed, we varied the parameter C to find its value which produced the highest classification rate.
Experiments setting
We used kernel density estimation and support vector machine methods to evaluate the proposed approach. We completed the experiment for each channel and record available in the European ST-T database. First, we trained the classifier based on a subset of ST episodes and normal ECG. Then we tested how well the feature values discriminated the two classes, ST episode and normal. When we formed the ST episode data, we used all the ischemic ST episodes except ST deviations data resulted from non-ischemic causes such as position related changes in the electrical axis of the heart. To preserve balance between ST episode and normal ECG data, we collected normal data from the beginning of each record as much as the amount of ST episode data. When dividing the data into training and test sets, we assigned one tenth of data to the training data, and the rest to the test data. In the cases of e0106 lead 0, e0110, e0136, e0170, e0304, e0601, and e0615 records, we constructed the training data of one third of all data and test data of two thirds because these records had much small ischemic ST episode data. To avoid ambiguous region between ischemic ST episode and normal ECG, we removed 10 seconds amount of ECG data from each side of the boundary.
When we classified a test set , four quantities were computed: true positive (TP), false negative (FN), false positive (FP), and true negative (TN). TP is a number of ischemic events correctly detected. FN is a number of erroneously rejected (missed) ischemic events. FP is a number of non-ischemic, that is, normal parts which the classifier erroneously detected as ischemic events. TN is a number of normal parts which our classifier correctly rejected as non-ischemic events [34]. These are numbers of corresponding yi points which were obtained by averaging three feature values of successive five beats in Algorithm 2. The sensitivity and specificity are expressed in a usual fashion, Se=TP/(TP + FN) and Sp=TN/(TN + FP) respectively [6].
We tested the classifiers by counting how many ST episodes were correctly caught, out of 367 episodes in the 85 records of European ST-T database. For an interval of ischemic ST episode data, we formed n test points from the data (Algorithm 2), and classified each test point and then counted numbers of two classes, “ischemic” and “normal”. If the number of class “ischemic” was larger than n/2, we declared the interval to be an ischemic ST episode. The experiments were completed for 367 ischemic ST episodes.
We compared the results of kernel density estimation (KDE) and support vector machine (SVM) methods with those formed by artificial neural network (ANN). The corresponding ANN classifier exhibits the following topology. The input layer has three nodes which accept feature1feature2 and feature3respectively. The output layer has two nodes which have target values and in the cases of “ischemia” and “normal” classes, respectively. We initialized bias weights as 0, and assigned random values between -1.0 and 1.0 to the weights of the network. The learning was carried out by running the backpropagation method [17] for 3000 iterations. We used a sigmoid activation function and set learning rate 0.01. We adopted various topologies of hidden layers such as and where the number in each parenthesis represents a number of nodes in the corresponding hidden layer. We used stochastic (incremental) gradient descent method to alleviate some drawbacks of the standard gradient descent method, see [17].
Results
KDE with various kernels
We can use various kernels in kernel density estimation. If we have training points and a test point y, the probability density at y is given as follows [35].
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
| (18) |
Here kG, kR, kE, kB, kTriw and kTria are constants, and is given as because we use three feature values. The indicator function is given as follows.
| (19) |
Table 1 shows classification results for various kernels. In all cases we used Daubechies8 wavelet to produce training and test sets. We took each bandwidth for class cl, ischemic or normal, and 1≤i≤3. The “detect” means how many ST episodes our classifier correctly detected, out of total 367 episodes. The “factor” in this table specifies how we multiplied on the to form the kernel bandwidth . We varied this factor from 0.1 to 3.0, and selected the one for which a sum of sensitivity and specificity values attains a maximum. Because the Gaussian kernel produced best results, in the sequel we will use the Gaussian kernel. Table 2 shows the results with respect to various kernel bandwidths.
Table 1.
Classification results with respect to various kernels
| | ||||||||
|---|---|---|---|---|---|---|---|---|
| Kernel | Factor | Se. | Sp. | TP | TN | FP | FN | Detect |
| Gaussian |
0.5 |
0.939 |
0.912 |
27600 |
21441 |
2075 |
1794 |
349 |
| Rectangular |
1.5 |
0.892 |
0.913 |
26209 |
21460 |
2056 |
3185 |
329 |
| Epanechnikov |
1.7 |
0.904 |
0.915 |
26583 |
21522 |
1994 |
2811 |
335 |
| Byweight |
2.0 |
0.912 |
0.916 |
26794 |
21533 |
1983 |
2600 |
333 |
| Triweight |
2.1 |
0.916 |
0.916 |
26923 |
21542 |
1974 |
2471 |
336 |
| Triangular | 1.8 | 0.908 | 0.917 | 26680 | 21554 | 1962 | 2714 | 334 |
Table 2.
