Abstract
Background:
There is a novel hypothesis in that antibodies may have specificity for two distinct antigens that have been named “dual specificity”. This hypothesis was evaluated for some defined therapeutic monoclonal antibodies (mAbs) such as Trastuzumab, Pertuzumab, Bevacizumab, and Cetuximab. In silico design and construction of expression vectors for trastuzumab monoclonal antibody also in this work were performed.
Materials and Methods:
First, in bioinformatics studies the 3D structures of concerned mAbs were obtained from the Protein Data Bank (PDB). Three-dimensional structural alignments were performed with SIM and MUSTANG softwares. AutoDock4.2 software also was used for the docking analysis. Second, the suitable genes for trastuzumab heavy and light chains were designed, synthesized, and cloned in the prokaryotic vector. These fragments individually were PCR amplified and cloned into pcDNA™ 3.3-TOPO® and pOptiVEC™ TOPO® shuttle vectors, using standard methods.
Results:
First, many bioinformatics tools and softwares were applied but we did not meet any new dual specificity in the selected antibodies. In the following step, the suitable expression cascade for the heavy and light chains of Trastuzumab therapeutic mAb were designed and constructed. Gene cloning was successfully performed and created constructs were confirmed using gene mapping and sequencing.
Conclusions:
This study was based on a recently developed technology for mAb expression in mammalian cells. The obtained constructs could be successfully used for biosimilar recombinant mAb production in CHO DG44 dihydrofolate reductase (DHFR) gene deficient cell line in the suspension culture medium.
Keywords: Dual specificity, recombinant monoclonal antibody, trastuzumab
INTRODUCTION
Over expression in ErbB family receptors of tyrosine kinase has been observed in several human malignancies. ErbB receptors are located in cell surface and have been used mainly as therapeutic purposes for control or reduction of the tumor malignancy growth. One successful way to disable of ErbB receptors has been the development of monoclonal antibody drugs such as Trastuzumab.[1–4] Trastuzumab (Herceptin) is a recombinant humanized IgG-1 that selectively targets the extra-cellular domain of the human epidermal growth factor receptor 2 (HER-2/neu) that has been developed by Genentech/Roche.[5,6] Trastuzumab inhibits the proliferation of tumor cells that overexpress HER-2 antigen.[7] The interaction between the antibodies and the HER-2 antigen is performed with high efficiency and specificity that is an essential prerequisite to use them for the targeted treatment of diseases.[8]
Recently, an antibody is discovered that can attach to two separate antigens.[9] This dual-specificity is in contradiction with the “lock and key” hypothesis about antibodies. This occurrence has two main reasons, first, this antibody, called dual specific bh1, has an extraordinary flexibility that is essential to create separated conformation for joining antigens, second, diverse amino acids are critical in connecting to antigen.[9] According to this information, a hypothesis was proposed and analyzed in silico. The related information of some defined therapeutic mAbs was used for investigation a new dual specificity in them.[10,11]
AutoDock is software for automated docking of flexible ligands to receptors. AutoDock is carried out for predict the bound conformations of a flexible ligand to a macromolecule target of the known structure. The technique combines simulated annealing for conformation searching with a rapid grid-based method of energy evaluation.[12,13] Autodock was used to dock the antibodies as previously mentioned with the HER2, epidermal growth factor receptor (EGFR), and vascular endothelial growth factor (VEGF) antigens. A total of 18 compounds were docked together and their binding modes were analyzed in terms of docking energy, hydrogen bonding, and hydrophobic interactions. Except the defined antigen-antibody interactions we could not find any new composition with the low-energy docked structure.
Moreover, due to the large demand for developing new applications for existing monoclonal antibodies (mAbs) or producing biosimilar biological drugs, the Fab amino acid sequences of trastuzumab were obtained. The appropriate secretory signal peptide and the best mRNA structure for nucleic acid codon of the light and heavy chain of trastuzumab were designed. Cloning were performed with pcDNA™ 3.3-TOPO® TA and pOptiVEC™ TOPO® TA shuttle vectors and suitable expression vectors were obtained for further studies.
