Summary
Centrioles are essential for the formation of cilia and flagella. They also form the core of the centrosome, which organizes microtubule arrays important for cell shape, polarity, motility and division. Here, we have used super-resolution 3D-structured illumination microscopy to analyse the spatial relationship of 18 centriole and pericentriolar matrix (PCM) components of human centrosomes at different cell cycle stages. During mitosis, PCM proteins formed extended networks with interspersed γ-Tubulin. During interphase, most proteins were arranged at specific distances from the walls of centrioles, resulting in ring staining, often with discernible density masses. Through use of site-specific antibodies, we found the C-terminus of Cep152 to be closer to centrioles than the N-terminus, illustrating the power of 3D-SIM to study protein disposition. Appendage proteins showed rings with multiple density masses, and the number of these masses was strongly reduced during mitosis. At the proximal end of centrioles, Sas-6 formed a dot at the site of daughter centriole assembly, consistent with its role in cartwheel formation. Plk4 and STIL co-localized with Sas-6, but Cep135 was associated mostly with mother centrioles. Remarkably, Plk4 formed a dot on the surface of the mother centriole before Sas-6 staining became detectable, indicating that Plk4 constitutes an early marker for the site of nascent centriole formation. Our study provides novel insights into the architecture of human centrosomes and illustrates the power of super-resolution microscopy in revealing the relative localization of centriole and PCM proteins in unprecedented detail.
Key words: 3D-SIM, Plk4, Centriole, Centrosome, Super-resolution microscopy
Introduction
Centrioles and basal bodies are closely related barrel-shaped structures that function in the formation of centrosomes and the assembly of cilia and flagella, respectively (Bettencourt-Dias and Glover, 2007; Bornens, 2012; Lüders and Stearns, 2007; Nigg and Raff, 2009). In human cells, centriole cylinders are built of nine triplet microtubules (MTs) and have approximate diameters of 150–200 nm and lengths of 400–450 nm (Bornens, 2012; Paintrand et al., 1992; Vorobjev and Chentsov, 1980). Each centrosome comprises two centrioles that are surrounded by an amorphous protein cloud, the pericentriolar material (PCM) (Bornens, 2002; Chrétien et al., 1997; Vorobjev and Chentsov, 1980). As indicated by proteomics analyses, the PCM is composed of more than 100 different proteins (Andersen et al., 2003; Jakobsen et al., 2011). Of the two centrioles, the older one (frequently referred to as the mother) carries distinctive distal and subdistal appendages (Paintrand et al., 1992), which confer important functional asymmetry (Bornens, 2012; Nigg and Stearns, 2011). In dividing cells, centrosomes organize the microtubule cytoskeleton throughout interphase and they assist in the formation of bipolar spindles in mitosis. In differentiated cells, basal bodies form either de novo or from the oldest, appendage-bearing mother centriole. These then serve as platforms for the formation of motile and immotile cilia that are essential for development and health of the organism (Bettencourt-Dias et al., 2011; Ishikawa and Marshall, 2011; Nigg and Raff, 2009).
In recent years, much progress has been made towards assembling a parts list of human centrosomes and key proteins important for centriole biogenesis, duplication, PCM recruitment and basal body functions have been identified (Carvalho-Santos et al., 2011; Dobbelaere et al., 2008; Strnad and Gönczy, 2008). Moreover, the importance of centrioles/basal bodies and centrosomes for human health and disease is well recognized (Bettencourt-Dias et al., 2011; Doxsey et al., 2005b; Nigg, 2002; Nigg and Raff, 2009). In contrast, although recent cryo-electron tomography has revealed important structural information on centriole architecture (Guichard et al., 2010; Li et al., 2012), definitive information about the spatial organization of the various components remains scarce. Conventional light microscopy has been widely used to determine the approximate localization of centrosomal proteins, but the dimensions of centrioles are close to the Abbe-Rayleigh diffraction limit of optical resolution, that is at best 200 nm in the lateral and 500 nm in the axial direction (Schermelleh et al., 2010). Immuno-electron microscopy (immuno-EM) affords higher resolution and has provided valuable information about the disposition of individual proteins within centrioles, basal bodies and centrosomes, but antigen preservation and accessibility are often problematic and co-localization studies challenging.
The recent development of super-resolution fluorescence microscopy techniques, such as structured illumination microscopy (SIM), stimulated emission depletion (STED) and single molecule localization approaches, makes it possible to analyze spatial relationships within subcellular structures and organelles below the diffraction limit with previously unattainable detail (Hell, 2007; Huang et al., 2010; Schermelleh et al., 2010; Toomre and Bewersdorf, 2010). In this study, we have used three-dimensional structured illumination microscopy (3D-SIM) (Gustafsson et al., 2008; Schermelleh et al., 2008) to determine the subcellular localization of key centriolar proteins in human U2OS and RPE-1 cells. This imaging approach allowed us to study organelles up to several µm deep inside fixed cells and simultaneously acquire optical serial sections for up to three different centrosomal proteins at a spatial resolution of ±120 nm in the x-,y- and ±300 nm in the z-direction (Gustafsson et al., 2008; Schermelleh et al., 2008). Our results reveal the relative localization of centriole and PCM proteins in unprecedented detail and provide novel insights into the architecture of human centrosomes.