Classification results of Gaussian kernels with respect to various bandwidths
| | |||||||
|---|---|---|---|---|---|---|---|
| Factor | Se. | Sp. | TP | TN | FP | FN | Detect |
| 0.2 |
0.943 |
0.867 |
27728 |
20399 |
3117 |
1666 |
353 |
| 0.3 |
0.944 |
0.893 |
27745 |
20996 |
2520 |
1649 |
352 |
| 0.4 |
0.942 |
0.905 |
27697 |
21279 |
2237 |
1697 |
351 |
| 0.5 |
0.939 |
0.912 |
27600 |
21441 |
2075 |
1794 |
349 |
| 0.6 |
0.934 |
0.915 |
27453 |
21526 |
1990 |
1941 |
343 |
| 0.7 |
0.929 |
0.916 |
27318 |
21550 |
1966 |
2076 |
338 |
| 0.8 | 0.924 | 0.916 | 27148 | 21529 | 1987 | 2246 | 337 |
Results for KDE, SVM and ANN with various wavelets
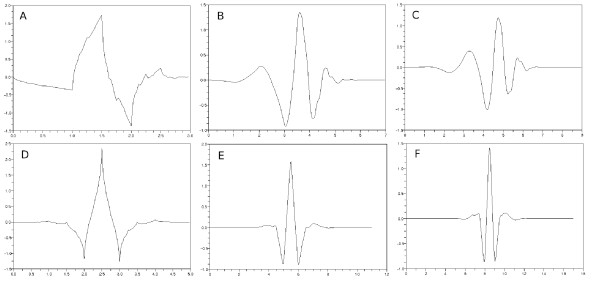
We examined the classifiers to find out how their performance depends on the mother wavelets which were used to produce training and test sets in Algorithm 1. We used 7 wavelets, Haar, Daubechies4, Daubechies8, Daubechies10, Coiflet6, Coiflet12 and Coiflet18 [22,36]. The number forming a part of the name of each wavelet designates the length of filter which characterizes corresponding wavelet. Figure 6 shows selected shapes of wavelet functions except for the Haar wavelet which is given as
Figure 6.
Shapes of various wavelets. (a) Daubechies4, (b) Daubechies8, (c) Daubechies10, (d) Coiflet6, (e) Coiflet12 and (f) Coiflet18.
| (20) |
Table 3 shows the classification results obtained for KDE. The kernel bandwidth is expressed as for each class cl and 1≤i≤3. We used Gaussian kernel.
Table 3.
Classification results for KDE with respect to various wavelets
| | |||||||
|---|---|---|---|---|---|---|---|
| Wavelet | Se. | Sp. | TP | TN | FP | FN | Detect |
| Haar |
0.915 |
0.893 |
25906 |
20245 |
2420 |
2418 |
339 |
| Daubechies4 |
0.936 |
0.906 |
27488 |
21130 |
2186 |
1886 |
343 |
| Daubechies8 |
0.939 |
0.912 |
27600 |
21441 |
2075 |
1794 |
349 |
| Daubechies10 |
0.942 |
0.916 |
27862 |
21585 |
1969 |
1710 |
348 |
| Coiflet6 |
0.934 |
0.900 |
28586 |
21837 |
2430 |
2027 |
349 |
| Coiflet12 |
0.932 |
0.914 |
27846 |
21757 |
2041 |
2045 |
349 |
| Coiflet18 | 0.937 | 0.919 | 27721 | 21612 | 1911 | 1859 | 349 |
Table 4 shows the classification results for the KDE with respect to various bandwidths and wavelets. The first column for each wavelet item represents the sum of sensitivity and specificity. The second column shows how many ST episodes were correctly detected. We used the kernel bandwidths for each class cl and 1≤i≤3. The sum of sensitivity and specificity becomes maximum when the bandwidth is around .
Table 4.
Classification results for KDE versus selected values of bandwidths and types of wavelets
| | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor | Haar | Daub4 | Daub8 | Daub10 | Coif6 | Coif12 | Coif18 | |||||||
| 0.2 |
1.770 |
348 |
1.797 |
348 |
1.811 |
353 |
1.817 |
355 |
1.794 |
356 |
1.812 |
360 |
1.825 |
354 |
| 0.3 |
1.797 |
350 |
1.825 |
350 |
1.837 |
352 |
1.843 |
356 |
1.822 |
354 |
1.833 |
357 |
1.848 |
353 |
| 0.4 |
1.808 |
345 |
1.838 |
344 |
1.847 |
351 |
1.856 |
353 |
1.833 |
354 |
1.844 |
355 |
1.857 |
354 |
| 0.5 |
1.808 |
339 |
1.842 |
343 |
1.851 |
349 |
1.859 |
348 |
1.834 |
349 |
1.846 |
349 |
1.856 |
349 |
| 0.6 |
1.806 |
336 |
1.840 |
339 |
1.849 |
343 |
1.857 |
343 |
1.832 |
345 |
1.843 |
346 |
1.854 |
346 |
| 0.7 |
1.800 |
331 |
1.836 |
336 |
1.846 |
338 |
1.853 |
338 |
1.829 |
340 |
1.837 |
341 |
1.849 |
344 |
| 0.8 | 1.792 | 325 | 1.829 | 334 | 1.839 | 337 | 1.848 | 335 | 1.824 | 334 | 1.831 | 339 | 1.842 | 340 |
Table 5 shows the classification results obtained for SVM. The parameter C controls the trade-off between the slack variable (ξi) penalty and the margin (wT·w) penalties. We examined the classification accuracy versus the values of C changing from 0.1 to 300.0 in step of 0.1, and selected the one that made the sum of sensitivity and specificity maximal.