MATERIALS AND METHODS
Bioinformatics studies
Three-dimesional structures of proteins were obtained from the Protein Data Bank (http://www.rcsb.org).[9,14–17] Three-dimensional structural alignments were performed with SIM and MUSTANG softwares.[18] AutoDock4.2 software[19] was used for the docking analysis. IMGT-3D structures-DB is an immunological structure database that was used for variable domains analysis (http://imgt.cines.fr).[20,21] For sequence analysis such as multiple sequences alignments the following centers were used (http://www.ebi.ac.uk and http://www.ncbi.nlm.nih.gov). RNA structure prediction was carried out with the use of (genebee.msu.su/services) site. For the gene design and codon usage preference, the sites of (http://slam.bs.jhmi.edu/gd/) and (http://www.kazusa.or.jp/codon/) were used.
Gene amplification
The full length heavy chain (HC) and light chain (LC) of Trastuzumab were obtained from PDB database (1N8Z) and drug bank database (AC. DB00072). Suitable secretory signal sequence of the leader KV3A9-Mouse IgG kappa chain V-III was selected from Swiss-Prot database (code P01661). Translation of amino acid was performed using the NCBI database according to codon preference for Chinese Hamster. The final designed sequences were used to construct the genes in the pUC57 prokaryotic vector. The genes syntheses were performed by the Jarf Company (Iran).
PCR were performed on prokaryotic constructs using Platinum® Taq DNA Polymerase High Fidelity kit (Invitrogen, Germany) using an Applied Biosystems (Thermal cycler 2720) instrument. The forward and reverse primers used to amplification of HC were as follows: 5’ TCTCG AGCAC CATGG AG 3’ and 5’ GAATT CTCAT CACTT GCC 3’ and the forward and reverse primers used to amplification of LC were as follows: 5’ AGCTA GCCAC CATGG AG 3’ and 5’ CGTAC GTCAT CAGCA CTC 3’. The reaction was prepared in a DNase/RNase-free vial according to the protocol provided with the kit. The PCR reaction was performed as follow: Initial denaturation: 1 min at 94°C. 30 cycles of; denaturation 30 s at 94°C, annealing: 30 s at 55°C and extension: (1 min for LC and 2 min for HC) at 68°C and final extension: 20 min at 68°C. The amplified fragments were purified using agarose gel Pure Link™ Quick Gel Extraction kit (invitrogen).
Cloning
PCR products related to the heavy and light chains were individually inserted into pcDNA™ 3.3-TOPO® and pOptiVEC™ TOPO® shuttle vectors (included in OptiCHO™ antibody express kit, Invitrogen). In order to increase the efficiency of expression, both trastuzumab monoclonal antibody heavy and light chain genes were cloned into each of the above vectors. Two different expression plasmids were obtained for each chain. Ligation reactions were transformed separately into the competent bacterial host (E.Coli TOP10, Invitrogen). Selection was performed in the LB-Amp solid medium. The alkaline lysis method was used for plasmid extraction in the mini-prep scale and the PureLink™ HiPure plasmid DNA purification kit (Invitrogen) was used for midi prep DNA extraction.
The accuracy of cloning was confirmed using restriction enzyme mapping and sequencing. For each set of cloning, four constructs were selected. The plasmids were extracted using PureLink™ HiPure plasmid DNA purification kit and were digested with the XhoI, EcoRI, NheI, BsiWI, and PvuI restriction endonuclease enzymes. For each reaction 8 μL of purified plasmid, 2 μL of buffer, 2 μL enzyme (1 μL of each one in double digestion) and 8 μL ddH2O were added and digestion were done 5’ in 37°C with FastDigest restriction enzymes (fermentas). Final confirmations of positive clones were performed by DNA sequencing (Kawsar Biotech Co., Iran).
