Abstract
Background:
There are significant limitations in repair of irrecoverable bone defects. Stem-cell therapy is a promising approach for the construction of bone tissue. Mesenchymal stem cells (MSCs) have been introduced as basic tools for bone tissue generation. Through MSCs, adipose-derived stem cells (ADSCs) are more interesting. Since the similarity of native osteoblasts and differentiated osteoblasts from ADSCs in terms of gene expression pattern is unknown, this study was designed to compare gene expression patterns of some genes involved in osteogenesis between human native osteoblasts and adipose-derived differentiated osteoblasts.
Materials and Methods:
Realtime qRT-PCR was used for studying the gene expression of osteocalcin, osteopontin, and core binding factor alpha 1 (Cbfa1) in human native osteoblasts and adipose derived osteogenic osteoblasts at days 7, 14, 21, and 28 of differentiation.
Results:
This study demonstrated that native osteoblasts and differentiated osteoblasts, cultured in common osteogenic medium, have significant differences in gene expression levels for osteocalcin and osteopontin. Compared to native osteoblasts, these genes are expressed lower in all four groups of differentiated osteoblastic cells. We also found, there is a progressive increase in cbfa1 expression over the differentiation period of ADSCs from day 7 to day 28.
Conclusions:
Our findings help for better assessment of adipose-derived differentiated cells as a source for cell-based therapy.
Keywords: Adipose-derived mesenchymal stem cell, Cbfa1, osteoblast, osteocalcin, osteopontin, quantitative real-time PCR
INTRODUCTION
Bone, a major organ for calcium homeostasis and mechanical supporter of soft tissues is known for the tissue high capacity in repair and regeneration.[1] As bone is rigid, partial or total breakages and other types of damages may occur. Different mechanisms of repairs dependent on the biophysical environment such as primary and direct bone repair are engaged to restore the damaged bone tissue.[2] In severe conditions in which there is a vast irrecoverable damage in bone, restoration and bone replacement will be inevitable. At present, autograft transplants are the finest methods for bone repair as this method will not trigger immune response. Nevertheless this method is limited for several reasons such as possible infection of the donor site and limitation of obtained bone marrow.[3] The alternative method is allograft bone transplantation. Possible trigger immune response and feasible transmission of infectional diseases are disadvantages of this method.[4] Another highly used method is metal transplant. Use of metals as substitutes for bone segmental defects makes a need for a second surgery to take it out. Beside possible immune response it can cause infection.[5] Up to now, several alloplasts as substitutes for lost bone have been introduced. For instance synthetic materials with mineral base have been asserted but none of them is principally appropriate and host tissues treat them as a foreign object.[6] One of promising and highly potential methods for functional repair of degenerated and damaged bone is engineering a new bone tissue with the use of host cells.[7] Recent studies on tissue engineering techniques have introduced mesenchymal stem cells as a basic tool for the construction of bone tissue.[8] A Mesenchymal stem cell is defined as a multipotent cell with indefinite self-renewal capability that can differentiate into a variety of cell types, including: Osteoblasts, chondrocytes, and adipocytes.[9] Mesenchymal stem cells can be acquired from different tissues such as bone marrow, fat, bone, and dental pulp.[10] Bone marrow, a well-established source of stem cells yields a limited number of mesenchymal stem cells. Besides, harvesting the bone marrow is painful. Adipose tissue is interesting for the tissue abundance and easy accessibility.[11] Therefore, adipose tissue may be a better candidate source. Adipose-derived stem cells cultured in osteogenic medium differentiate into osteoblast-like cells.[12]
Studies have shown the different genes are involved in regulation of osteogenesis.[13] Although, media supplemented with dexamethasone, ascorbic acid, and β-glycerol phosphate induce the osteoblast phenotype of adipose-derived stem cells (ADSCs), but the amount of the similarity of native osteoblasts and differentiated osteoblasts in expression of bone-specific genes is not clear. To explore changes in gene expression during osteogenic differentiation of ADSCs and compare these cells with native osteoblasts some genes representative of osteogenesis have been chosen. This is a discovery-based study in which three genes involved in osteogenesis have been chosen. We selected osteocalcin, osteopontin, and cbfa1 genes regarding their importance in tissue engineering of bone. Osteocalcin is the most abundant noncollagenous protein that constitutes 1–2% of the total matrix proteins. Osteocalcin is a vitamin K-dependent protein and exclusively secreted by osteoblasts.[14] Osteopontin, another major noncollagenous bone proteins associated with bone remodeling and is highly interested in tissue engineering.[15] Cbfa1, a key transcription factor involved in osteoblast differentiation and is essential for subsequent bone matrix mineralization.[16] In this study qRT-PCR was used to obtain gene expression patterns of osteocalcin, osteopontin, and cbfa1 in native osteoblasts and differentiated osteoblasts from ADSCs on days 7, 14, 21, and 28.