Table 5.
Classification results for SVM for various wavelets
| | ||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelet | C | Se. | Sp. | TP | TN | FP | FN | Detect |
| Haar |
291.3 |
0.924 |
0.907 |
26163 |
20547 |
2118 |
2161 |
345 |
| Daubechies4 |
242.9 |
0.937 |
0.923 |
27527 |
21519 |
1797 |
1847 |
349 |
| Daubechies8 |
245.5 |
0.941 |
0.923 |
27658 |
21712 |
1804 |
1736 |
355 |
| Daubechies10 |
174.3 |
0.943 |
0.927 |
27894 |
21838 |
1716 |
1678 |
349 |
| Coiflet6 |
288.2 |
0.933 |
0.918 |
28571 |
22284 |
1983 |
2042 |
348 |
| Coiflet12 |
52.8 |
0.929 |
0.918 |
27757 |
21858 |
1940 |
2134 |
348 |
| Coiflet18 | 23.4 | 0.936 | 0.927 | 27692 | 21805 | 1718 | 1888 | 352 |
Table 6 shows the classification results obtained by ANN. The number in parenthesis represents the number of nodes in the corresponding hidden layer. The first, second and third column express sensitivity, specificity and the “detect” respectively. We experimented 10 times, and averaged the results because we obtained different results each time due to the random initialization of weights.
Table 6.
Results for ANN classifiers with respect to various wavelets and sizes of hidden layers
| | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wavelet | ||||||||||||
| Haar |
0.851 |
0.920 |
311.8 |
0.866 |
0.916 |
319.2 |
0.867 |
0.917 |
319.1 |
0.867 |
0.916 |
320.7 |
| Daub4 |
0.866 |
0.932 |
304.2 |
0.881 |
0.930 |
313.7 |
0.880 |
0.932 |
315.2 |
0.881 |
0.931 |
317.8 |
| Daub8 |
0.864 |
0.931 |
307.2 |
0.878 |
0.929 |
317.9 |
0.875 |
0.931 |
319.4 |
0.880 |
0.932 |
319.4 |
| Daub10 |
0.866 |
0.939 |
312.5 |
0.882 |
0.935 |
321.8 |
0.885 |
0.935 |
325.5 |
0.882 |
0.937 |
325.1 |
| Coif6 |
0.848 |
0.930 |
311.2 |
0.868 |
0.920 |
319.3 |
0.863 |
0.923 |
319 |
0.872 |
0.927 |
321 |
| Coif12 |
0.855 |
0.936 |
310.5 |
0.868 |
0.933 |
318.2 |
0.866 |
0.935 |
319.6 |
0.874 |
0.935 |
319.8 |
| Coif18 | 0.874 | 0.938 | 311.8 | 0.883 | 0.937 | 319.2 | 0.884 | 0.936 | 320.2 | 0.886 | 0.937 | 323.5 |
Tables 35 and 6 show that the Daubechies8 and Daubechies10 wavelets give us superior results. The shapes of these two wavelets are similar to typical ECG waveforms [37,38]. From now on, we use the Daubechies8 wavelet exclusively.
Effects of baseline wandering in ECG
Table 7 shows the classification results by KDE, SVM and ANN when we did not remove baseline wandering in ECG. If we compare this table with the Tables 3, 5 and 6, we get to know it is essential to remove baseline wandering in Algorithm 1. In the Table 7, we selected the kernel bandwidths in the KDE classifier as , . We used the ANN classifier with sizes of layers expressed as . The results of ANN were obtained by averaging results for 10 repetition of the experiments. The parameter C of the SVM classifier was 297.9.
Table 7.
