Summary
Salamander limb regeneration is dependent upon tissue interactions that are local to the amputation site. Communication among limb epidermis, peripheral nerves, and mesenchyme coordinate cell migration, cell proliferation, and tissue patterning to generate a blastema, which will form missing limb structures. An outstanding question is how cross-talk between these tissues gives rise to the regeneration blastema. To identify genes associated with epidermis-nerve-mesenchymal interactions during limb regeneration, we examined histological and transcriptional changes during the first week following injury in the wound epidermis and subjacent cells between three injury types; 1) a flank wound on the side of the animal that will not regenerate a limb, 2) a denervated limb that will not regenerate a limb, and 3) an innervated limb that will regenerate a limb. Early, histological and transcriptional changes were similar between the injury types, presumably because a common wound-healing program is employed across anatomical locations. However, some transcripts were enriched in limbs compared to the flank and are associated with vertebrate limb development. Many of these genes were activated before blastema outgrowth and expressed in specific tissue types including the epidermis, peripheral nerve, and mesenchyme. We also identified a relatively small group of transcripts that were more highly expressed in innervated limbs versus denervated limbs. These transcripts encode for proteins involved in myelination of peripheral nerves, epidermal cell function, and proliferation of mesenchymal cells. Overall, our study identifies limb-specific and nerve-dependent genes that are upstream of regenerative growth, and thus promising candidates for the regulation of blastema formation.
Key words: Regeneration, Limb, Axolotl, Wound healing
Introduction
All animals regenerate some of their tissues by physiological turnover, yet only a select few can regenerate appendages. Vertebrates accomplish this feat by generating a blastema, a mass of lineage-restricted progenitor cells at the end of an amputation stump (Kragl et al., 2009). The cellular and molecular processes that coordinate blastema formation are poorly understood, likely because it is a complex process, requiring tissues of an anatomically complex amputation stump to coordinate wound healing, progenitor cell recruitment, cell proliferation, and tissue patterning. A major hurdle towards understanding appendage regeneration is to identify the necessary processes for regeneration and the molecular mechanisms by which these processes regulate blastema formation. For example, some cellular processes like inflammation, epidermal migration, and cell proliferation are common to all injury types, so it is necessary to devise experimental strategies that can distinguish pathways specific to general injury processes from those required for appendage regeneration.
The blastema of a regenerating salamander limb is a classic paradigm for studying appendage regeneration because it is an accessible experimental system that regenerates a morphologically complex structure. A critical tissue interaction that is necessary for blastema formation occurs between the wound epithelium (WE), which forms from rapid migration of adjacent epidermis, and the underlying mesenchymal stump cells. Blastema formation is inhibited if the WE is disrupted, either by suturing full thickness skin over the amputation stump (Mescher, 1976; Tassava and Garling, 1979), irradiation (Thornton, 1958), surgical removal (Thornton, 1957), or implantation of the limb stump into the body cavity (Goss, 1956) or dorsal fin to disrupt epidermal migration (Stocum and Dearlove, 1972). The WE gradually thickens after amputation to generate the apical epithelial cap (AEC). The AEC is a signaling center which supports mesenchymal cell proliferation (Boilly and Albert, 1990; Globus and Vethamany-Globus, 1985), promotes tissue histolysis (Singer and Salpeter, 1961), and regulates cell migration (Thornton, 1960b; Thornton, 1960a; Thornton and Steen, 1962; Thornton and Thornton, 1965). Molecules expressed in the AEC include; the transcription factors msx2, dlx3, id2, id3, hes1, sp9 (Satoh et al., 2008), the secreted signaling molecules wnt5a, wnt5b (Ghosh et al., 2008), fgf1, fgf2, fgf8, fgf10, the extracellular matrix molecules collagen type XII, collagen type IV, lamb1, and the enzymes mmp3/10b, mmp9 (Campbell and Crews, 2008). However, these studies were not designed to uncover molecules that are vital to the early function of the WE or that regulate its relationship with peripheral nerves and blastemal cells.
Innervation of the limb stump is necessary for regeneration. Transecting the spinal nerves that innervate the forelimb just prior to or shortly after limb amputation will block blastema formation (Singer, 1952). It is unknown why denervation leads to loss of regeneration in axolotls, but the favored hypothesis is that nerves provide trophic factors that support cell proliferation of the blastema, which is lost upon denervation. Several factors have been proposed as the trophic factor (Dungan et al., 2002; Globus et al., 1991; Mescher et al., 1997; Mullen et al., 1996; Satoh et al., 2008; Wang et al., 2000), but none have been clearly demonstrated to be the factor (Stocum, 2011). Furthermore, it is unclear if the nerve exerts its effects on the mesenchyme, epidermis, or Schwann cells. Part of the difficulty in identifying the exact mechanism of the nerve's influence is because nerve fibers quickly invade throughout the distal mesenchyme and wound epidermis after amputation (Singer, 1949; Taban, 1949; Thornton, 1954). Limbs containing only motor nerves that do not innervate the epidermis can regenerate, demonstrating that direct innervation of the epidermis is not necessary for limb regeneration (Sidman and Singer, 1960; Thornton, 1960b). Taken together, nerves need to invade the amputation stump to support cell proliferation, but the exact relationship between the nerves with the mesenchyme and epidermis is unclear. Identification of the downstream targets of the nerve during limb regeneration may reveal the relationship of the nerve with the amputation stump.
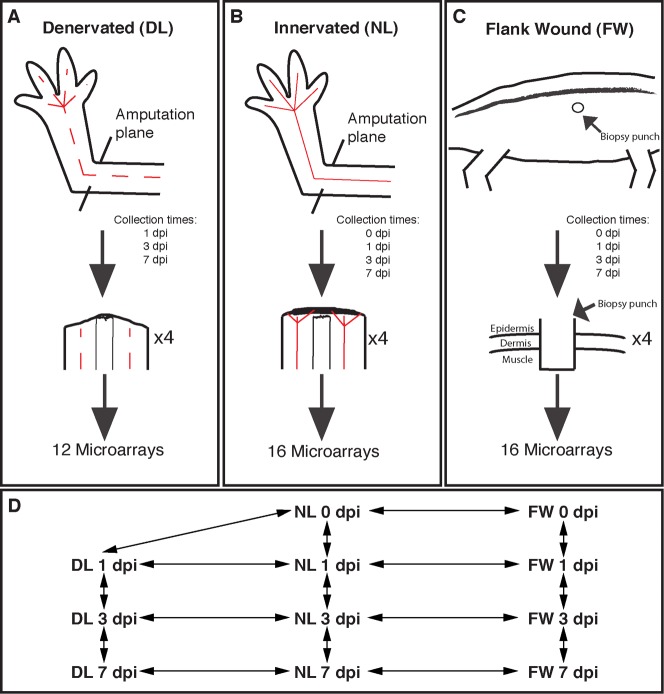
Although we do not yet understand the relationship between the epidermis, nerve, and mesenchymal cells at the molecular level, it is clear that the function of each tissue is dependent upon the presence of the other tissues and these interactions take place locally at the amputation plane. For this reason, the goal of this study was to identify a specific set of genes that are expressed in the WE and cells directly beneath the WE of the amputated limb stump early after injury. Our strategy was to use a custom microarray platform developed for the axolotl (Huggins et al., 2012) to compare gene expression differences over the first week after injury between an injured tissue that will not form a limb (flank wound), an example of aborted limb regeneration (denervated limb), and a regenerating limb (Fig. 1A–C). Using statistical approaches to identify significant transcript abundance differences between regenerating limbs and non-regenerating tissues (Fig. 1D), we were able to identify a regeneration-specific gene expression profile. A flank injury located outside the limb field was chosen because site-specific differences in the skin are known to mediate limb and flank morphology (Rinn et al., 2006; Satoh et al., 2007; Tank, 1984; Tank, 1987). Denervated limbs were chosen because they are an example of aborted limb regeneration – they present limb-specific gene expression patterns but do not generate a blastema. Lastly, gene expression during the first seven days after injury was chosen to identify genes expressed at the onset of AEC formation and blastema cell proliferation. In the following paragraphs we describe histological changes that take place over the first seven days in each injury type and then overlay transcriptional patterns of limb-enriched and nerve-dependent genes in the amputation stump.
Fig. 1. Experimental design of the microarray analysis.
(A) Cartoon showing where the limb was amputated in denervated limbs. Denervated nerve tracks are represented by a dashed red line. The bottom cartoon shows the regressing limb around the bone. (B) Cartoon showing an innervated limb with a solid line representing the nerves. The bottom cartoon shows the innervation of the amputation stump, thickening of the WE, and the beginning of cell accumulation underneath the WE. (C) Cartoon showing where the flank wound was administered on the flank of the animal. The bottom cartoon shows how deep the flank wound enters into the axolotl flank. (D) Schematic showing the 16 contrasts made in the analysis of the microarray. Notice that comparisons were performed over time and between treatments.
Results
Histology and BrdU analysis of injured innervated limbs, denervated limbs, and flank wounds
We histologically characterized normal innervated limbs (NL), denervated limbs (DL), and flank wounds (FW) over the first seven days post injury (dpi) in order to examine differences between each injury response at the cellular level (Fig. 2). Masson's Trichrome staining revealed that the structure of the uninjured skin in NL, DL, and FW were similar with one another (data not shown). Uninjured epithelium consisted of an outer apical layer of epithelial cells, an interstitial layer of mucous secreting Leydig cells interspersed with keratinocytes, and a basal layer of germinative basal keratinocytes (Fox, 1986; Kelly, 1966). The underlying uninjured dermis consisted of mucous and granular glands interspersed with a loose network of fibroblasts that overlies muscle (Seifert et al., 2012).