RESULTS
The defined structure of variable domains of four recombinant mAbs approved for cancer therapy namely Trastuzumab (1N8Z),[14] Pertuzumab (1S78),[15] Bevacizumab (1BJ1)[16] and Cetuximab (1YY9),[17] and a new developed mAb namely dual specific bH1 (3BE1, 3BDY)[9] were used in bioinformatics studies. These structures firstly were analyzed with IMGT/3D structure tools. Amino acids of rmAbs involved in hydrogen bonds with the antigen in light and heavy chains were obtained from IMGT/3Dstructure-DB, http://imgt.cines.fr, and their comparisons were shown in Table 1.
Table 1.
Amino acids of rmAbs involved in hydrogen bonds with the antigens, from IMGT/3Dstructure-DB, http://imgt.cines.fr
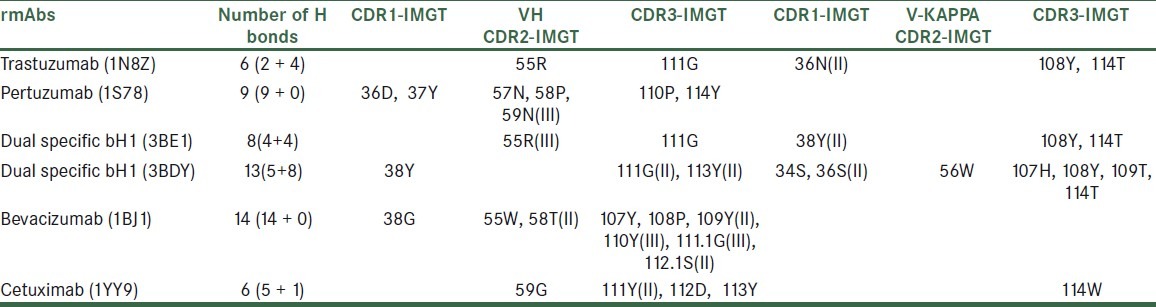
The light and heavy chain CDR loops of Trastuzumab, Pertuzumab, Bevacizumab, and Cetuximab were obtained and the residues involved in antigen binding sites were compared [Figure 1]. The main amino acids that are involved in antigen binding site in these antibodies also were investigated using DeepView/Swiss-PDBViewer 3.7. The main amino acids that are involved in antigen binding in the light and heavy chain CDR loops of Trastuzumab and Bevacizumab were shown in scaled ball and stick format [Figure 2]. AutoDock4.2 software was used for the docking analysis of each antibody with other related antigens with the aim of achieving a dual specificity in them. For example, cetuximab was docked with erbB2 and VEGF antigens and Pertuzumab was docked with HER2 and VEGF antigens, but we did not meet any new specificity in these antibodies.
Figure 1.
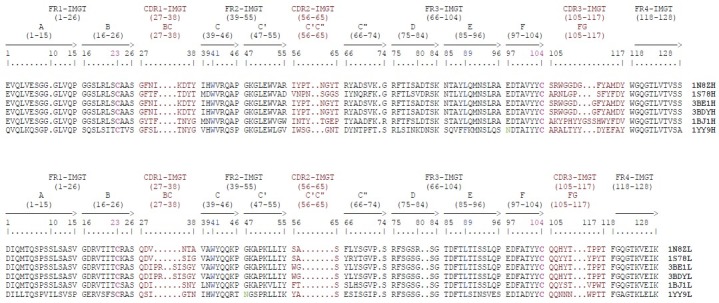
The VH and V-KAPPA domains of the Trastuzumab, Pertuzumab, Dual specific bH1, Bevacizumab, and Cetuximab
Figure 2.