Classification results for KDE, SVM and ANN without removal of baseline wandering
| | |||||||
|---|---|---|---|---|---|---|---|
| Se. | Sp. | TP | TN | FP | FN | Detect | |
| KDE |
0.852 |
0.837 |
22132 |
17211 |
3342 |
3839 |
328 |
| SVM |
0.870 |
0.835 |
22605 |
17161 |
3392 |
3366 |
326 |
| ANN | 0.785 | 0.827 | 20388.8 | 17005.7 | 3547.3 | 5582.2 | 296.1 |
If we use unsuitable wavelet scale like the one in Figure 3 to remove baseline wandering, it becomes difficult to obtain good results. As the sampling frequency was 250 Hz, we selected the wavelet scale in Algorithm 1. Table 8 shows the classification results when wrong wavelet scales were selected. The kernel bandwidth setting in KDE and layer composition of ANN classifier were same as the Table 7. The middle row of wavelet scale 8 in Table 8 was our choice in Algorithm 1. Each entry in the row of wavelet scale 8 has counterparts in “Daubechies8” rows in Tables 3, 5 and 6.
Table 8.
Classification results for KDE, SVM and ANN with incorrectly selected wavelet scales to remove baseline wandering
| | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
KDE |
SVM |
ANN |
|||||||
| Se. | Sp. | Detect | Trade-off | Se. | Sp. | Detect | Se. | Sp. | Detect | |
| scale 6 |
0.851 |
0.785 |
319 |
288.2 |
0.842 |
0.818 |
318 |
0.748 |
0.864 |
277.5 |
| scale 7 |
0.931 |
0.905 |
344 |
232.8 |
0.932 |
0.915 |
349 |
0.876 |
0.933 |
320.7 |
| scale 8 |
0.939 |
0.912 |
349 |
245.5 |
0.941 |
0.923 |
355 |
0.875 |
0.931 |
319.4 |
| scale 9 |
0.929 |
0.906 |
347 |
110.4 |
0.930 |
0.918 |
350 |
0.859 |
0.918 |
320.7 |
| scale 10 | 0.921 | 0.896 | 340 | 97.5 | 0.916 | 0.907 | 343 | 0.838 | 0.896 | 313.2 |
Effects of simulated noise
We examined performance of the classifiers when we added simulated noise into the original ECG signal. We modeled the noise as the sum of wandering baseline and AC power line 60 Hz noise.
Let us assume we have original signal data, ecg(i). First, we compute mean value and standard deviation of the ECG signal as and . Then we form a new signal ecg″(i) by
| (21) |
where a is an amplification factor and b is an angular frequency of the added baseline. Here Samp_Freq means sampling frequency which was 250 Hz in our case. We varied a from 0.1 to 1.0 in step of 0.1, and selected b to be equal to 2, 4 or 6.
Figure 7 shows the original ECG and its noise-impacted version. Tables 9, 10 and 11 show the experimental results for the noisy ECG signal. The first, second and third column in each b item represent the sensitivity, specificity and the “detect” respectively. The kernel bandwidth is set as for the KDE classifier. The layer composition of the ANN classifier was . The first column in each b item in Table 10 includes the trade-off parameter C which produced best results.
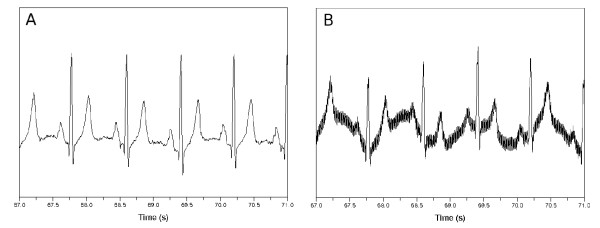
Figure 7.
ECG signal affected by synthetic noise. (a) Original signal. (b) Noise-affected signal when a is 1.0 and b is 6.0.
Table 9.
Classification results for KDE versus varying intensity of noise
| | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| a | b=2 | b=4 | b=6 | ||||||
| 0.1 |
0.937 |
0.903 |
346 |
0.934 |
0.902 |
346 |
0.933 |
0.904 |
346 |
| 0.2 |
0.933 |
0.893 |
346 |
0.927 |
0.892 |
341 |
0.916 |
0.879 |
337 |
| 0.3 |
0.929 |
0.884 |
342 |
0.915 |
0.873 |
340 |
0.908 |
0.856 |
338 |
| 0.4 |
0.925 |
0.864 |
346 |
0.903 |
0.852 |
344 |
0.885 |
0.834 |
327 |
| 0.5 |
0.916 |
0.858 |
346 |
0.882 |
0.848 |
333 |
0.871 |
0.817 |
333 |
| 0.6 |
0.906 |
0.866 |
343 |
0.872 |
0.832 |
329 |
0.858 |
0.803 |
321 |
| 0.7 |
0.891 |
0.856 |
340 |
0.859 |
0.815 |
325 |
0.848 |
0.794 |
322 |
| 0.8 |
0.887 |
0.850 |
339 |
0.852 |
0.799 |
325 |
0.838 |
0.793 |
317 |
| 0.9 |
0.881 |
0.849 |
341 |
0.837 |
0.798 |
312 |
0.837 |
0.778 |
301 |
| 1.0 | 0.870 | 0.844 | 335 | 0.832 | 0.787 | 317 | 0.824 | 0.779 | 319 |
Table 10.