MATERIALS AND METHODS
Written informed consent was gained from female adult donors undergoing liposuction operation and brain surgical operations. The study was approved by the ethics committee of the university.
Osteoblast cell culture
Primary human bone cell cultures were prepared from calvaria bone specimens obtained from three female donors (mean ± SD age 28 ± 6 years) after surgical operations for medical reasons. For isolation of osteoblasts from bone fragments in the explantation method, bone fragments were cut into pieces and rinsed in phosphate buffered saline (PBS) and kept in Dulbecco's modified Eagle's medium (DMEM) with 10% FCS, glutamine (58.5 μg/mL), penicillin (100 U/mL), streptomycin (100 μg/mL), 10–7M dexamethasone and 10 mM β-Glycerophosphate. Cultures were initiated within 3 h and fed twice a week.
Isolation and culture of human adipose-derived stem cells
Human adipose tissues were obtained from abdominal region of three healthy female adult donors undergoing liposuction operation (mean ± SD age 28 ± 6 years). The adipose tissue was first rinsed in PBS and then treated with 0.075% collagenase type I (Sigma) at 37°C for about 40 min, manually shaked every 10 min. Equal volume of Dulbeco's Modified Eagle's Medium (DMEM, Gipco) supplemented with fetal calf serum (FCS, Gipco) was used to stop the digestion process. Centrifugation at 1400 rpm for 10 min was performed to pellet cells. Cells then plated on culture flasks and cultured in growth media [DMEM supplemented with 10% FBS, 100 μg/mL penicillin-streptomycin (Gipco)] and incubated in an incubator with 5% Co2 at 37°C. Medium was refreshed every 3 days until cells were reached 85% confluency. Cells from passage 3 were used for osteogenic differentiation.
Osteogenic induction
In order to induce osteogenic differentiation of stem cells, approximately 2.5×105 ADSCs were plated in a 10 mm culture dish and cultured in osteogenic medium containing DMEM-high glucose, 10% FCS, 100 μg/mL penicillin-streptomycin, 50 μg/mL L-ascorbic acid-2-phosphates (Sigma), L-glutamine (Sigma), 10–7 M dexamethasone (Sigma), and 10 mM β-glycerophosphate (Sigma). The medium was replaced every 2–3 days. 2.5×105 cells on days 7, 14, and 21 were plated in 10 mm culture dishes for longer osteogenic treatment and 3×106 ells at days 7, 14, 21, and 28 of cell culture were used for RNA extraction (N=3 for each group).
Von Kossa staining
Von Kossa staining was used for confirmation of osteogenic differentiation. Cells in flasks were rinsed with PBS and then were fixed in 4% paraformaldehyde for 20 min. The cells were incubated in 5% silver nitrate in dark and then exposed to light for 1 h. After von Kossa staining, appearance of black nodules in extracellular matrix indicates the presence of minerals.
Isolation of total RNA
Fresh cell pellets with about 3×106 cells in each pellet of flask were used for RNA extraction using RNasy plus kit (Qiagen, Germany) according to manufacturer's protocol. Contaminating DNA was removed from total RNA by the gDNA eliminator spin column supplied in the kit. Spectrophotometer was used for quantification of total RNA at 260-nm wavelength and the quality of total RNA was confirmed by the 28S/18S rRNA ratio after running samples on 1% agarose gel. All steps performed under the strict RNase free condition.
Quantitative real-time PCR
Synthesis of first strand cDNA from RNA samples was performed by the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas). For each reverse transcription reaction, 6 μg of RNA and 1 μL of oligo (dT) primer was used according to manufacturer's instructions.