The light and heavy chain CDR loops of Trastuzumab (1N8Z) and Bevacizumab (1BJ1). The main amino acids that are involved in antigen binding are shown in the scaled ball and stick format
Design the heavy and light chains of trastuzumab was carried out according to the information from drug bank database. The secretory signal peptide, kozak sequence, and appropriate restriction enzyme sites were incorporated into upstream of each gene. InterPro database was used for verification of selected secretory signal peptide. The sequenced information obtained bioinformatically helped us to construct the genes in the pUC57 prokaryotic vector. In the next step, initially trastuzumab LC gene was amplified using PCR and subsequently purified. The purified band was cloned into pcDNA ™ 3.3-TOPO® and pOptiVEC ™-TOPO® shuttle vectors individually. The resulting constructs were named pSLC1-14 and pSLO1-14 for the cloned LC in pcDNA and pOptiVEC vectors, respectively. Second, the HC gene was amplified by PCR, purified and ligated in order to obtain recombinant plasmids pSHC1-11 and pSHO1-13 for the cloned HC in pcDNA and pOptiVEC vectors, respectively.
All constructs were checked on agarose gel initially and then four constructs of each set were selected for supplementary analyses. Gene mapping was carried out on four pSLC (i.e., pSLC 1, 2, 3, 4) and four pSLO (i.e pSLO 1, 2, 3, 7) plasmids. Each construct was digested either with NheI or NheI and BsiWI or NheI and PvuI. As expected we should see fragment sizing 6127 and 5122 bp after digesting pSLC and pSLO plasmids with NheI respectively. On the other hand pSLC and pSLO digestion with NheI and BsiWI gave two bands of (720 [insert] and 5407 bp) or (720 and 4402 bp) length, respectively. PvuI has a unique site in these plasmids. These constructs digestion with PvuI and NheI gave two (1478 and 4649 bp) or (1264 and 3858 bp) fragments, respectively [Figures 3a and b].
Figure 3.
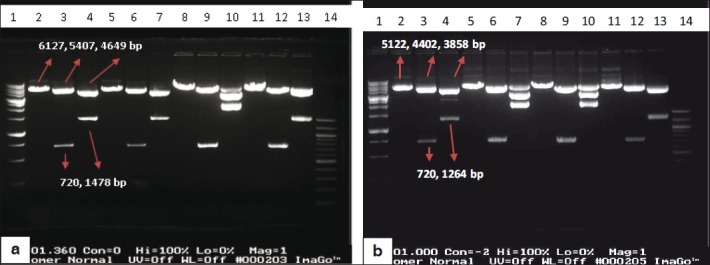
(a) Restriction digestion of pSLC1, 2, 3 and 4 constructs. Lanes 2, 3 and 4 pSLC1 construct; lanes 5, 6 and 7 pSLC2 construct; lanes 8, 9 and 10 pSLC3 construct and lanes 11, 12 and 13 pSLC4 construct that all of them were digested with 1. NheI, 2. NheI and BsiWI, 3. NheI and PvuI respectively. Lane 1: 1Kbp DNA ladder (Bioron, Australia) and lane 14: 100 bp DNA ladder (BioLabs, USA). (b) Restriction digestion of pSLO1, 2, 3 and 7 constructs. Lanes 2, 3 and 4 pSLO1 construct; lanes 5, 6 and 7 pSLO2 construct; lanes 8, 9 and 10 pSLO3 construct and lanes 11, 12 and 13 pSLO7 construct that all of them were digested with 1. NheI, 2. NheI and BsiWI, 3. NheI and PvuI respectively. Lane 1: 1Kbp DNA ladder (Bioron) and lane 14: 100 bp DNA ladder (BioLabs)
In order to confirm cloning of the trastuzumab HC gene in the pOptiVEC vector, four corresponding plasmids (i.e., pSHO2, 5, 8, 9) were extracted and digested with EcoRI or (EcoRI and XhoI) or (XhoI and PvuI) restriction enzymes [Figure 4a]. The expected fragment sizes were 6837 bp or (1430 bp [insert] and 5407 bp) or (1478 bp and 5359 bp) for pSHC constructs, respectively. The expected fragment sizes for pSHO were 5832 or (1430 and 4402) or (1264 and 4568 bp), respectively [Figure 4b].