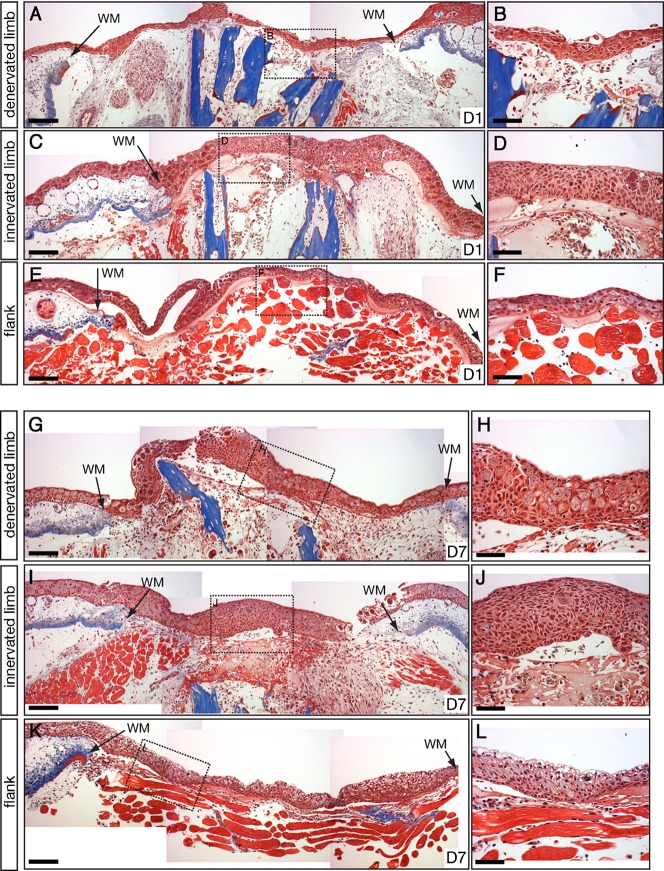
Fig. 2. Histology of NL, DL, and FW.
Masson's trichrome staining of sections of NL, DL, and FW at 1 dpi (A–F) and 7 dpi (G–L). Area of magnified images on right are boxed in images on left. (A,B) Denervated limb at 1 dpi showing injury closure by the WE and the hemostatic response under the WE. (C,D) Innervated limb 1 dpi showing high similarity to the denervated limb. Normal epidermis and dermis can be seen outside the wound margins (WM). (E,F) FW at 1 dpi showing that the WE has closed the wound directly over the muscle and that a small hemostatic response is taking place. (G–L) Injuries at 7 dpi showing the thickening of the WE in DL (G,H), NL (I,J), and FW (K,L). Scale bar in A,C,E,G,I,K = 200 µm. Scale bar in B,D,F,H,J,L = 100 µm.
For all three injury types, the wound re-epithelialized within 24 hours after injury by migration of surrounding epidermis, generating a WE comprised of Leydig cells and keratinocytes (Fig. 2A–F). Underneath the WE was an accumulation of plasma and blood cells, with more blood in NL and DL versus FW, possibly because amputation severed major vasculature in the limb (Fig. 2B,D,F). Additionally, the FW was almost exclusively composed of muscle, while the limbs included bone, peripheral nerves, vasculature and muscle. In all three cases, the WE appeared to behave similarly during the first 24 hours after injury, although the extent of the hemostatic response and complexity of the underlying tissue is greater in the amputated limb compared to the flank.
By 7 dpi, the WE had thickened in all three injury types, but a distinct mound of epidermal cells was apparent in the middle of NL and DL WE, which was not present in FW (Fig. 2G–L). The epidermal mound may represent the maturation of the WE into the AEC, which was likely due to continuous cell migration from the wound margins rather than cell proliferation within the WE because BrdU-positive cells were evident at the margins of NL and DL (Fig. 3A,D), but not within the center of the WE (Fig. 3B,E) (Chalkley, 1954; Hay and Fischman, 1961). The WE of the FW was in direct contact with the subjacent muscle with little muscle dedifferentiation (Fig. 2K,L). In contrast, muscle fibers and peripheral nerves were becoming disorganized due to degeneration in both NL and DL (Fig. 2G,I). Limbs also had more plasma, red blood cells, and inflammatory cells. All together, these results suggest similar processes were taking place in each injury type, but tissue histolysis was more complete by seven days after injury in limbs versus FW.
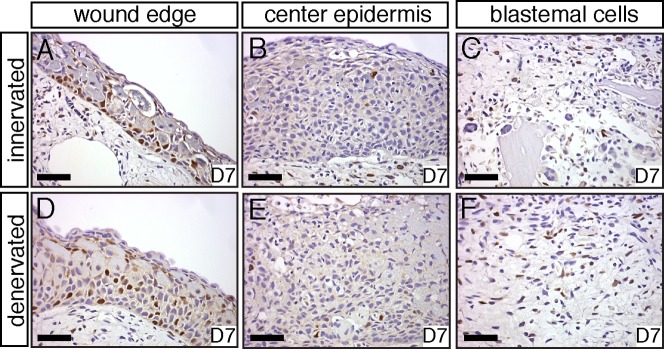
Fig. 3. BrdU staining of sections of injured limbs.
(A–C) BrdU staining of NL at 7 dpi. (A,D) Cell proliferation is present in the epidermis near the wound edge in both NL and DL. (B,E) Little DNA synthesis is present in the WE in NL and DL. (C,F) Some DNA synthesis is present in the limb mesenchyme of both NL and DL at 7 dpi. Scale bar in A–F = 100 µm.
Taken together our data show that both limbs exhibited a hemostatic response and were histologically similar, containing a thickened WE, osteoclasts surrounding the bone, and histolysed tissues (Fig. 2A,C,G,I). The WE was lying directly over the bone in DL, while cells were present between the WE and bone in NL, suggesting that blastema growth was beginning within NL. BrdU analysis showed that DNA synthesis was taking place within the WE margin and mesenchyme (Fig. 3A,C,D,F) of both NL and DL, which is in accordance with previous studies showing that mesenchymal cells and epidermal cells enter S phase and divide in both denervated and innervated limbs (Maden, 1978). Loss of cell cycling in the mesenchyme of denervated limbs likely takes place after 7 dpi in the large-sized animals used in this study. These findings demonstrate that the time frame chosen for our study encompassed blastema formation rather than blastema outgrowth.
Commonly changed genes following injury
In order to characterize transcription during regeneration, transcript abundances were estimated from total RNA collected from the WE and a few subjacent cells from all three treatments (Fig. 1A–C). A total of 6684 probe sets yielded expression estimates that differed significantly as a function of RNA source and sample time (supplementary material Table S1). These genes were parsed to identify similarities and differences between injury types. First, probe sets that changed significantly from baseline to 1 dpi, 1 dpi to 3 dpi, or from 3 dpi to 7 dpi in each injury were identified to examine the commonalities between the injuries. We found that transcription was more similar between NL, DL, and FW than it was different with 1840 genes up-regulated and 1667 genes down-regulated in all three injuries (Fig. 4A,B). This high degree of similarity suggested the presence of a general wound-healing response regardless of whether or not a limb will regenerate. The list of genes that presented higher transcript abundances above baseline was significantly enriched for genes that annotate to gene ontology terms associated with processes known to take place during mammalian skin wound healing including immune system response (n = 187), macrophage activation (n = 36), and response to stimulus (n = 124) (supplementary material Table S2). Down-regulated genes belonged to ontology categories including lipid metabolic process (n = 110), chromosome segregation (n = 30), metabolic process (n = 604), and response to stress (n = 48). These results suggest that many of the same processes that take place during mammalian wound healing also occur in the axolotl following injury. Indeed, our histological analysis supports this result as well as an in-depth study on flank wound healing in the axolotl, which demonstrated that inflammation and a hemostatic response occurs in axolotls, but is dampened compared to mammals (Seifert et al., 2012). Although the set of genes common to all three injuries provides insight about wound closure, inflammation, and immunity, we focus in the following paragraphs on gene expression patterns that associate specifically with limb regeneration.
Fig. 4. Summary of differentially regulated genes during limb regeneration.
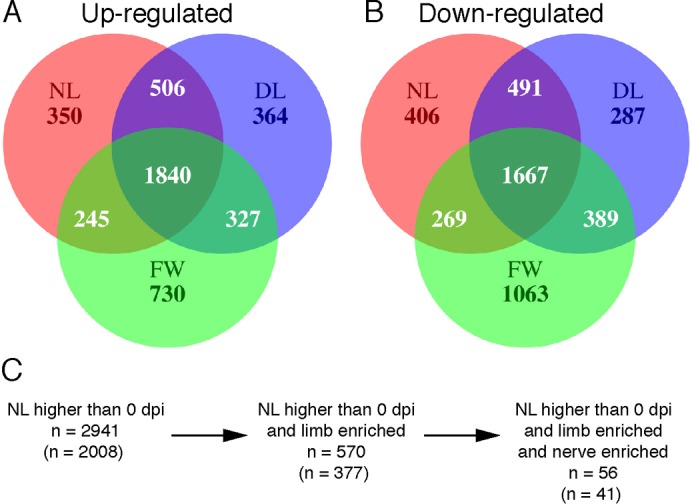
(A,B) Venn diagram showing the number of probe sets that measured significantly higher (A) and lower (B) transcript abundances in injured tissues at either 1 dpi versus baseline, 3 dpi versus 1 dpi, or 7 dpi versus 3 dpi. The total number of differentially regulated genes is represented for each injury type. (C) A schematic representing the progression from the total number of probe sets with higher transcript abundance in injured NL tissues compared with baseline (red circle in A) to the identification of limb-specific and nerve-dependent genes during the first 7 dpi. Numbers outside parentheses represent the total number of probe sets identified and the numbers within parentheses represent unique probe sets that have presumptive human orthologs.