Classification results for SVM with varying intensity of the simulated noise
| | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b=2 | b=4 | b=6 | |||||||||
| 0.1 |
79.0 |
0.936 |
0.920 |
350 |
254.6 |
0.940 |
0.915 |
352 |
147.4 |
0.936 |
0.916 |
349 |
| 0.2 |
143.0 |
0.929 |
0.914 |
347 |
168.5 |
0.929 |
0.904 |
349 |
33.9 |
0.922 |
0.888 |
339 |
| 0.3 |
84.1 |
0.923 |
0.910 |
348 |
50.9 |
0.915 |
0.891 |
341 |
72.3 |
0.902 |
0.875 |
337 |
| 0.4 |
124.5 |
0.919 |
0.899 |
348 |
51.3 |
0.903 |
0.869 |
341 |
52.0 |
0.888 |
0.848 |
330 |
| 0.5 |
65.1 |
0.909 |
0.895 |
347 |
86.7 |
0.882 |
0.859 |
333 |
79.4 |
0.872 |
0.828 |
328 |
| 0.6 |
96.7 |
0.903 |
0.887 |
343 |
67.3 |
0.879 |
0.844 |
330 |
71.6 |
0.865 |
0.809 |
323 |
| 0.7 |
119.3 |
0.888 |
0.880 |
339 |
27.2 |
0.868 |
0.832 |
329 |
36.6 |
0.856 |
0.796 |
313 |
| 0.8 |
201.8 |
0.893 |
0.863 |
334 |
24.9 |
0.851 |
0.829 |
320 |
27.1 |
0.848 |
0.785 |
310 |
| 0.9 |
278.2 |
0.888 |
0.857 |
335 |
16.7 |
0.841 |
0.816 |
308 |
71.0 |
0.850 |
0.777 |
315 |
| 1.0 | 211.7 | 0.879 | 0.859 | 324 | 58.2 | 0.835 | 0.806 | 311 | 20.3 | 0.837 | 0.776 | 317 |
Table 11.
Classification results for ANN with varying intensity of the simulated noise
| | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| a | b=2 | b=4 | b=6 | ||||||
| 0.1 |
0.880 |
0.929 |
322.2 |
0.870 |
0.921 |
320.8 |
0.874 |
0.922 |
320.3 |
| 0.2 |
0.867 |
0.928 |
322.8 |
0.844 |
0.922 |
315.1 |
0.840 |
0.905 |
314.2 |
| 0.3 |
0.863 |
0.929 |
319.3 |
0.837 |
0.906 |
313.4 |
0.811 |
0.895 |
303.4 |
| 0.4 |
0.856 |
0.917 |
320.5 |
0.823 |
0.887 |
312.3 |
0.784 |
0.874 |
300.6 |
| 0.5 |
0.830 |
0.918 |
320.1 |
0.811 |
0.874 |
309.1 |
0.778 |
0.850 |
300.3 |
| 0.6 |
0.818 |
0.916 |
314.9 |
0.803 |
0.858 |
299.2 |
0.762 |
0.833 |
292 |
| 0.7 |
0.803 |
0.910 |
309.6 |
0.759 |
0.865 |
281.9 |
0.761 |
0.811 |
283.2 |
| 0.8 |
0.814 |
0.898 |
308.6 |
0.753 |
0.839 |
288.2 |
0.745 |
0.801 |
280.7 |
| 0.9 |
0.798 |
0.892 |
303.4 |
0.723 |
0.838 |
273.9 |
0.728 |
0.795 |
261.7 |
| 1.0 | 0.777 | 0.890 | 297.1 | 0.724 | 0.807 | 265.3 | 0.728 | 0.788 | 269.3 |
Comparison with others’ works
To compare our approach with others’ works, we tested the classifiers on 10 selected records, e0103, e0104, e0105, e0108, e0113, e0114, e0147, e0159, e0162 and e0206. Table 12 shows results of comparison. The papers by Papaloukas et al. [10], Goletsis et al. [39], Exarchos et al. [13] and Murugan et al. [15] in Table 12 dealt with the 10 records.
Table 12.
Results of comparative analysis
We used the Daubechies8 wavelet in Algorithm 1 to analyze the ECG waveform, and took the kernel bandwidths for the KDE classifier with Gaussian kernel. We used the SVM classifier with C=281.1.
Discussion
Table 1 showed how the classification results were dependent on various kernel functions in kernel density estimation. Gaussian kernel produced best results.
Tables 3, 4, 5 and 6 show how the classification results depend on mother wavelets used in Algorithm 1. Daubechies8 and Daubechies10 wavelets were best. Because we implemented wavelet transform program with the use of matrix multiplication, we selected Daubechies8 wavelet to reduce computational burden. Daubechies10 wavelet did not produce much better classification accuracy than Daubechies8 wavelet.