Figure 4.
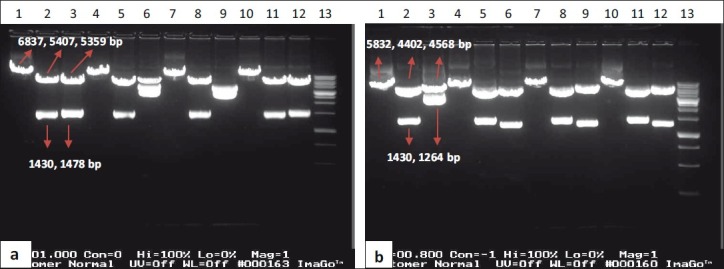
(a) Restriction digestion of pSHC1, 5, 8 and 12 constructs. Lanes 1, 2 and 3 pSHC1 construct; lanes 4, 5 and 6 pSHC5 construct; lanes 7, 8 and 9 pSHC8 construct and lanes 10, 11 and 12 pSHC12 construct that all of them were digested with 1. EcoRI, 2. EcoRI and XhoI, 3. XhoI and PvuI respectively. Lane 13: 1Kbp DNA ladder (Bioron, Australia). (b) Restriction digestion of pSHO2, 5, 8 and 9 constructs. Lanes 1, 2 and 3 pSHO2 construct; lanes 4, 5 and 6 pSHO5 construct; lanes 7, 8 and 9 pSHO8 construct and lanes 10, 11 and 12 pSHO9 construct that all of them were digested with 1. EcoRI, 2. EcoRI and XhoI, 3. XhoI and PvuI respectively. Lane 13: 1Kbp DNA ladder (Bioron, Australia)
Results showed that SLC1, 2, 4 and SLO1, 7 constructs were cloned completely in the direct direction but the SLC3 and SLO2, 3 constructs (containing the LC gene) were cloned in a wrong orientation [Figure 3]. At the other hand, two SHC1, 12 constructs and three SHO5, 8, 9 constructs that were containing the HC gene were cloned completely and in the direct direction [Figure 4].
For final confirmation; pSLC1, 2, 4 and pSLO1, 7 plasmids were sequenced using CMV forward sequencing primer and each sequence was aligned with the designed LC gene sequence using ClustalW software. pSHC1, 12 and pSHO5, 8, 9 were also sequenced using CMV sequencing primer as forward and TK polyA sequencing primer and EMCV IRES sequencing primer as reverse primers respectively. Multiple alignments were carried out using ClustalW software versus parental HC and fidelity of the clones was confirmed this way.
DISCUSSION
Concerning the dual specificity of different mAbs studied, we did not find any bifunctional property using bioinformatic tools. In this regard, the recent crystal structure analysis of an antibody that has shown to have binding sites for two distinct proteins with high affinity namely, dual specific bH1 (3BE1, 3BDY) had to be explained. In our point of view, this is either a very rare phenomena or if more thoroughly investigated more examples could be found.
We designed heavy and light chains of trastuzumab monoclonal antibody according to our bioinformatics data and cloned them into pcDNA™ 3.3-TOPO® and pOptiVEC™ TOPO® TA shuttle vectors. In total, we obtained five correct constructs for LC and five correct constructs for the HC. These constructs were checked in various ways to ensure the right orientation and DNA sequence. The properties of these vectors allow to produce high levels of recombinant proteins rapidly and with less effort than is required using conventional vectors. These constructs will be valuable for other purposes, such as expression in mammalian cell lines for Ab production. We are in the process of testing protein expression properties of these clones.