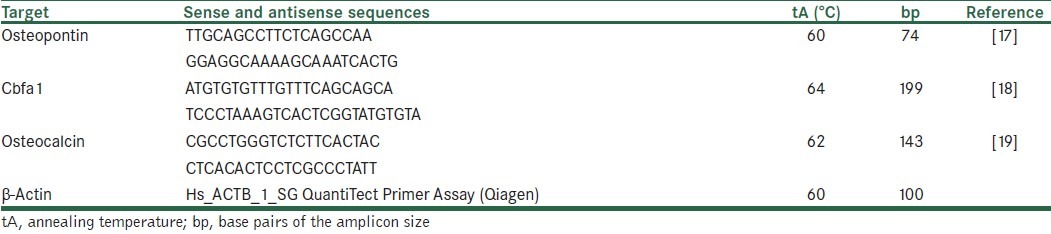
Quantitative real-time PCR was performed using Rotogene thermocycler system (Corbet). Reactions were performed in 20 μL volume. In each reaction, 12.5 μL SYBR Green PCR master mix (Qiagen), specific primer set [Table 1], and 2 μL of cDNA were used. Beta actin gene was used as an internal control to standardize real-time RT-PCR results. Two-step real-time RT-PCR procedure was used for osteopontin and beta actin, in which 60°C (15 s) was used as annealing and extension temperatures. Three-step real-time RT-PCR procedure was used for osteocalcin and cbfa1 with annealing temperature of 64°C and extension temperature of 72°C. Fivefold serial dilutions of osteoblast cDNA samples were used for construction of standard curve. Triplicate measurements were used for each sample. Samples without RNA and reverse transcriptase were used as negative controls.
Table 1.
Primer sequences, annealing temperature, product size, and the references used for quantitative real-time PCR analysis of the genes under study
Statistical analysis
Dunett t test was used in which native osteoblasts treated as a control group and all other groups were compared against it (%95 confidence interval). Besides, Friedman test was used to measure the expression pattern of interested genes.
RESULTS
Isolation and characterization of human native osteoblasts and ADSCs
After about 7–10 days, native osteoblasts isolated form bone fragments [Figure 1a] and after about 14 days petri dishes were filled by cells [Figure 1b].
Figure 1.
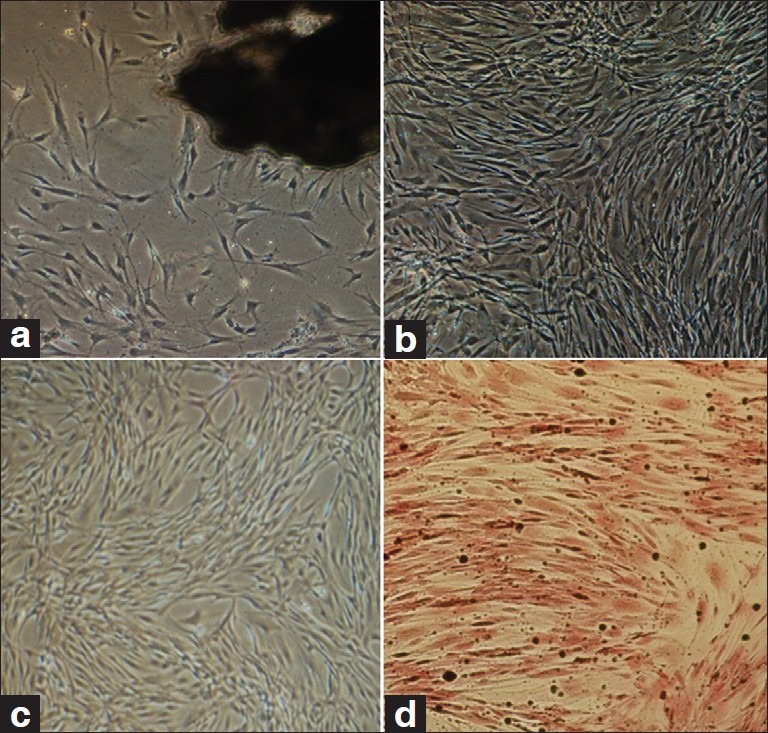
(a) Explant culture of calvarial bone, cells start to isolate from bone segment (dark mass), (b) Native osteoblasts, one passage after isolation, (c) ADSCs. MSCs from third passage with spindle-shaped morphology, (d) Von Kossa staining of differentiated osteoblasts (day 14 of differentiation). Black nodules show deposition of mineralized extracellular matrix
After isolation of ADSCs, uniform cells were appeared after three passages [Figure 1c]. To confirm the differentiation of MSCs from adipose tissue, ADSCs were induced by an osteogenic medium and examined by Von Kossa staining. Histologic detection of calcium depositions demonstrated osteogenic differentiation [Figure 1d].