Limb-enriched gene expression patterns
Overall, 2941 probe sets measured higher transcript abundances above baseline during the first seven days in NL (supplementary material Table S1). This list was filtered to identify genes with higher transcript levels in NL versus FW at 0, 1, 3, or 7 dpi; this yielded a list of 570 injury-induced, limb-enriched genes, of which 377 annotate to unique presumptive human orthologs (Fig. 4C; supplementary material Table S1). Annotation of gene ontologies for these probe sets identified a substantial number of genes involved in biological processes linked to limb development including developmental process (n = 125), mesoderm development (n = 70), and ectoderm development (n = 70) (supplementary material Table S3). In fact, mutations in 31 of these unique limb-enriched genes manifest human or mouse developmental limb defects when genetically disrupted, strongly suggesting that these genes play pivotal roles in vertebrate limb formation (Table 1). Furthermore, comparing our list to other genomic screens of limb regeneration (Campbell et al., 2011; Monaghan et al., 2009) we identified 73 genes that were commonly identified as highly expressed in amputated limbs (supplementary material Table S1).
Table 1. List of up-regulated, limb-enriched genes that cause limb defects in humans or mice when mutated.
Each of the 377 up-regulated, limb-enriched genes was queried against OMIM and Pubmed to identify published examples demonstrating that gene mutations cause congenital limb defects. Fold change differences between NL and FW are shown on the right.
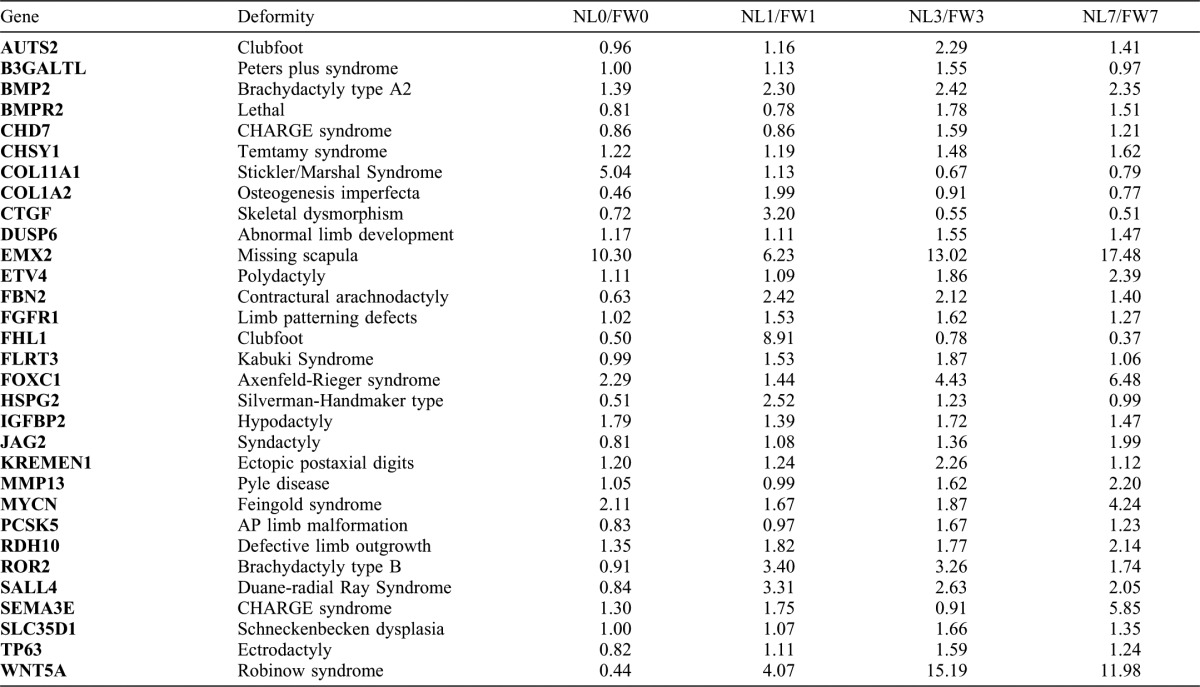
Key regulatory genes involved in signaling pathways known to be necessary for limb development and limb regeneration were found in the limb-enriched list including genes integral to β-catenin-independent Wnt/planar cell polarity signaling (prickle1, prickle2, wnt5a, fzd2, fzd8, and ror2) (Stoick-Cooper et al., 2007), retinoic acid signaling (aldh1a3, crabp1, crabp2, and rdh10) (Blum and Begemann, 2012), insulin growth factor signaling (ctgf [igfbp8], cyr61 [igfbp10], igfbp2, igfbp3, htra1, and kazald1 [igfbp-rP10]) (Chablais and Jazwinska, 2010), FGF signaling (dusp6, fgfr1, and pdlim7) (Lee et al., 2009), and BMP signaling (bmp2, id3, bmp2r) (Guimond et al., 2010). Overall, this list supports the hypothesis that some gene expression programs used in development are re-deployed during limb regeneration (Muneoka and Sassoon, 1992). Surprisingly, genes associated with limb patterning and growth were up-regulated before considerable increases in cell proliferation and blastemal outgrowth, suggesting that patterning of the limb blastema may occur in parallel or prior to blastema growth.
Limb-enriched and nerve-dependent gene expression patterns
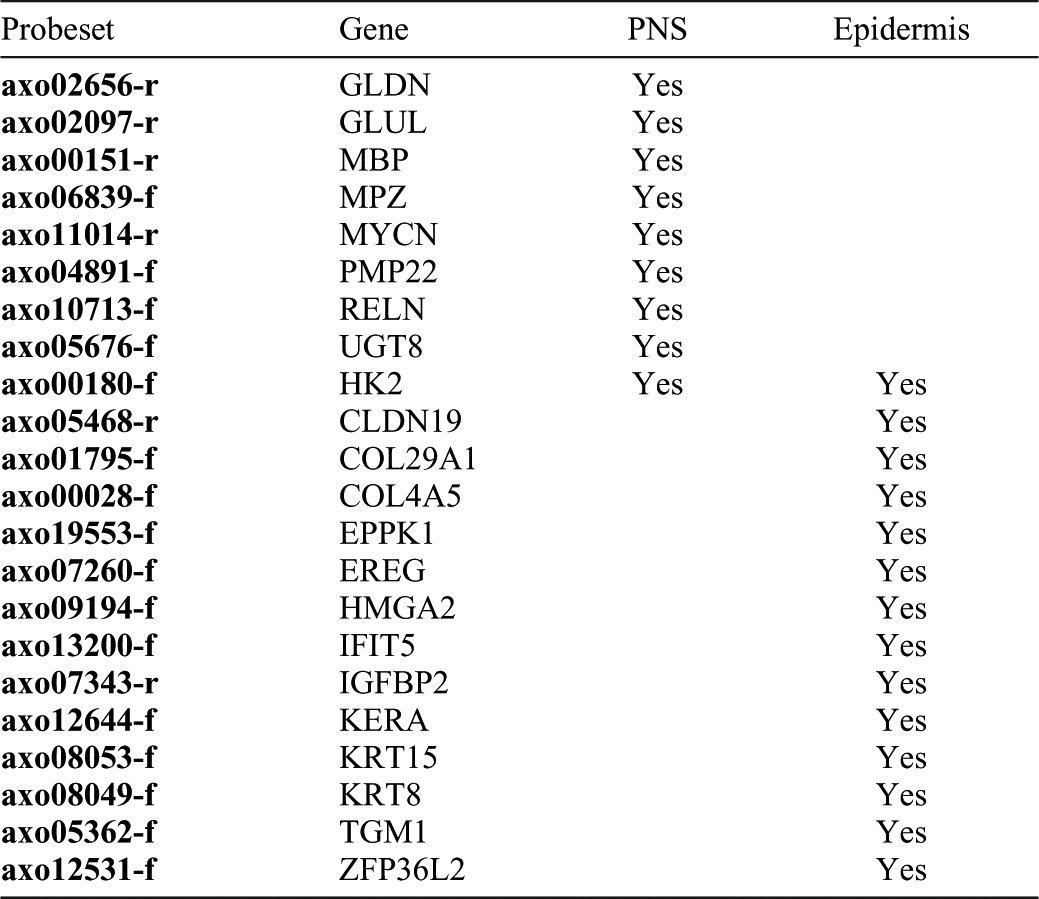
To identify nerve-dependent genes, limb-enriched genes were filtered to identify probe sets that measured higher transcript levels in NL versus DL at 1, 3, or 7 dpi. This strategy identified a short list of 56 genes (41 unique transcripts with presumptive human orthologs) that were up-regulated after injury, had higher transcript abundance in NL versus FW, and had higher transcript abundance in NL versus DL (Fig. 4C; supplementary material Table S1). This list was significantly enriched for genes that annotate to developmental process (n = 19), ectoderm development (n = 6), cell cycle (n = 8), and neurological system process (n = 7) ontology terms (supplementary material Table S4). Further annotation through PubMed searches showed that 14 of the 40 genes are important in epithelial function (Table 2), including genes important in maintaining the structure of the epithelia (krt8, kera, krt15, cldn19, col29a1, eppk1, and tgm1), epithelial cell growth factors (ereg), and transcription factors involved in keratinocyte growth and differentiation (zfp36l2, ifit5), suggesting that these genes are necessary for the maturation of the WE into the AEC. We also identified 9 out of 40 genes that are highly expressed in the peripheral nervous system of vertebrates including four genes that are highly abundant in peripheral nervous system myelin Schwann cells (mbp, pmp22, gldn, and mpz) (Table 2). Overall, this list of genes suggests that denervation affects maturation of the WE and behavior of Schwann cells within the first week of regeneration.