Since we are using different vectors from the original patent holder and will use different cell system as well as culture condition we may be able to apply for biosimilar trastuzumab production. Manufacturers of biotechnological/biological products frequently make changes to manufacturing processes of products both during development and after approval. These modifications would not adversely impact the safety and efficacy of the drug product. As a consequence, such change may result in an evolution of quality profile during the product lifecycle.[22]
ACKNOWLEDGEMENTS
We thank Dr. Rasaei and Dr Salmanian for their helpful discussions during the early part of the work. We also thank Dr. Mehdi Maleki for his suggestions regarding the work and for reviewing the manuscript. This study was supported by a grant from Kawsar Human Genetics Research Center.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Zhang H, Berezov A, Wang Q, Zhang G, Drebin J, Murali R, et al. ErbB receptors: From oncogenes to targeted cancer therapies. J Clin Invest. 2007;117:2051–8. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garrett JT, Rawale S, Allen SD, Phillips G, Forni G, Morris JC, et al. Novel engineered trastuzumab conformational epitopes demonstrate in vitro and in vivo antitumor properties against HER-2/neu. J Immunol. 2007;178:7120–31. doi: 10.4049/jimmunol.178.11.7120. [DOI] [PubMed] [Google Scholar]
- 3.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat Rev Drug Discov. 2007;6:349–56. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 4.Ross JS, Fletcher JA, Linette GP, Stec J, Clark E, Ayers M, et al. The Her-2/neu gene and protein in breast cancer 2003: Biomarker and target of therapy. Oncologist. 2003;8:307–25. doi: 10.1634/theoncologist.8-4-307. [DOI] [PubMed] [Google Scholar]
- 5.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A. 1992;89:4285–9. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–57. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 7.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 8.Magdelaine-Beuzelin C, Kaas Q, Wehbi V, Ohresser M, Jefferis R, Lefranc MP, et al. Structure-function relationships of the variable domains of monoclonal antibodies approved for cancer treatment. Crit Rev Oncol Hematol. 2007;64:210–25. doi: 10.1016/j.critrevonc.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Bostrom J, Yu SF, Kan D, Appleton BA, Lee CV, Billeci K, et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–4. doi: 10.1126/science.1165480. [DOI] [PubMed] [Google Scholar]
- 10.Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC. Monoclonal antibody successes in the clinic. Nat Biotechnol. 2005;23:1073–8. doi: 10.1038/nbt0905-1073. [DOI] [PubMed] [Google Scholar]
- 11.Foote J. Immunology. Isomeric antibodies. Science. 2003;299:1327–8. doi: 10.1126/science.1082717. [DOI] [PubMed] [Google Scholar]
- 12.Satyanarayanajois S, Villalba S, Jianchao L, Lin GM. Design, synthesis, and docking studies of peptidomimetics based on HER2-herceptin binding site with potential antiproliferative activity against breast cancer cell lines. Chem Biol Drug Des. 2009;74:246–57. doi: 10.1111/j.1747-0285.2009.00855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: Applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–60. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 15.Franklin MC, Carey KD, Vajdos FF, Leahy DJ, De Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell. 2004;5:317–28. doi: 10.1016/s1535-6108(04)00083-2. [DOI] [PubMed] [Google Scholar]
- 16.Muller YA, Chen Y, Christinger HW, Li B, Cunningham BC, Lowman HB, et al. VEGF and the Fab fragment of a humanized neutralizing antibody: Crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface. Structure. 1998;6:1153–67. doi: 10.1016/s0969-2126(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Schmitz KR, Jeffrey PD, Wiltzius JJ, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–11. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: A multiple structural alignment algorithm. Proteins. 2006;64:559–74. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 19.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefranc MP. IMGT Collier de Perles for the variable (V), constant (C), and groove (G) domains of IG, TR, MH, IgSF, and MhSF. Cold Spring Harb Protoc. 2011;2011:643–51. doi: 10.1101/pdb.ip86. [DOI] [PubMed] [Google Scholar]
- 21.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–12. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.European medicines agency: Concept paper on the revision of the guideline on similar biological medicinal products containing biotechnologyderived proteins as active substance: Quality issues. EMA/CHMP/BWP/617111/2010. www.ema.europa.eu. [Google Scholar]