Quantitative real-time PCR
Normalization of the study carried out for all of selected genes and preliminary experiments were done to gain expected PCR products. Specific melting curves were confirmed by the observation of single band on agarose gel [Figure 2]. No primer dimer was observed for target genes after 40 PCR cycles. For all markers, efficiency of amplification ranged from 0.92 to 1.01 and this showed PCR is optimized. These efficiencies allowed comparison between genes [Figure 3].
Figure 2.
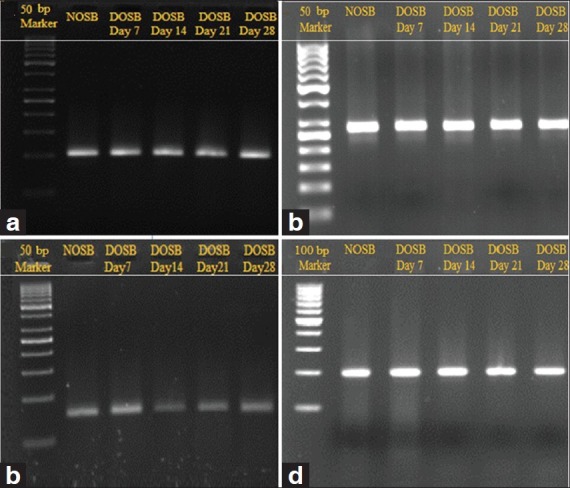
Confirmation of real-time PCR results by observation of a single band of targeted products on agarose gel, (a) Beta actin (100 bp), (b) Osteocalcin (143 bp), (c) Osteopontin (74 bp), (d) Cbfa1(199bp). NOSB: normal osteoblast. DOSB: differentiated osteoblast
Figure 3.
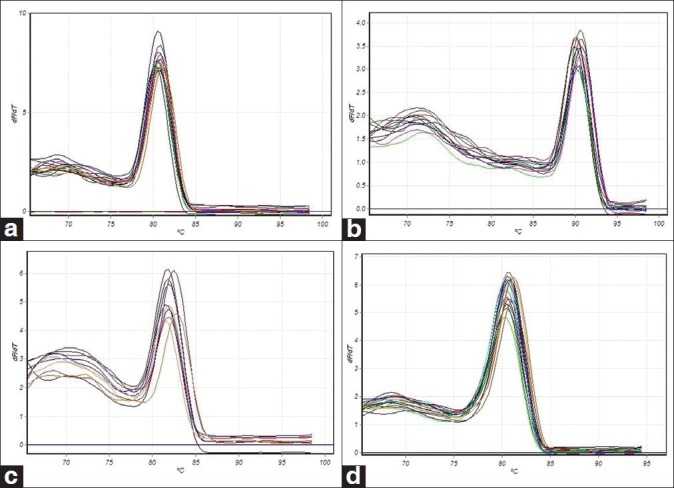
Melting curve analysis. (a) Beta actin, (b) Osteocalcin, (c) Osteopontin, (d) Cbfa1. Normally plotted with fluorescence on the Y-axis and temperature on the X-axis
Osteocalcin expression
Quantitative real-time RT-PCR results demonstrated there is a significant difference in the expression levels of osteocalcin between native osteoblasts and differentiated osteoblast-like cells in all days in which osteocalcin is upregulated in control group (P <0.005 when normalized with beta actin). Mean difference for downregulated osteocalcin in sample groups for days 7, 14, 21, and 28 in comparison to control group (native osteoblasts) are –86833, –84800, –88500, and –84800 respectively [Figure 4]. There is no meaningful difference between groups at days 7, 14, 21, and 28.
Figure 4.