Table 2. List of up-regulated, limb-enriched, and nerve-dependent genes (n = 41) that play a role in epithelial function (n = 14) or peripheral nerve development or myelination (n = 9).
Only one of four significant probe sets that represent EPPK1 is shown.
Localization of up-regulated, limb-enriched, and nerve-dependent transcripts
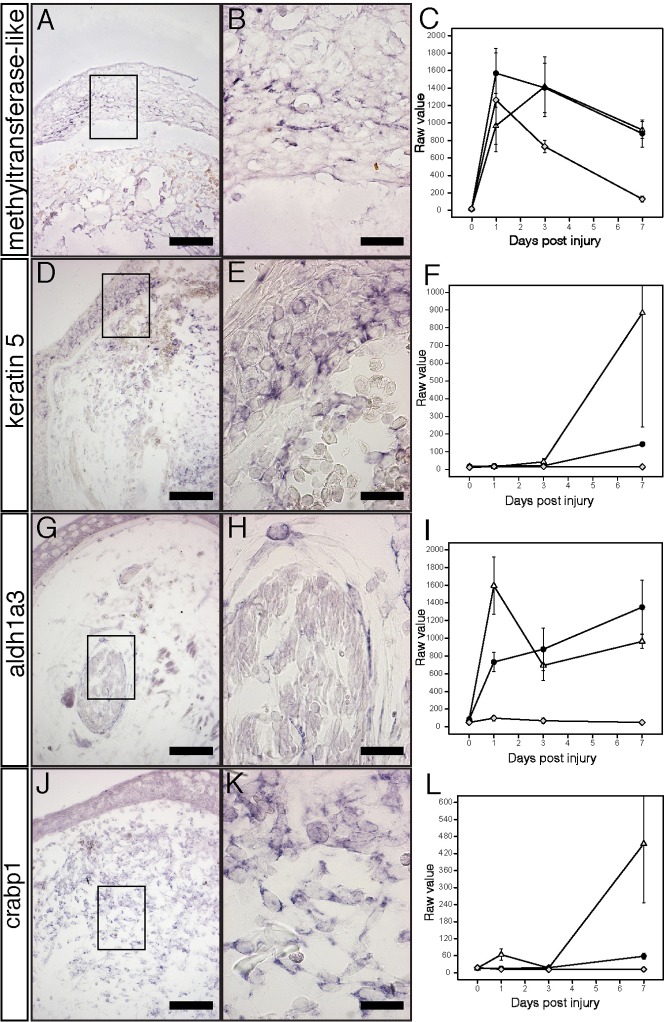
In situ hybridization was used to localize mRNA expression of limb-enriched and nerve-dependent genes at 7 dpi. The results show considerable variation in the location of transcripts among WE keratinocytes, blastema cells, and peripheral nerves (Fig. 5). For example, a putative S-adenosylmethionine-dependent methyltransferase (axo23458-r) was expressed in keratinocytes of the WE and not the underlying mesenchyme of 7 dpi limbs (Fig. 5A,B). This methyltransferase-like gene was highly expressed in each injury type at 1 dpi, but expression was sustained at higher levels in NL and DL at 7 dpi (Fig. 5C). This gene was identified in other genomic screens of limb regeneration (supplementary material Table S1), making it a promising candidate for its involvement in WE function after injury.
Fig. 5. In situ hybridization (ISH) of limb-enriched genes in NL 7 dpi limbs.
(A,B) ISH staining of methyltransferase-like (axo23458-r) showing specific staining in the WE. Close-up of boxed area can be seen in B. (C) Transcriptional profile of methyltransferase-like showing strong up regulation in all injury types, but sustained expression in limbs. Y axis is the raw microarray value with error bars indicating ± SEM. Grey diamond indicates FW. Black circle indicates DL. White triangle indicates NL. (D,E) ISH of krt5 (axo06032-f) showing expression in the WE and underlying mesenchyme. (F) Transcriptional profile of krt5 showing expression only in innervated limbs. (G,H) ISH of aldh1a3 (axo07976-r) showing specific staining in cells surrounding and within peripheral nerve bundles. (I) Transcriptional profile of aldh1a3 showing that mRNA expression is only in limbs. (J,K) ISH of crabp1 (axo10015-r) showing strong staining in mesenchymal cells throughout the early blastema. (L) Transcriptional profile of crabp1 showing that mRNA expression is only expressed in innervated limbs. Scale bar in A,D,G,J = 200 µm. Scale bar in B,E,H,K = 50 µm.
A transcript highly similar to human krt5 was expressed in WE keratinocytes and the underlying mesenchyme and cartilage cells (Fig. 5D,E). In mice, krt5 and its binding partner krt15, are markers for salivary gland epithelial progenitor cells, which show decreased cell proliferation and krt5 expression upon removal of parasympathetic innervation (Knox et al., 2010). In our study, krt15 was limb-enriched and nerve-dependent, and krt5 was eight times higher in NL versus DL at 7 dpi (Fig. 5F), although highly variable estimates among replicates yielded a p-value below our statistical cutoff (P = 0.019). This strongly suggests that krt5 and krt15 are limb-specific and nerve-dependent gene candidates. In support of these results, a newt type II cytokeratin that is highly similar to our presumptive krt5 (blastn; 83% identical), is transcribed in the mesenchyme and WE of regenerating newt limbs (Ferretti et al., 1991; Ferretti and Ghosh, 1997). Together, these data suggest important roles for krt5 and krt15 in the blastema and more generally, nerve-mesenchyme-epidermis interactions that typify a normal regenerative response.
In situ hybridization also showed that genes associated with retinoic acid signaling were expressed in regenerating limbs (Fig. 5G–L). Aldh1a3, a retinaldehyde dehydrogenase that synthesizes retinoic acid during development and adulthood, was exclusively expressed in a subset of cells within peripheral nerve bundles 7 dpi (Fig. 5G,H) and did not rise above baseline levels in FW (Fig. 5I), likely because nerve bundles are only present in the limb samples. We also found that crabp1, an intracellular retinoic acid binding protein that regulates RA nuclear signaling, was up-regulated from baseline exclusively in the mesenchyme of the limb blastema and was both limb-specific and nerve-dependent (Fig. 5J–L). These expression patterns may explain why RA is necessary for appendage regeneration (Blum and Begemann, 2012; Maden, 1998) and can re-specify pattern in the regenerating axolotl limb (Maden, 1982). Further investigation is needed to identify whether aldh1a3 expressing cells are producing RA, signal to crabp1-expressing blastemal cells, and if this process is necessary for regeneration. Overall, our ISH analyses show that the genes identified in our study are expressed in three tissues that mediate blastema formation; the epidermis, mesenchyme, and peripheral nerve. It also suggests that each of these tissues is affected by denervation prior to blastema formation.
Differential expression between innervated and denervated limbs
Previous studies have shown that gene expression between innervated and denervated limbs is often quantitatively changed rather than absolutely (Monaghan et al., 2009). In order to address this possibility, differentially regulated genes were identified between innervated and denervated limbs regardless of expression changes from baseline at 1, 3, and 7 dpi. Comparing NL and DL at 1 dpi identified a small list of 25 unique genes (supplementary material Table S1) that presented higher transcript abundances in NL and were significantly enriched for genes that annotate to developmental, neurological, and systems process ontology terms (Table 3). This list includes genes associated with microvascular morphogenesis (krit1), blood coagulation (f5), retinoic acid binding (crabp1), extracellular matrix structure (col29a1), myelin synthesis and structure (mbp, pmp22, mpz), axon guidance (reln, homer1), and axon development (gldn). The genes that presented higher transcript abundances in denervated limbs included three genes, xdh, alox12b, and alox15b, that enriched 2 ontology terms, respiratory electron chain transport and generation of precursor metabolites and energy. Other genes in this list are predicted to function in the regulation of apoptosis (btg3), neurogenesis and pluropotency (rbbp9), and neurodegeneration (yars). These results show that denervation significantly alters transcription as early as 24 hours post amputation and we note that several of these differentially expressed genes were also found in our lists of limb-enriched and nerve-dependent genes.
Table 3. List of statistically over-represented biological process terms identified from genes with higher transcript abundance in NL versus DL.
The numbers reference the observed number of genes in each process.