Tables 2 and 4 indicate that the choice of kernel bandwidths was reasonable. When we took the kernel bandwidths for class cl, 1≤i≤3, we obtained best results except for the case of Coiflet18 wavelet. Even in the case, the best parameter was close enough to . We maintained this choice in Tables 7, 8 and 9. In this way, we could automatically select 6 kernel bandwidths, and this exempted us from choosing any numerical parameters.
The SVM classifiers in Tables 5, 7 and 8 produced better results than the KDE classifiers, but they required us to determine optimal value of the parameter C. Whenever we used different wavelets on the same data set in Table 5, we had to choose different trade-off parameter C. This was also the case in Table 8 where we intentionally selected incorrect wavelet scales to remove baseline wandering.
Order of magnitude of feature3was very different from feature1and feature2. When we produced the feature values using Daubechies8 wavelet in Algorithm 1, mean values of , and were 7.327, 7.613 and 0.004, respectively. Thus we had to normalize the feature values to use them in classification. Even if the orders of magnitude of feature1, feature2 and feature3were very different, the equation of kernel density estimation included a term . Furthermore each operand in the sum comes in the form of , normalization by kernel bandwidth. We thought these would be helpful to overcome the difference of order of magnitude between feature1, feature2 and feature3. This was a main driving force to adopt the kernel density estimation.
We implemented the KDE and ANN classifier in C language for ourselves. For SVM classifier, we used libsvm library [33]. We compiled the programs with gcc and g++ without using any SIMD (single instruction multiple data) math library. Total amount of ECG text files which we used in our analysis was 200.4 MB. This amount is just about voltage information not including time information. When we ran our programs to process the ECG text files in Pentium4 3.2 GHz CPU, it took 243.0 seconds until the procedures of removing baseline wandering and detecting time positions in Algorithm 1 were completed. This was when we used Daubechies8 wavelet. Feature extraction in Algorithm 2, required 0.6 seconds. It took 1.2 seconds for the KDE classifier to process all the files. The SVM classifier required 0.8 seconds for the same job. The ANN classifier with layer composition required 94.7 seconds.
We compared our QRS detection algorithm with Hamilton and Tompkins’ algorithm [40] which was implemented in C language as an open source software [41]. We supplanted the portion from DWT of fecg(n) to making idx_QRS_Peak(n) in Algorithm 1, with the Hamilton and Tompkins’ program. When we ran the modified program to process the procedures of removing baseline wandering and detecting time positions, it took 59.5 seconds which was approximately four times faster than ours. However the KDE classifier produced the results of being (0.904, 0.891, 329). These results are somewhat inferior compared to the results in Table 2.
Conclusions
The ST segment deviation in ECG can be an indicator of myocardial ischemia. If we can predict an ischemic syndrome as early as possible, we will be able to prevent more severe heart disease such as myocardial infarction [8,9].
To detect ischemic ST episode, we adopted a method directly using morphological features of ECG waveforms. We did not use weight tuning methods such as artificial neural network or decision tree because we wanted to show explicitly which features of ECG waveforms were meaningful to detect ischemic ST episodes. In this regard, we calculated three feature values for each heart beat. They were area between QRS offset and T-peak points, normalized and signed sum from QRS offset to effective zero voltage point, and slope from QRS onset to offset point. After calculating these feature values for each heart beat, we averaged the values of successive five beats because we wanted to reduce outlier effects. The order of magnitude of the third feature value, the slope from QRS onset to offset point, was very different from the other two feature values. To take care of this problem, we considered classification method by kernel density estimation.
We described how we removed baseline wandering in ECG, and detected time positions of QRS complexes by the discrete wavelet transform. Since our classifier selects automatically kernel bandwidths in kernel density estimation, virtually it does not require any numerical parameter which operator should provide. In the tests, SVM with optimal parameters showed just a slightly better classification accuracy than the proposed method, but finding those parameters is a heavy burden compared with the proposed method. We can conclude that overall our proposed method is efficient enough and has more advantages than existing methods.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Mr. Park implemented the whole algorithm, and wrote the manuscript. Dr. Pedrycz proofread it and provided useful technical comments. Dr. Jeon designed the experiments and checked the validity of the proposed methods. All authors read and approved the final manuscript.
Contributor Information
Jinho Park, Email: jinho@gist.ac.kr.
Witold Pedrycz, Email: wpedrycz@ualberta.ca.
Moongu Jeon, Email: mgjeon@gist.ac.kr.
Acknowledgements
This work was supported by the systems biology infrastructure establishment grant provided by Gwangju Institute of Science & Technology, Korea.