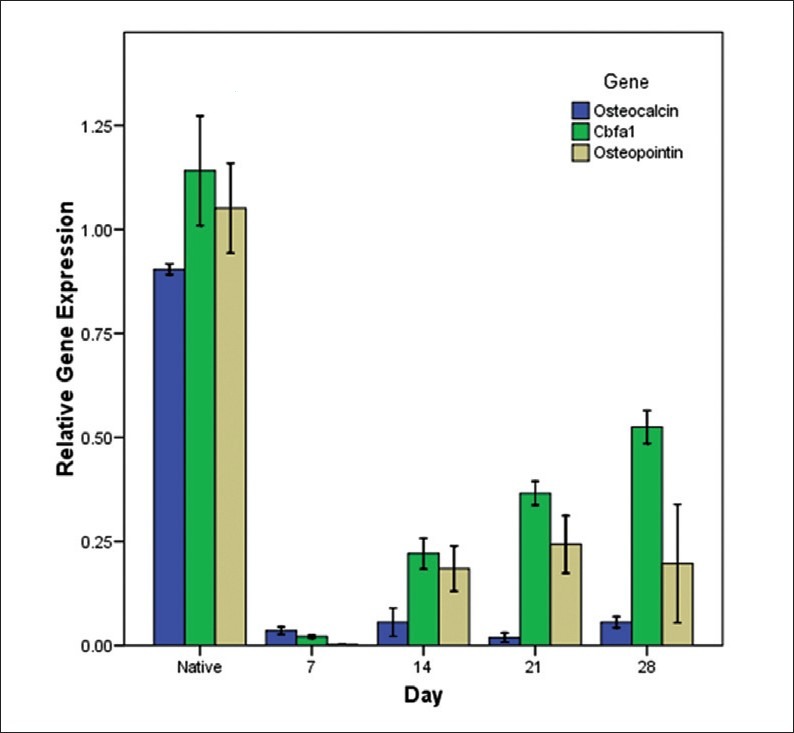
Relative gene expression analysis for osteocalcin, osteopontin, and cbfa1 in different time intervals
Osteopontin expression
Quantitative real-time RT-PCR results showed a considerable upregulation of the osteopontin gene in osteoblast cells in comparison with differentiated cells in days 7, 14, 21, and 28 (P <0.005 when normalized with h beta actin). Osteopontin is downregulated in sample groups (day 7, 14, 21, and 28) in comparison to control groups (native osteoblasts) by a mean difference of –1.04860, –88600, –80800, and –85400, respectively. Osteopontin is downregulated in differentiated cells at day 7 in comparison to other groups but there is no significant difference between groups at days 14, 21, and 28 [Figure 4].
Cbfa1 expression
Real-time RT-PCR results demonstrated Cbfa1 is upregulated in control group (native osteoblast) in comparison to sample groups at days 7, 14, 21, and 28 (P <0.005 when normalized with h beta actin) with mean difference of –1.12000, –92000, –77500, and –61600. Interestingly, there is a steady increase in gene expression pattern of Cbfa1 in adipose derived osteoblastic cells (P ≥0.005 when normalized with h actin) [Figure 4].
DISCUSSION
ADSCs could be a suitable source for cell based therapy of degenerative bone and skeletal defects. In this paper, we evaluated the expression levels of osteocalcin, osteopontin and cbfa1 in normal osteoblasts and adipose-derived differentiated osteoblasts at days 7, 14, 21, and 28. In our study, significant down-regulation of osteocalcin in ADSCs differentiated osteoblasts compared to native osteoblasts was observed. Longer treatment of ADSCs, up to 28 days in osteogenic medium had not significant effect on gene expression level of osteocalcin in ADSCs differentiated osteoblasts. In a study, Shui and colleagues showed osteoblasts differentiated from human bone marrow stromal cells (BMSC) express osteocalcin in an additive time-dependent manner as osteoblastic differentiation progresses from day 2 to day 15.[20] It should be noted that semiquantitative RT-PCR was used for studying the gene expression of osteocalcin. In another study Monnipha demonstrated bone marrow-derived human mesenchymal stem cells express osteocalcin gene on day 14.[21] In this study, gene expression was investigated from day 4 to day 28 of osteoblastic differentiation using RT-PCR. In another study by Monaco et al., They found continuous changes in osteocalcin mRNA abundance from porcine bone marrow-derived osteoblastic cells from day 2 to day 28 of differentiation. Supporting our data, they showed the gene expression level of osteocalcin insignificantly changes in porcine adipose-derived osteoblastic cell from day 2 to day 28 of differentiation.[22] These studies used the same supplemented osteogenic medium as we used. Osteocalcin a key component of bone plays an important role in bone mineralization and calcium homeostasis and is a major indicator for differentiation of osteoblast progenitor cells.