More genes were differentially expressed at Day 3 than Day 1 (n = 52 unique genes) (supplementary material Table S1) and these enriched different biological process terms between innervated and denervated limbs (Table 3). Genes that were expressed more highly in innervated limbs enriched carbohydrate metabolism, transport, hematopoeisis, and B-cell immunity biological process terms. In addition, this list included genes associated with: (1) Schwann cells and neurons (gfpt2, tuba1a, glul, lnx1, marveld2), (2) extracellular matrix structure and synthesis (ugdh, slc23d2), (3) regulation of epithelial-mesenchymal transition (EMT) (fam3c, hmga2), (4) regulation of skeletal development (wsb1, tpp3), (5) FGF-signaling of angiogenesis (cav1) and limb development (pdlim7), and (6) regulation of epidermal cell differentiation and proliferation (tgm1, ovol2, lmo7, ehf, ereg, sorbs3, eppk1). The results show that a diverse group of developmentally important genes are differentially regulated between innervated and denervated limbs by 3 dpi.
The largest number of differentially expressed genes was discovered for 7 dpi (n = 103 unique genes) (supplementary material Table S1), and again, these enriched different biological process terms between innervated and denervated limbs (Table 3). Approximately 50% of the 103 genes that were expressed more highly in innervated limbs annotated to cell cycle and mitosis-related gene ontologies. These genes encode regulators of the cell cycle (ccna2, ccnb1, ccnb2, ccnb3, cdc2, tk1, ube2c, uhrf1), chromosome condensation and DNA repair (smc2, smc4, ncpag, c16orf75, pcna), genome replication (mcm2, mcm3, mcm4, mcm6, mcm7), and chromosome segregation (fbxo5, mad2l1, aurka, aurkb, aspm, kif11). In addition, 36 genes significantly enriched the nucleic acids metabolic process term, and the developmental process term was also enriched with genes associated with cell proliferation and differentiation (bcn1, zfhx4, krt15, krt8, crabp1, lingo1, lnx1, hmgb3, tk1). This strong signature of cell proliferation and DNA synthesis was missing in the list of genes that were significantly up-regulated >1.5 fold from baseline levels in NL (supplementary material Table S2). This suggests that cell cycle components are affected in a quantitative manner by denervation and they only begin to increase above baseline levels at 7 dpi in NL. This is in support of our histological analysis (Fig. 3), which showed that cell proliferation was present in both DL and NL at 7dpi, suggesting that proliferation dynamics are just beginning to diverge at this time (Fig. 3). Overall, the results show that denervation has a major effect on the transcription of proliferation-associated genes that are likely required for blastema growth.
Technical and biological replication of microarray results
To validate and extend the Affy microarray results, expression values were estimated using the nCounter platform. This analysis used 24 RNA samples from the Affymetrix analysis plus a newly generated set of 24 biological replicates. Custom Nanostring capture probes were designed for 50 genes (supplementary material Table S5) and fold change estimates were obtained between Day 0, 1, and 7 time points for innervated and denervated limbs. The correlation of fold change for the technical replicate samples was uniformly high across times and treatments (r = 0.95–0.97) (supplementary material Table S6). Thus, the Affy and nCounter platforms yielded precise estimates of fold change when the same RNA samples were processed. Precise estimates of fold change were also obtained between the Affy and nCounter platforms for biological replicates, and also between the two sets of 24 samples processed on the nCounter instrument. However, relative to the high correlation between technical replicates, the correlations between biological replicates were relatively lower for the Day1 and Day7 comparisons (r = 0.80–0.87) and lower still for all comparisons to Day 0 (r = 0.59–0.72). These results suggest that more variation is present between biological replicates than technical replicates and that the abundance of transcripts is most variable among samples that were collected at the time of limb amputation. This may be because animals were at different stages of the molting cycle at the time of collection. Overall, replication of the microarray results was high using the nCounter platform, demonstrating the reliability of each platform.
Discussion
A recent comparison of transcription between innervated and denervated limbs of the Mexican axolotl provided the first global, transcriptional description of the limb regeneration program (Monaghan et al., 2009). That study used a small format microarray (∼4500 probes with 3271 presumptive human orthologs) to detail gene expression of whole blastemas at 5 days and 14 days after limb amputation. However, a more comprehensive analysis of gene expression was needed in concert with an earlier and more precise tissue-sampling scheme to thoroughly investigate the transcriptomics of blastema formation. To this end, we investigated transcription within axolotl epithelium and subjacent cells during the first week of limb regeneration with the primary goal of identifying a core set of genes that are likely to be necessary for limb regeneration. To meet this goal, we devised a strategy that allowed us to subtract out genes common to all injury responses as well as to identify genes that are uniquely expressed in limbs. We further selected genes specific to limbs that regenerate (NL) rather than regress (DL) to identify genes associated with blastema formation. Our study is the most detailed molecular analysis of limb regeneration to date and is the first to identify genes specific to the limb regeneration process by comparing the general wound healing response outside a limb field. Overall, the genes identified here will be useful as tissue specific markers for regenerating limbs and candidates for regulating blastema formation.
At both histological and transcriptional levels, we show that the initial injury response is similar between NL, DL, and FW. The time to re-epithelialization was within 1 dpi and many of the same genes were differentially regulated in NL, DL, and FW. Interestingly, many of these injury-response genes are similarly regulated in mammals, suggesting conservation of some aspects of wound healing among tetrapods.
Previous studies have shown that there are fundamental differences between limb skin and flank skin. When forelimb skin is replaced with grafts of flank skin in newts and axolotls, limb regeneration is defective (Tank, 1984; Tank, 1987). We compared gene expression differences between NL and FW and identified transcripts with limb-specific expression patterns. For example, the homeobox-containing transcription factor, emx2, was highly expressed in uninjured limb skin and was up-regulated after injury only in limb samples (supplementary material Table S1). Mouse emx2 null mutants fail to form a scapula during development (Table 1) (Capellini et al., 2010; Pellegrini et al., 2001) and newt emx2 is expressed in a graded proximodistal manner mainly in the epidermis of regenerating newt limbs (Beauchemin et al., 1998). Emx2 and other limb-specific genes identified in our study (supplementary material Table S1) may regulate limb-specific patterning events during regeneration.
Our analysis also identified a connection between salamander limb-enriched genes and orthologs that are associated with limb deformities in mammals (Table 1). For example, numerous genes involved in the Wnt/Planar cell polarity (PCP) signaling pathway were up-regulated in DL and NL, but remained at baseline levels in FW. Activation of PCP signaling by Wnt5 ligand through Vangl and Ror2 activation regulates limb bud elongation during mammalian development (Gao et al., 2011) and Wnt5a activity is necessary for axolotl limb regeneration (Ghosh et al., 2008). WNT/PCP signaling is thought to stabilize cellular polarity in epithelium of developing limbs, organize directional cell migration, and regulate directional cell proliferation (Wang et al., 2011). Overall, it is clear that activation of Wnt signaling through Wnt5a is necessary for limb outgrowth, but the key problem is to identify the property of salamander limbs that allows this pathway to re-activate after injury while not being induced after a flank injury. It is possible that sustained expression of genes like emx2 into post-embryonic and larval stages allows accessibility of this important signaling pathway in adult axolotls.
Genes involved in other important signaling pathways were also up-regulated specifically in limb samples. For example, genes associated with retinoic acid (RA) signaling were dynamically expressed in limbs after injury. Retinoic acid is an important signaling molecule involved in the development and regeneration of limbs; disruption of this pathway disrupts limb formation (Blum and Begemann, 2012; Kikuchi et al., 2011; Maden, 1998; Maden, 2007). We found that crabp1 was only up-regulated in NL and was expressed exclusively in the limb mesenchyme. In contrast, we found that crabp2 was up-regulated in NL, DL, and FW at 7 dpi. Our findings are in accordance with previous studies showing that CRABP protein is up-regulated during regeneration, although it is unclear whether these studies were detecting CRABP1 or CRABP2 (Maden et al., 1989; McCormick et al., 1988). CRABPs are intracellular RA binding proteins that are thought to shuttle RA to the nucleus to regulate RA-mediated transcription, which may explain why we observe their expression during limb regeneration (Noy, 2000). We also found that aldh1a3 and rdh10, enzymes involved in the synthesis of RA during development, were up-regulated in NL and DL at 1 dpi and aldh1a3 was expressed specifically in cells resembling perineural fibroblasts in peripheral nerve bundles. Altogether, our data suggest that RA signaling is a dynamic process during limb regeneration and identifies the genes that may mediate the necessity of RA during epimorphic regeneration (Blum and Begemann, 2012; Kikuchi et al., 2011).
Beyond signaling pathways, structural proteins showed very specific transcriptional profiles in regenerating tissues. Numerous keratins (krt5, krt8, krt15, and krt13) and keratin-associated molecules (eppk1, tgm1, kera) were up-regulated after injury and were enriched in limbs. Furthermore, some genes like krt8 and krt15 were highly nerve-dependent. Keratins are components of intermediate filaments that protect the structural integrity of cells, but have recently been implicated in other cellular processes including cell motility, cell signaling, cell growth, and cancer metastasis (Karantza, 2011; Windoffer et al., 2011). Although previous studies in newts have identified keratins NvKII, krt8, and krt18 in mesenchymal and WE cells during limb regeneration (Ferretti et al., 1991; Ferretti and Ghosh, 1997) and knockdown of krt8 and krt18 in newt blastemal cells in vitro decreased DNA synthesis (Corcoran and Ferretti, 1997), our understanding of these proteins during regeneration remains poor. Functional testing is necessary to determine if keratin proteins play solely a supportive, structural role during regeneration or whether they are mediating cell signaling to promote growth or patterning. Together, the highly limb-specific and nerve-dependent expression patterns of the keratin genes strongly suggest that they are integral to the formation of the blastema.