References
- Kusumoto FM. Cardiovascular Pathophysiology. North Carolina: Hayes Barton Press; 2004. [Google Scholar]
- Brownfield J, Herbert M. EKG Criteria for Fibrinolysis: What’s Up with the J Point? Western Journal of Emergency Medicine. 2008;9:40–42. [PMC free article] [PubMed] [Google Scholar]
- Rabbani H, Mahjoob MP, Farahabadi E, Farahabadi A, Dehnavi AM. Ischemia detection by electrocardiogram in wavelet domain using entropy measure. Journal of Research in Medical Sciences. 2011;16(11):1473–1482. [PMC free article] [PubMed] [Google Scholar]
- Lemire D, Pharand C, Rajaonah J, Dube B, LeBlanc AR. Wavelet time entropy, T wave morphology and myocardial ischemia. IEEE Transactions on Biomedical Engineering. 2000;47(7):967–970. doi: 10.1109/10.846692. [DOI] [PubMed] [Google Scholar]
- Pang L, Tchoudovski I, Braecklein M, Egorouchkina K, Kellermann W, Bolz A. Real time heart ischemia detection in the smart home care system. Proceedings IEEE Engineering Medicine Biology Society. 2005. pp. 3703–3706. [DOI] [PubMed]
- Tonekabonipour H, Emam A, Teshnelab M, Shoorehdeli MA. Ischemia prediction via ECG using MLP and RBF predictors with ANFIS classifiers. Seventh International Conference on Natural Computation. 2011. pp. 776–780.
- Stamkopoulos T, Diamantaras K, Maglaveras N, Strintzis M. ECG analysis using nonlinear PCA neural networks for ischemia detection. IEEE Transactions on Signal Processing. 1998;46(11):3058–3067. doi: 10.1109/78.726818. [DOI] [Google Scholar]
- Maglaveras N, Stamkopoulos T, Pappas C, Strintzis MG. An adaptive backpropagation neural network for real-time ischemia episodes detection: development and performance analysis using the European ST-T database. IEEE transactions on bio-medical engineering. 1998;45:805–813. doi: 10.1109/10.686788. [DOI] [PubMed] [Google Scholar]
- Afsar FA, Arif M, Yang J. Detection of ST segment deviation episodes in ECG using KLT with an ensemble neural classifier. Physiological measurement. 2008;29(7):747–760. doi: 10.1088/0967-3334/29/7/004. [DOI] [PubMed] [Google Scholar]
- Papaloukas C, Fotiadis DI, Likas A, Michalis LK. An ischemia detection method based on artificial neural networks. Artificial Intelligence in Medicine. 2002;24(2):167–178. doi: 10.1016/S0933-3657(01)00100-2. [DOI] [PubMed] [Google Scholar]
- Andreao RV, Dorizzi B, Boudy J, Mota JCM. ST-segment analysis using hidden Markov Model beat segmentation: application to ischemia detection. Computers in Cardiology. 2004. pp. 381–384.
- Faganeli J, Jager F. Automatic distinguishing between ischemic and heart-rate related transient ST segment episodes in ambulatory ECG records. Computers in Cardiology. 2008. pp. 381–384.
- Exarchos TP, Tsipouras MG, Exarchos CP, Papaloukas C, Fotiadis DI, Michalis LK. A methodology for the automated creation of fuzzy expert systems for ischaemic and arrhythmic beat classification based on a set of rules obtained by a decision tree. Artificial Intelligence in Medicine. 2007;40(3):187–200. doi: 10.1016/j.artmed.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Garcia J, Sörnmo L, Olmos S, Laguna P. Automatic detection of ST-T complex changes on the ECG using filtered RMS difference series: application to ambulatory ischemia monitoring. IEEE Transactions on Biomedical Engineering. 2000;47(9):1195–1201. doi: 10.1109/10.867943. [DOI] [PubMed] [Google Scholar]
- Murugan S, Radhakrishnan S. Rule Based Classification Of Ischemic ECG Beats Using Ant-Miner. International Journal of Engineering Science and Technology. 2010;2(8):3929–3935. [Google Scholar]
- Bakhshipour A, Pooyan M, Mohammadnejad H, Fallahi A. Myocardial ischemia detection with ECG analysis, using Wavelet Transform and Support Vector Machines. In. 17th Iranian Conference of Biomedical Engineering (ICBME) 2010. pp. 1–4.
- Mitchell TM. Machine Learning. New York: McGraw-Hill; 1997. [Google Scholar]
- Taddei A, Distante G, Emdin M, Pisani P, Moody GB, Zeelenberg C, Marchesi C. The European ST-T database: standard for evaluating systems for the analysis of ST-T changes in ambulatory electrocardiography. European Heart Journal. 1992;13(9):1164–1172. doi: 10.1093/oxfordjournals.eurheartj.a060332. [DOI] [PubMed] [Google Scholar]
- Goldberger AL, Amaral LAN, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Mietus JE, Moody GB, Peng CK, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation. 2000;101(23):e215–e220. doi: 10.1161/01.CIR.101.23.e215. [Circulation Electronic Pages: http://circ.ahajournals.org/cgi/content/full/101/23/e215 PMID:1085218 [ doi:10.1161/01.CIR.101.23.e215] [DOI] [PubMed] [Google Scholar]
- Jané R, Laguna P, Thakor NV, Caminal P, Adaptive baseline wander removal in the ECG. Comparative analysis with cubic spline technique. Computers in Cardiology. 1992. pp. 143–146.