For osteopontin, adipose-derived osteogenic osteoblast compared to native osteoblasts exhibited a significant overexpression of osteopontin mRNAs. Although we found an increase in the osteopontin expression pattern from day 7 to day 21 of differentiation but a decrease was observed from day 21 to day 28. There is a meaningful difference between day 7 and day 14 of differentiation. Monaco et al. evaluated the expression pattern of osteopontin is osteoblastic-differentiated cells from porcine bone marrow and adipose tissues. By evaluation of the osteopontin expression level in bone-marrow-derived differentiated cells they found a decrease from day 7 to day 14 followed by a pike at day 21 and another decrease from day 21 to day 28 of differentiation. They also found an steady increase in osteopontin expression pattern from day 7 to day 21 and a decrease from day 21 to day 28 of differentiation of adipose derived osteoblastic cells.[22] Osteopontin is an early bone marker and has an important and critical role in bone remodeling.[13]
We also found a steady increase in the Cbfa1 expression over the differentiation period from day 7 to day 28. In this study, we found Cbfa1 compared to osteocalcin and osteopontin is the only gene that has significant increase in gene expression during 28 days of differentiation. In a contradictory study performed by Shui et al., they found there was no significant increase in mRNA and protein levels of cbfaI during osteoblastic differentiation of human BMSCs.[20] In other studies performed on rodent samples, they found mRNA and protein levels of cbfa1 change during osteoblastic differentiation.[23–26] Sudhakar et al. found level of cbfaI mRNA regulates differentiation of osteoblastic progenitors. This factor directly stimulates most of osteoblast specific genes.[13]
In different studies, different sources of MSCs have been compared for their chondrogenic and osteogenic potentials. For instance Bernardo et al. showed fetal or adult bone-derived MSCs, exhibit a higher chondrogenic potential than MSCs from fetal lung and placenta.[27] Anker and colleagues showed different potential of MSCs isolated from fetal bone marrow, lung, liver and spleen to differentiate into osteoblast and adipocytes.[28] Monaco et al. compared adipose and bone marrow porcine stem cells and found there are some differences in morphological and transcriptome characteristics of these cells. Different osteogenic potential of different sources for mesenchymal stem cells may be connected to some of contradict data acquired in this study compared to others.
Up to now effect of different osteogenic inducers such as strontium, vitamin D and retinoic acid on expression of osteoblast specific genes have been considered. Probably they will cause an earlier expression of some genes and enhance osteogenesis.[21,29,30] We suggest the effect of osteogenic inducers on expression pattern of different osteoblast specific and bone-related genes will be evaluated to help researchers to choose the best osteogenic supplement for differentiation of MSCs.
We also recommend that effect of different scaffolds such as D, D, L, L-polylactide (PLLA), collagen I/III, and polygalactin-910/polydioxanone (PGPD) and OPF hydrogels on expression pattern of bone-related genes will be considered.[31–34] Regarding this, the best condition for cell growth, adhesion, propagation, and differentiation will be provided.
CONCLUSION
Our results show that
-
I)
Native osteoblasts and adipose-derived differentiated osteoblasts on days 7, 14, 21, and 28 have significant differences in the gene expression level for osteocalcin
-
II)
There is a steady increase in gene expression pattern of cbfa1 in adipose-derived osteoblastic cells.
-
III)
Differentiated osteoblasts on days 7, 14, 21, and 28 express cbfa1 lower than native osteoblast.
-
IV)
Compared to native osteoblasts expression of osteopontin is downregulated in differentiated osteoblastic cells at all considered days.
Based on these observations, our data suggest that treatment of ADSCs (up to 28 days of differentiation) is not enough for gaining the similar transcriptomic pattern of osteogenesis-related genes and the same osteoblastic properties. Our findings help for better assessment of adipose-derived differentiated cells as a source for cell-based therapy of degenerative bone and skeletal defects.