Other limb-enriched genes were more quantitatively different than FW rather than being expressed exclusively in the limb. For example, two possible salamander-specific genes, sodefrin-like (axo22108-r) and methyltransferase-like (axo23458-r), were up-regulated in NL, DL, and FW, exclusively in the epidermis (Fig. 5A,B; data not shown), but expression was only maintained in NL and DL. This suggests that these molecules are not limb-specific, although sustained expression in the limb WE may impose some necessary function to the WE during limb regeneration. Regardless, the fact that these genes seem to be unique to salamanders (Campbell et al., 2011) and show strong and specific expression in the WE warrants further functional studies.
A surprising result was the observation that myelin-associated genes were up-regulated and both limb-specific and nerve-dependent. Myelinated peripheral nerves permeate throughout the uninjured limb, but only naked sensory nerve fibers are found in uninjured epidermis of animals (Boulais and Misery, 2008). Hence, our tissue collection scheme did not sample myelinated nerve fibers in uninjured samples, yet injured NL and DL samples contained transected nerve bundles located just proximal to the WE. This likely explains why mRNA levels of myelin-associated genes increased above baseline in NL at 1 dpi. The fact that myelin-associated gene mRNA did not increase in DL suggests that expression of these genes was lost following denervation. A similar phenomenon takes place in mammals, where peripheral nerve fiber transection down-regulates expression of myelin-associated genes in distal Schwann cells (Hall, 2005). This result is interesting because it suggests that Schwann cells are affected early after denervation, which may have detrimental effects on downstream blastema formation. In newts, the protein Anterior Gradient 2 is expressed in Schwann cells after limb amputation and supplemental Anterior Gradient 2 can partially rescue regeneration in the denervated state (Kumar et al., 2007). Others have shown that denervation in axolotls induces peripheral nerves to become inhibitory to limb regeneration, suggesting that they may secrete inhibitory factors (Irvin and Tassava, 1998; Tassava and Olsen-Winner, 2003). It will be critical in future experiments to determine if the response of Schwann cells to denervation is the cause of a loss of blastema formation.
The proliferation of blastema cells is known to be a target of the nerve during limb regeneration (Stocum, 2011). In order to increase our sensitivity for identifying proliferation-associated genes during regeneration, we directly compared NL to DL without comparing samples to baseline or FW. This analysis showed that by 7 dpi, approximately 50% of the genes that were higher in NL versus DL were associated with the cell cycle, supporting the notion that the cell cycle is the primary target of denervation. Most of these genes were only different at 7 dpi, suggesting that our study identified the genes likely upstream of the cell proliferation effect of denervation. This result highlights that the limb-enriched and nerve-enriched genes we identified in our study are excellent candidates for regulating the increase in cell proliferation that is characteristic to limb regeneration. Overall, our study used a focused approach to identify the genes that are likely necessary for limb regeneration and showed that many of these genes are expressed in specific tissues and before considerable outgrowth takes place in the limb. The identification of these genes is an important advance in our ability to tease apart the cellular and molecular mechanisms that drive regeneration and will be a useful resource for regeneration researchers that may be looking for specific genes to analyze during early blastema formation.
Materials and Methods
Animals and surgical procedures
Axolotls were obtained from the Ambystoma Genetic Stock Center, Lexington, KY and raised to 7–10 cm snout to vent length. Animal care and use procedures were approved by the University of Florida IACUC (Application Number 201101534). Denervations were performed by anesthetizing animals in 0.01% benzocaine, making a small incision at the shoulder to expose the brachial nerves entering each forelimb, and severing the nerve bundles with surgical scissors. Limb amputations were performed at the mid-stylopod and the humerus was trimmed to make the amputation plane flush. Full thickness excisional wounds were performed along the flank of anesthetized animals using a 4 mm biopsy punch tool.
Histology and BrdU analysis
Tissues were processed for paraffin embedding, sectioned at 5 µm, and stained according to previous methods (Seifert et al., 2012) except that limb samples were decalcified in 10% EDTA for 3 days with daily changes at 4°C before histological processing. For DNA synthesis analysis, animals were injected with bromodeoxyuridine (BrdU) (conc. = 100 mg/g) 24 hours before tissue collection, harvested 24 hours later, and processed for paraffin embedding. After sectioning, sections were de-paraffinized, blocked for endogenous peroxidase activity in 3% H2O2 in methanol for 10 mins, rehydrated, treated for antigen retrieval in pH 6.0 sodium citrate buffer in a microwave for 25 mins, rinsed in water, incubated in 37°C 2N HCl for 15 mins, rinsed thoroughly in water, rinsed with TBS, blocked with rabbit serum, blocked for endogenous avidin and biotin, incubated with primary antibody rat anti-BrdU (1:500, Accurate Scientific), washed, incubated with biotinylated secondary anti-rat (1:400, Vector Scientific), washed and visualized using Vector ABC horseradish peroxidase and DAB reagents according to manufacturer's instructions. Tissue sections were counterstained with Hematoxylin (Vector).
Tissue collection for microarray analysis
One day prior to limb amputation, 12 axolotls were anesthetized and their forelimbs denervated. Approximately 24 hours later, these same axolotls with denervated limbs and 16 additional axolotls with innervated limbs were anesthetized and administered amputations at the mid-stylopod of both forelimbs (Fig. 1A,B). The epidermis adjacent to the amputation plane was taken from the arms of each of four individuals that were not denervated the day before; these served as Day 0 samples for the innervated and denervated limbs. To obtain a sufficient amount of RNA for microarray analysis, both forelimb samples from each individual were pooled to yield independent, replicate samples. An additional 16 axolotls were then anesthetized and a full thickness excisional wound was performed along the flank of each animal (Fig. 1C). After 1, 3, and 7 days post injury, the wound epithelium was removed and any cells that were adhered to the epithelium were included in the sample. Two tissues were pooled from each individual to obtain 4 replicate samples for each time point and tissue type.
RNA isolation and microarray analysis
Total RNA was isolated from all 44 samples and each was processed for hybridization to 44 independent and custom A. mexicanum (Amby_002) Affymetrix GeneChips (Huggins et al., 2012). Microarray results from FW samples are summarized in a different manuscript (Seifert et al., 2012), but the entire dataset can be found at the Gene Expression Omnibus (Accession number GSE37198). This Amby_002 GeneChip contains approximately ∼20,000 perfect match probesets. The probesets were designed using A. mexicanum expressed sequence tag contigs from Sal-Site (Smith et al., 2005). The GeneChips were processed by the University of Kentucky Microarrray Core Facility and expression values were extracted using RMA (Irizarry et al., 2003) and Affymetrix Expression Console software. The resulting data were subjected to one way Analysis of Variance using JMP Genomics version 4.1 and statistical estimates were defined to make 16 comparisons between groups (Fig. 1D). A gene was identified as differentially expressed if it passed a false discovery rate of ≤ 0.05 and had a fold change of ≥ 2, or passed a false discovery rate ≤ 0.001 and a fold change of ≥ 1.5. Genes that were defined as differentially expressed were analyzed further using pair-wise comparisons at a significance threshold of P<0.003 and a fold change cutoff of 1.5 for the comparison of interest. This threshold was determined using a Bonferroni correction to adjust for 16 pairwise comparisons at an alpha level of 0.05. The R package, VennDiagram was utilized to generate Venn diagrams (Chen and Boutros, 2011). Significant genes were annotated with gene ontology information from Panther (http://www.pantherdb.org) and gene lists were compiled and compared to identify biological processes that were statistically over-represented. For all analyses, the 11,131 probesets on the Ambystoma GeneChip that could be mapped to human orthologs in the Panther database were used to generate expected values (i.e., as the background). The count threshold was set to three and the significance threshold was set to P<0.05. The lists of significant biological process terms were manually inspected to remove redundant terms.
Cloning and RNA probe production
Axolotl genes were cloned using gene-specific primers designed using sequences collected from the Ambystoma Gene Collection (Smith et al., 2005). Total RNA was isolated from 7 dpi limb tissue and used to make cDNA template (iScript; BioRad). Genes were amplified as follows: krt5 primers were 5′ GAG GGA GCA GGT TCT GTG AG 3′ and 5′ ATC ACC CAG CCA GAA GAA TG 3′; aldh1a3 primers were 5′ CCT GCA TTG TGT TTG CTG AC 3′ and 5′ TGT CAG AGC CGG ATA ATT CA 3′; crabp1 primers were 5′ AGG AGT CCC CTG ACT TGG AG 3′ and 5′ TGC CAC CAC AAA TGA TGA GT 3′. PCR products were gel isolated, cloned into pGEM-T Easy Vectors (Promega), and sequence verified. methyltransferase-like primers 5′ TAA TAC GAC TCA CTA TAG GGA GAC AGC TCT GTG GAT CTG GTC A 3′ and 5′ ATT TAG GTG ACA CTA TAG AAG AGT CTC TAA GGT GCG GCT TGT T 3′ were used to make a PCR template that was used to generate a digoxygenin-labelled RNA probe using a Roche RNA labeling kit.