- Sörnmo L. Time-varying digital filtering of ECG baseline wander. Medical and Biological Engineering and Computing. 1993;31(5):503–508. doi: 10.1007/BF02441986. [DOI] [PubMed] [Google Scholar]
- Rao RM, Bopardikar AS. Wavelet transforms - introduction to theory and applications. Reading. Massachusetts: Addison-Wesley-Longman; 1997. [Google Scholar]
- Oppenheim AV, Schafer RW, Buck JR. Discrete-time signal processing. Englewood Cliffs, New Jersey. Prentice-Hall: Inc; 1989. [Google Scholar]
- Aspiras TH, Asari VK. Analysis of spatiotemporal relationship of multiple energy spectra of EEG data for emotion recognition. The Fifth International Conference on Information Processing. 2011. pp. 572–581.
- Discrete wavelet transformation frequencies. [ http://visual.cs.utsa.edu/research/projects/davis/visualizations/wavelet/frequency-bands]
- Arvinti B, Toader D, Costache M, Isar A. Electrocardiogram baseline wander removal using stationary wavelet approximations. 12th International Conference on Optimization of Electrical and Electronic Equipment. 2010. pp. 890–895.
- Censi F, Calcagnini G, Triventi M, Mattei E, Bartolini P, Corazza I, Boriani G. Effect of high-pass filtering on ECG signal on the analysis of patients prone to atrial fibrillation. Ann Ist Super Sanita. 2009;45(4):427–431. doi: 10.1590/s0021-25712009000400012. [DOI] [PubMed] [Google Scholar]
- Vila J, Presedo J, Delgado M, Barro S, Ruiz R, Palacios F. SUTIL: intelligent ischemia monitoring system. International Journal of Medical Informatics. 1998;47(6):193–214. doi: 10.1016/s1386-5056(97)00095-6. [DOI] [PubMed] [Google Scholar]
- Bishop CM. Pattern Recognition and Machine Learning. New York LLC: Springer Science + Business Media; 2006. [Google Scholar]
- Scott DW, Sain SR. Handbook of Statistics, Volume 24. North-Holland Publishing Co; 2005. Multi-dimensional Density Estimation; pp. 229–261. [Data Mining and Data Visualization] [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern Classification. Wiley-Interscience: New York; 2000. [Google Scholar]
- Turlach BA. Bandwidth Selection in Kernel Density Estimation: A Review. CORE and Institut de Statistique. 1993. pp. 1–33.
- Chang CC, Lin CJ. LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3):27:1–27:27. [Software available at, http://www.csie.ntu.edu.tw/cjlin/libsvm] [Google Scholar]
- Jager F, Moody GB, Taddei A, Mark RG. Performance measures for algorithms to detect transient ischemic ST segment changes. Computers in Cardiology. 1991. pp. 369–372.
- Izenman AJ. Modern multivariate statistical techniques: regression, classification, and manifold learning. Springer texts in statistics. New York: Springer; 2008. [Google Scholar]
- Chau FT, Liang YZ, Gao J, Shao XG. Chemometrics: from basics to wavelet transform. Chemical analysis. New Jersey: John Wiley; 2004. [Google Scholar]
- Martis RJ, Chakraborty C, Ray AK. An Integrated ECG Feature Extraction Scheme Using PCA and Wavelet Transform. 2009 Annual IEEE India Conference. 2009. pp. 1–4.
- Mahmoodabadi S, Ahmadian A, Abolhasani M, Eslami M, Bidgoli J. ECG Feature Extraction Based on Multiresolution Wavelet Transform. Conference Proceedings of the International Conference of IEEE Engineering in Medicine and Biology Society. 2005. pp. 3902–3905. [DOI] [PubMed]
- Goletsis Y, Papaloukas C, Fotiadis DI, Likas A, Michalis LK. Automated ischemic beat classification using genetic algorithms and multicriteria decision analysis. IEEE Transaction Biomedical Engineering. 2004;51(10):1717–1725. doi: 10.1109/TBME.2004.828033. [DOI] [PubMed] [Google Scholar]
- Hamilton PS, Tompkins WJ. Quantitative investigation of QRS detection rules using the MIT/BIH arrhythmia database. IEEE Transactions on Biomedical Engineering. 1986;33(12):1157–1165. doi: 10.1109/tbme.1986.325695. [DOI] [PubMed] [Google Scholar]
- EP Limited: Open Source Software. [ http://www.eplimited.com/software.htm]