ACKNOWLEDGMENTS
The authors wish to thank Mohammad Kazemi and Farzaneh Jafari for technical assistances. We also appreciate Mohammad Salehi for his help in statistical analysis.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Rebelatto CK, Aguiar AM, Moretão MP, Senegaglia AC, Hansen P, Barchiki F, et al. Dissimilar differentiation of mesenchymal stem cells from bone marrow, umbilical cord blood, and adipose tissue. Exp Biol Med (Maywood) 2008;233:901–13. doi: 10.3181/0712-RM-356. [DOI] [PubMed] [Google Scholar]
- 2.Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 3.Gazdag AR, Lane JM, Glaser D, Forster RA. Alternatives to autogenous bone graft: Efficacy and indications. J Am Acad Orthop Surg. 1995;3:1–8. doi: 10.5435/00124635-199501000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Ikada Y. Challenges in tissue engineering. J R Soc Interface. 2006;3:589–601. doi: 10.1098/rsif.2006.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CC, Chueh JY, Tseng H, Huang HM, Lee SY. Preparation and characterization of biodegradable PLA polymeric blends. Biomaterials. 2003;24:1167–73. doi: 10.1016/s0142-9612(02)00466-0. [DOI] [PubMed] [Google Scholar]
- 6.Kaigler D, Mooney D. Tissue engineering's impact on dentistry. J Dent Educ. 2001;65:456–62. [PubMed] [Google Scholar]
- 7.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–6. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 8.Aubin JE. Bone stem cells. J Cell Biochem Suppl. 1998;30-31:73–82. [PubMed] [Google Scholar]
- 9.Presnell SC, Petersen B, Heidaran M. Stem cells in adult tissues. Semin Cell Dev Biol. 2002;13:369–76. doi: 10.1016/s1084952102000939. [DOI] [PubMed] [Google Scholar]
- 10.Salgado AJ, Oliveira JT, Pedro AJ, Reis RL. Adult stem cells in bone and cartilage tissue engineering. Curr Stem Cell Res Ther. 2006;1:345–64. doi: 10.2174/157488806778226803. [DOI] [PubMed] [Google Scholar]
- 11.Guilak F, Awad HA, Fermor B, Leddy HA, Gimble JM. Adipose-derived adult stem cells for cartilage tissue engineering. Biorheology. 2004;41:389–99. [PubMed] [Google Scholar]
- 12.Fraser JK, Wulur I, Alfonso Z, Hedrick MH. Fat tissue: An underappreciated source of stem cells for biotechnology. Trends Biotechnol. 2006;24:150–4. doi: 10.1016/j.tibtech.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Kirkham GR, Cartmell SH. Ashammakhi N, editor. Genes and proteins involved in the regulation of osteogenesis, in Topics in tissue engineering, R.R.E.C. 2007 [Google Scholar]
- 14.Lian JB, Stein GS, Stewart C, Puchacz E, Mackowiak S, Aronow M, et al. Osteocalcin: Characterization and regulated expression of the rat gene. Connect Tissue Res. 1989;21:61–8. doi: 10.3109/03008208909049996. discussion 69. [DOI] [PubMed] [Google Scholar]
- 15.McKee MD, Nanci A. Osteopontin: An interfacial extracellular matrix protein in mineralized tissues. Connect Tissue Res. 1996;35:197–205. doi: 10.3109/03008209609029192. [DOI] [PubMed] [Google Scholar]
- 16.Ziros PG, Basdra EK, Papavassiliou AG. Runx2: Of bone and stretch. Int J Biochem Cell Biol. 2008;40:1659–63. doi: 10.1016/j.biocel.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Wang-Rodriguez J, Urquidi V, Rivard A, Goodison S. Elevated osteopontin and thrombospondin expression identifies malignant human breast carcinoma but is not indicative of metastatic status. Breast Cancer Res. 2003;5:R136–43. doi: 10.1186/bcr620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Araki S, Mezawa M, Sasaki Y, Yang L, Li Z, Takai H, et al. Parathyroid hormone regulation of the human bone sialoprotein gene transcription is mediated through two cAMP response elements. J Cell Biochem. 2009;106:618–25. doi: 10.1002/jcb.22039. [DOI] [PubMed] [Google Scholar]
- 19.Burkhardt JK, Halama D, Frerich B, Gaunitz F. Real-time RT-PCR discriminating mRNA encoding osteocalcin from unspecific targets. Anal Bioanal Chem. 2009;393:1351–5. doi: 10.1007/s00216-008-2528-4. [DOI] [PubMed] [Google Scholar]
- 20.Shui C, Spelsberg TC, Riggs BL, Khosla S. Changes in Runx2/Cbfa1 expression and activity during osteoblastic differentiation of human bone marrow stromal cells. J Bone Miner Res. 2003;18:213–21. doi: 10.1359/jbmr.2003.18.2.213. [DOI] [PubMed] [Google Scholar]