In situ hybridization
Limbs were collected 7 dpi and fixed overnight in 4% PFA at 4°C, mounted in optimal cutting medium, sectioned at 20 µm, dried for two hours, and processed for in situ hybridizations on the same day according to previously published methods (David Parichy, personal communication). Proteinase k treatment consisted of 10 minutes at 10 µg/ml concentration. Probe concentration was 0.5 µg/ml in hybridization solution at 55°C overnight. Anti-DIG antibody was incubated at 1:5000 dilution at 4°C overnight. Stained sections were mounted in 80% glycerol and images captured on a Nikon Eclipse 6600 upright compound microscope using a Cool-Snap Pro true color camera.
Nanostring nCounter development and analysis
Transcript abundance estimates obtained from the Ambystoma Affy GeneChip were compared to estimates obtained from the Nanostring nCounter System. The nCounter is a moderate throughput gene expression analysis instrument that estimates the number of RNA transcripts from samples of total RNA or lysed tissues. Nanostring staff designed capture probes for 50 genes from the Affy GeneChip (supplementary material Table S5) and processed 48 RNA samples. Twenty-four of the RNA samples corresponded to the same replicate Day 0, Day 1, and Day 7 RNA samples that were used in the Affymetrix experiment. The second group of 24 samples corresponded to a new set of replicate D0, D1, and D7 samples. The count data for all genes were normalized to the counts of two capture probes that were consistently expressed across the innervated and denervated treatments. Technical correlation of gene expression was examined between the Affymetrix and Nanostring platforms by calculating Pearson's correlation coefficient (r) across all 50 genes for fold change estimates obtained using the same RNA samples. Biological correlation of gene expression was examined between the Affymetrix and Nanostring platforms, and between the two sets of replicates processed on the Nanostring platform. Again, Pearson's correlation coefficient (r) was calculated across all 50 genes for fold change estimates obtained between different RNA samples.
Supplementary Material
Acknowledgments
The axolotls were obtained from the Ambystoma Genetic Stock Center at the University of Kentucky, which is funded by the National Science Foundation [DBI-0951484 to S.R.V.]. The Ambystoma GeneChip was generated under National Institute of Health [R24-RR016344 to S.R.V.] and Army Research Office [W911NF-09-1-0305 to S.R.V.]. The work was supported by National Institute of Health [RC2-NS069480 to S.R.V. and M.M.] and funding from The Regeneration Project at University of Florida to S.R.V., M.M., D.M.G., and J.R.M.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Beauchemin M., Del Rio–Tsonis K., Tsonis P. A., Tremblay M., Savard P. (1998). Graded expression of Emx-2 in the adult newt limb and its corresponding regeneration blastema. J. Mol. Biol. 279, 501–511 10.1006/jmbi.1998.1782 [DOI] [PubMed] [Google Scholar]
- Blum N., Begemann G. (2012). Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development 139, 107–116 10.1242/dev.065391 [DOI] [PubMed] [Google Scholar]
- Boilly B., Albert P. (1990). In vitro control of blastema cell proliferation by extracts from epidermal cap and mesenchyme of regenerating limbs of axolotls. Dev. Genes Evol. 198, 443–447 10.1007/BF00399054 [DOI] [PubMed] [Google Scholar]
- Boulais N., Misery L. (2008). The epidermis: a sensory tissue. Eur. J. Dermatol. 18, 119–127. [DOI] [PubMed] [Google Scholar]
- Campbell L. J., Crews C. M. (2008). Molecular and cellular basis of regeneration and tissue repair: wound epidermis formation and function in urodele amphibian limb regeneration. Cell. Mol. Life Sci. 65, 73–79 10.1007/s00018-007-7433-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L. J., Suárez–Castillo E. C., Ortiz–Zuazaga H., Knapp D., Tanaka E. M., Crews C. M. (2011). Gene expression profile of the regeneration epithelium during axolotl limb regeneration. Dev. Dyn. 240, 1826–1840 10.1002/dvdy.22669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellini T. D., Vaccari G., Ferretti E., Fantini S., He M., Pellegrini M., Quintana L., Di Giacomo G., Sharpe J., Selleri L.et al. (2010). Scapula development is governed by genetic interactions of Pbx1 with its family members and with Emx2 via their cooperative control of Alx1. Development 137, 2559–2569 10.1242/dev.048819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F., Jazwinska A. (2010). IGF signaling between blastema and wound epidermis is required for fin regeneration. Development 137, 871–879 10.1242/dev.043885 [DOI] [PubMed] [Google Scholar]
- Chalkley D. T. (1954). A quantitative histological analysis of forelimb regeneration in Triturus viridescens. J. Morphol. 94, 21–70 10.1002/jmor.1050940103 [DOI] [Google Scholar]
- Chen H., Boutros P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran J. P., Ferretti P. (1997). Keratin 8 and 18 expression in mesenchymal progenitor cells of regenerating limbs is associated with cell proliferation and differentiation. Dev. Dyn. 210, 355–370 [DOI] [PubMed] [Google Scholar]
- Dungan K. M., Wei T. Y., Nace J. D., Poulin M. L., Chiu I.–M., Lang J. C., Tassava R. A. (2002). Expression and biological effect of urodele fibroblast growth factor 1: relationship to limb regeneration. J. Exp. Zool. 292, 540–554 10.1002/jez.10077 [DOI] [PubMed] [Google Scholar]
- Ferretti P., Ghosh S. (1997). Expression of regeneration-associated cytoskeletal proteins reveals differences and similarities between regenerating organs. Dev. Dyn. 210, 288–304 [DOI] [PubMed] [Google Scholar]
- Ferretti P., Brockes J. P., Brown R. (1991). A newt type II keratin restricted to normal and regenerating limbs and tails is responsive to retinoic acid. Development 111, 497–507. [DOI] [PubMed] [Google Scholar]
- Fox H. (1986). The skin of amphibia. Biology Of The Integument, Vol. 2, Vertebrates (ed. Bereiter–Hahn J, Matoltsy A G, Sylvia Richards K.), pp. 78–148 Berlin; Heidelberg: Springer-Verlag. [Google Scholar]
- Gao B., Song H., Bishop K., Elliot G., Garrett L., English M. A., Andre P., Robinson J., Sood R., Minami Y.et al. (2011). Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev. Cell 20, 163–176 10.1016/j.devcel.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Roy S., Séguin C., Bryant S. V., Gardiner D. M. (2008). Analysis of the expression and function of Wnt-5a and Wnt-5b in developing and regenerating axolotl (Ambystoma mexicanum) limbs. Dev. Growth Differ. 50, 289–297 10.1111/j.1440-169X.2008.01000.x [DOI] [PubMed] [Google Scholar]
- Globus M., Vethamany–Globus S. (1985). In vitro studies of controlling factors in newt limb regeneration. Regulation Of Vertebrate Limb Regeneration (ed. Sicard R E.), pp. 106–127 New York: Oxford University Press. [Google Scholar]
- Globus M., Smith M. J., Vethamany–Globus S. (1991). Evidence supporting a mitogenic role for substance P in amphibian limb regeneration. Involvement of the inositol phospholipid signaling pathway. Ann. N. Y. Acad. Sci. 632, 396–399 10.1111/j.1749-6632.1991.tb33135.x [DOI] [PubMed] [Google Scholar]
- Goss R. J. (1956). Regenerative inhibition following limb amputation and immediate insertion into the body cavity. Anat. Rec. 126, 15–27 10.1002/ar.1091260103 [DOI] [PubMed] [Google Scholar]
- Guimond J.–C., Lévesque M., Michaud P.–L., Berdugo J., Finnson K., Philip A., Roy S. (2010). BMP-2 functions independently of SHH signaling and triggers cell condensation and apoptosis in regenerating axolotl limbs. BMC Dev. Biol. 10, 15 10.1186/1471-213X-10-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. (2005). The response to injury in the peripheral nervous system. J. Bone Joint Surg. Br. 87–B, 1309–1319 10.1302/0301-620X.87B10.16700 [DOI] [PubMed] [Google Scholar]
- Hay E. D., Fischman D. A. (1961). Origin of the blastema in regenerating limbs of the newt Triturus viridescens: An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev. Biol. 3, 26–59 10.1016/0012-1606(61)90009-4 [DOI] [PubMed] [Google Scholar]
- Huggins P., Johnson C. K., Schoergendorfer A., Putta S., Bathke A. C., Stromberg A. J., Voss S. R. (2012). Identification of differentially expressed thyroid hormone responsive genes from the brain of the Mexican Axolotl (Ambystoma mexicanum). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 155, 128–135 10.1016/j.cbpc.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer–Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Irvin B. C., Tassava R. A. (1998). Effects of peripheral nerve implants on the regeneration of partially and fully innervated urodele forelimbs. Wound Repair Regen. 6, S–382–S–387 10.1046/j.1524-475X.1998.60414.x [DOI] [PubMed] [Google Scholar]
- Karantza V. (2011). Keratins in health and cancer: more than mere epithelial cell markers. Oncogene 30, 127–138 10.1038/onc.2010.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. E. (1966). The Leydig cell in larval amphibian epidermis. Fine structure and function. Anat. Rec. 154, 685–699 10.1002/ar.1091540314 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G., Poss K. D. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404 10.1016/j.devcel.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox S. M., Lombaert I. M. A., Reed X., Vitale–Cross L., Gutkind J. S., Hoffman M. P. (2010). Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science 329, 1645–1647 10.1126/science.1192046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M., Knapp D., Nacu E., Khattak S., Maden M., Epperlein H. H., Tanaka E. M. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- Kumar A., Godwin J. W., Gates P. B., Garza–Garcia A. A., Brockes J. P. (2007). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 10.1126/science.1147710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Hami D., De Val S., Kagermeier–Schenk B., Wills A. A., Black B. L., Weidinger G., Poss K. D. (2009). Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 331, 270–280 10.1016/j.ydbio.2009.05.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M. (1978). Neurotrophic control of the cell cycle during amphibian limb regeneration. J. Embryol. Exp. Morphol. 48, 169–175. [PubMed] [Google Scholar]
- Maden M. (1982). Vitamin A and pattern formation in the regenerating limb. Nature 295, 672–675 10.1038/295672a0 [DOI] [PubMed] [Google Scholar]
- Maden M. (1998). Retinoids as endogenous components of the regenerating limb and tail. Wound Repair Regen. 6, S–358–S–365 10.1046/j.1524-475X.1998.60411.x [DOI] [PubMed] [Google Scholar]
- Maden M. (2007). Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci. 8, 755–765 10.1038/nrn2212 [DOI] [PubMed] [Google Scholar]
- Maden M., Ong D. E., Summerbell D., Chytil F. (1989). The role of retinoid-binding proteins in the generation of pattern in the developing limb, the regenerating limb and the nervous system. Development 107 Suppl, 109–119. [DOI] [PubMed] [Google Scholar]
- McCormick A. M., Shubeita H. E., Stocum D. L. (1988). Cellular retinoic acid binding protein: detection and quantitation in regenerating axolotl limbs. J. Exp. Zool. 245, 270–276 10.1002/jez.1402450307 [DOI] [PubMed] [Google Scholar]
- Mescher A. L. (1976). Effects on adult newt limb regeneration of partial and complete skin flaps over the amputation surface. J. Exp. Zool. 195, 117–127 10.1002/jez.1401950111 [DOI] [PubMed] [Google Scholar]
- Mescher A. L., Connell E., Hsu C., Patel C., Overton B. (1997). Transferrin is necessary and sufficient for the neural effect on growth in amphibian limb regeneration blastemas. Dev. Growth Differ. 39, 677–684 10.1046/j.1440-169X.1997.t01-5-00003.x [DOI] [PubMed] [Google Scholar]
- Monaghan J. R., Epp L. G., Putta S., Page R. B., Walker J. A., Beachy C. K., Zhu W., Pao G. M., Verma I. M., Hunter T.et al. (2009). Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol. 7, 1 10.1186/1741-7007-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen L. M., Bryant S. V., Torok M. A., Blumberg B., Gardiner D. M. (1996). Nerve dependency of regeneration: the role of Distal-less and FGF signaling in amphibian limb regeneration. Development 122, 3487–3497. [DOI] [PubMed] [Google Scholar]
- Muneoka K., Sassoon D. (1992). Molecular aspects of regeneration in developing vertebrate limbs. Dev. Biol. 152, 37–49 10.1016/0012-1606(92)90154-9 [DOI] [PubMed] [Google Scholar]
- Noy N. (2000). Retinoid-binding proteins: mediators of retinoid action. Biochem. J. 348, 481–495 10.1042/0264-6021:3480481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M., Pantano S., Fumi M. P., Lucchini F., Forabosco A. (2001). Agenesis of the scapula in Emx2 homozygous mutants. Dev. Biol. 232, 149–156 10.1006/dbio.2001.0159 [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Bondre C., Gladstone H. B., Brown P. O., Chang H. Y. (2006). Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2, e119 10.1371/journal.pgen.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh A., Gardiner D. M., Bryant S. V., Endo T. (2007). Nerve-induced ectopic limb blastemas in the Axolotl are equivalent to amputation-induced blastemas. Dev. Biol. 312, 231–244 10.1016/j.ydbio.2007.09.021 [DOI] [PubMed] [Google Scholar]
- Satoh A., Graham G. M. C., Bryant S. V., Gardiner D. M. (2008). Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum). Dev. Biol. 319, 321–335 10.1016/j.ydbio.2008.04.030 [DOI] [PubMed] [Google Scholar]
- Seifert A. W., Monaghan J. R., Voss S. R., Maden M. (2012). Skin regeneration in adult axolotls: a blueprint for scar-free healing in vertebrates. PLoS ONE 7, e32875 10.1371/journal.pone.0032875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman R. L., Singer M. (1960). Limb regeneration without innervation of the apical epidermis in the adult newt, Triturus. J. Exp. Zool. 144, 105–109 10.1002/jez.1401440202 [DOI] [Google Scholar]
- Singer M. (1949). The invasion of the epidermis of the regenerating forelimb of the urodele, Triturus, by nerve fibers. J. Exp. Zool. 111, 189–209 10.1002/jez.1401110204 [DOI] [PubMed] [Google Scholar]
- Singer M. (1952). The influence of the nerve in regeneration of the amphibian extremity. Q. Rev. Biol. 27, 169–200 10.1086/398873 [DOI] [PubMed] [Google Scholar]
- Singer M., Salpeter M. (1961). Regeneration in vertebrates: the role of the wound epithelium. In Growth In Living Systems. (ed. Zarrow M. X.), pp. 277–311 New York: Basic Books, Inc. [Google Scholar]
- Smith J. J., Putta S., Walker J. A., Kump D. K., Samuels A. K., Monaghan J. R., Weisrock D. W., Staben C., Voss S. R. (2005). Sal-Site: integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics 6, 181 10.1186/1471-2164-6-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocum D. L. (2011). The role of peripheral nerves in urodele limb regeneration. Eur. J. Neurosci. 34, 908–916 10.1111/j.1460-9568.2011.07827.x [DOI] [PubMed] [Google Scholar]
- Stocum D. L., Dearlove G. E. (1972). Epidermal-mesodermal interaction during morphogenesis of the limb regeneration blastema in larval salamanders. J. Exp. Zool. 181, 49–61 10.1002/jez.1401810106 [DOI] [Google Scholar]
- Stoick–Cooper C. L., Weidinger G., Riehle K. J., Hubbert C., Major M. B., Fausto N., Moon R. T. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479–489 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- Taban C. (1949). les fibres nerveuses et l'épithélium dans l'édification des regenerats de pattes (in situ ou induites) chez le triton. Arch. Sci. 2, 553–561. [Google Scholar]
- Tank P. W. (1984). The influence of flank dermis on limb regeneration in the newt, Notophthalmus viridescens. J. Exp. Zool. 229, 143–153 10.1002/jez.1402290115 [DOI] [Google Scholar]
- Tank P. W. (1987). The effect of nonlimb tissues on forelimb regeneration in the axolotl, Ambystoma mexicanum. J. Exp. Zool. 244, 409–423 10.1002/jez.1402440308 [DOI] [Google Scholar]
- Tassava R. A., Garling D. J. (1979). Regenerative responses in larval axolotl limbs with skin grafts over the amputation surface. J. Exp. Zool. 208, 97–109 10.1002/jez.1402080111 [DOI] [PubMed] [Google Scholar]
- Tassava R. A., Olsen–Winner C. L. (2003). Responses to amputation of denervated Ambystoma limbs containing aneurogenic limb grafts. J. Exp. Zoolog. A Comp. Exp. Biol. 297, 64–79 10.1002/jez.a.10263 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1954). The relation of epidermal innervation to limb regeneration in Amblystoma larvae. J. Exp. Zool. 127, 577–601 10.1002/jez.1401270307 [DOI] [Google Scholar]
- Thornton C. S. (1957). The effect of apical cap removal on limb regeneration in Amblystoma larvae. J. Exp. Zool. 134, 357–381 10.1002/jez.1401340209 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1958). The inhibition of limb regeneration in urodele larvae by localized irradiation with ultraviolet light. J. Exp. Zool. 137, 153–179 10.1002/jez.1401370108 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1960a). Influence of an eccentric epidermal cap on limb regeneration in Amblystoma larvae. Dev. Biol. 2, 551–569 10.1016/0012-1606(60)90054-3 [DOI] [PubMed] [Google Scholar]
- Thornton C. S. (1960b). Regeneration of asensory limbs of Ambystoma larvae. Copeia 1960, 371–373 10.2307/1439783 [DOI] [Google Scholar]
- Thornton C. S., Steen T. P. (1962). Eccentric blastema formation in aneurogenic limbs of Ambystoma larvae following epidermal cap deviation. Dev. Biol. 5, 328–343 10.1016/0012-1606(62)90017-9 [DOI] [PubMed] [Google Scholar]
- Thornton C. S., Thornton M. T. (1965). The regeneration of accessory limb parts following epidermal cap transplantation in urodeles. Cell. Mol. Life Sci. 21, 146–148 10.1007/BF02141984 [DOI] [PubMed] [Google Scholar]
- Wang B., Sinha T., Jiao K., Serra R., Wang J. (2011). Disruption of PCP signaling causes limb morphogenesis and skeletal defects and may underlie Robinow syndrome and brachydactyly type B. Hum. Mol. Genet. 20, 271–285 10.1093/hmg/ddq462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Marchionni M. A., Tassava R. A. (2000). Cloning and neuronal expression of a type III newt neuregulin and rescue of denervated, nerve-dependent newt limb blastemas by rhGGF2. J. Neurobiol. 43, 150–158 [DOI] [PubMed] [Google Scholar]
- Windoffer R., Beil M., Magin T. M., Leube R. E. (2011). Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J. Cell Biol. 194, 669–678 10.1083/jcb.201008095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.