- 21.Sila-Asna M, Bunyaratvej A, Maeda S, Kitaguchi H, Bunyaratavej N. Osteoblast differentiation and bone formation gene expression in strontium-inducing bone marrow mesenchymal stem cell. Kobe J Med Sci. 2007;53:25–35. [PubMed] [Google Scholar]
- 22.Monaco E, Sobreira de Lima A, Bionaz M, Maki A, Wilson SM, Hurley WL, et al. Morphological and transcriptomic comparison of adipose and bone marrow derived porcine stem cells. Open Tissue Eng Regen Med. 2009;2:20–33. [Google Scholar]
- 23.Banerjee C, Javed A, Choi JY, Green J, Rosen V, van Wijnen AJ, et al. Differential regulation of the two principal Runx2/Cbfa1 n-terminal isoforms in response to bone morphogenetic protein-2 during development of the osteoblast phenotype. Endocrinology. 2001;142:4026–39. doi: 10.1210/endo.142.9.8367. [DOI] [PubMed] [Google Scholar]
- 24.Prince M, Banerjee C, Javed A, Green J, Lian JB, Stein GS, et al. Expression and regulation of Runx2/Cbfa1 and osteoblast phenotypic markers during the growth and differentiation of human osteoblasts. J Cell Biochem. 2001;80:424–40. doi: 10.1002/1097-4644(20010301)80:3<424::aid-jcb160>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Sudhakar S, Katz MS, Elango N. Analysis of type-I and type-II RUNX2 protein expression in osteoblasts. Biochem Biophys Res Commun. 2001;286:74–9. doi: 10.1006/bbrc.2001.5363. [DOI] [PubMed] [Google Scholar]
- 26.Sudhakar S, Li Y, Katz MS, Elango N. Translational regulation is a control point in RUNX2/Cbfa1 gene expression. Biochem Biophys Res Commun. 2001;289:616–22. doi: 10.1006/bbrc.2001.6033. [DOI] [PubMed] [Google Scholar]
- 27.Bernardo ME, Emons JA, Karperien M, Nauta AJ, Willemze R, Roelofs H, et al. Human mesenchymal stem cells derived from bone marrow display a better chondrogenic differentiation compared with other sources. Connect Tissue Res. 2007;48:132–40. doi: 10.1080/03008200701228464. [DOI] [PubMed] [Google Scholar]
- 28.Anker PS, Noort WA, Scherjon SA, Kleijburg-van der Keur C, Kruisselbrink AB, van Bezooijen RL, et al. Mesenchymal stem cells in human second-trimester bone marrow, liver, lung, and spleen exhibit a similar immunophenotype but a heterogeneous multilineage differentiation potential. Haematologica. 2003;88:845–52. [PubMed] [Google Scholar]
- 29.Khanna-Jain R, Vuorinen A, Sándor GK, Suuronen R, Miettinen S. Vitamin D(3) metabolites induce osteogenic differentiation in human dental pulp and human dental follicle cells. J Steroid Biochem Mol Biol. 2010;122:133–41. doi: 10.1016/j.jsbmb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Wan DC, Shi YY, Nacamuli RP, Quarto N, Lyons KM, Longaker MT. Osteogenic differentiation of mouse adipose-derived adult stromal cells requires retinoic acid and bone morphogenetic protein receptor type IB signaling. Proc Natl Acad Sci U S A. 2006;103:12335–40. doi: 10.1073/pnas.0604849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrell E, Byrne EM, Fischer J, O’Brien FJ, O’Connell BC, Prendergast PJ, et al. A comparison of the osteogenic potential of adult rat mesenchymal stem cells cultured in 2-D and on 3-D collagen glycosaminoglycan scaffolds. Technol Health Care. 2007;15:19–31. [PubMed] [Google Scholar]
- 32.Jäger M, Feser T, Denck H, Krauspe R. Proliferation and osteogenic differentiation of mesenchymal stem cells cultured onto three different polymers in vitro. Ann Biomed Eng. 2005;33:1319–32. doi: 10.1007/s10439-005-5889-2. [DOI] [PubMed] [Google Scholar]
- 33.Zavan B, Giorgi C, Bagnara GP, Vindigni V, Abatangelo G, Cortivo R. Osteogenic and chondrogenic differentiation: Comparison of human and rat bone marrow mesenchymal stem cells cultured into polymeric scaffolds. Eur J Histochem. 2007;51(Suppl 1):1–8. [PubMed] [Google Scholar]
- 34.Takamoto T, Hiraoka Y, Tabata Y. Enhanced proliferation and osteogenic differentiation of rat mesenchymal stem cells in collagen sponge reinforced with different poly(ethylene terephthalate) fibers. J Biomater Sci Polym Ed. 2007;18:865–81. doi: 10.1163/156856207781367738. [DOI] [PubMed] [Google Scholar]