Results
To explore the utility of 3D-SIM for studying the architecture of the human centrosome, we first focused on proteins that were expected to undergo cell cycle-dependent changes in amount and disposition, thus providing an opportunity to validate our methodology. Subsequently, we extended our studies to multiple aspects of centrosome organization, including the relationship between centrioles and PCM, the disposition of appendages and the biogenesis of centrioles.
Proteins involved in cell cycle-dependent MT nucleation
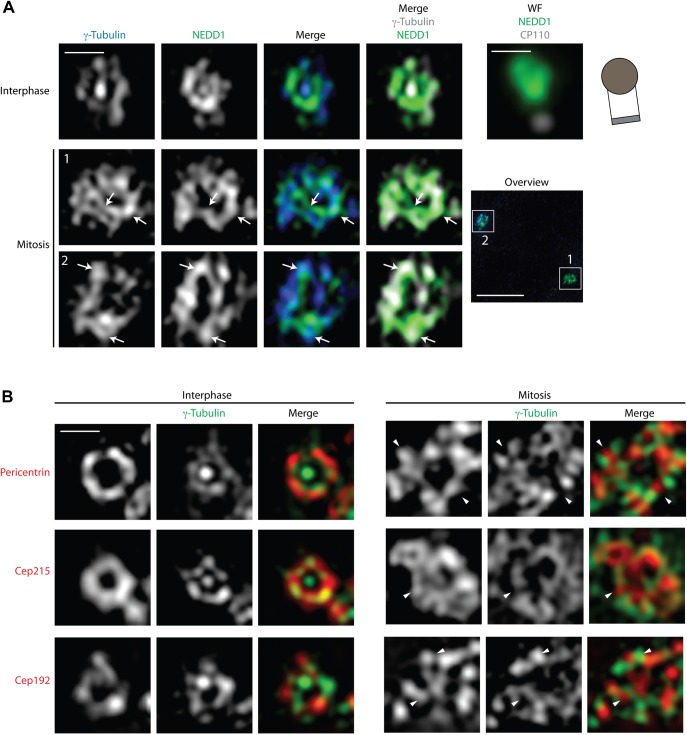
Centrosomes function as MT-organizing centers (MTOCs) throughout the cell cycle (Doxsey et al., 2005a; Lüders and Stearns, 2007). This function depends on γ-Tubulin ring complexes (γTuRCs) that are recruited to PCM through the docking protein NEDD1/GCP-WD (Haren et al., 2009; Lüders et al., 2006). Importantly, increased MT nucleation at the onset of mitosis is accompanied by both recruitment and exchange of PCM components (Blagden and Glover, 2003; Casenghi et al., 2003; Palazzo et al., 1999). An early pioneering study using deconvolution of fluorescence images had revealed a reticular lattice of Pericentrin and γ-Tubulin that expanded as cells approached mitosis (Dictenberg et al., 1998). Here, we examined the disposition of γ-Tubulin, NEDD1, and several PCM proteins in U2OS cells, both during interphase and early mitosis using 3D-SIM (Fig. 1; for comparison with conventional wide-field imaging, see supplementary material Fig. S1A). In these experiments, as in all subsequent ones, wide-field microscopy was used to select centrosomes for analysis and, where appropriate, schematics are drawn to help with the interpretation of images (e.g. Fig. 1A, upper panel). Furthermore, whenever required, an additional centriolar marker was acquired in the far-red channel by wide-field microscopy to reveal the orientation of centrosomes (e.g. Fig. 1A, upper panel; for details, see supplementary material Fig. S1B and Materials and Methods). In interphase cells, both γ-Tubulin and NEDD1 localized in ring-shaped patterns around the mother and, to a lesser extent, the daughter centriole adjacent to the mother (Fig. 1A, upper panel). Interestingly, both proteins were also detected as dots at the center of the ring surrounding the mother centriole, suggesting that these proteins also localize within the lumen of, at least, the mother centriole. This is consistent with immuno-EM data on the localization of γ-Tubulin (Fuller et al., 1995) and GCP6 (Bahtz et al., 2012). After U2OS cells entered mitosis (as indicated by centrosome separation), γ-Tubulin and NEDD1 showed a much broader distribution throughout the PCM and both proteins formed web-like arrays (Fig. 1A, lower panels). Staining showed a substantial degree of overlap (marked by arrows), consistent with the expected close proximity between γ-Tubulin and NEDD1, even though co-localization was not complete.
Fig. 1. γ-Tubulin localizes in close proximity to centriole walls in interphase but within an extended PCM meshwork in mitosis.
U2OS cells were fixed and stained with the indicated antibodies. Representative 3D-SIM optical mid-sections through centrosomes are shown for different cell cycle stages (Scale bars 0.5 µm). (A) γ-Tubulin and NEDD1 were stained in interphase and mitosis, respectively. Arrows point to regions of signal overlap. Correlative widefield image (WF) of green (NEDD1) and Cy5 (CP110) channels and schematic illustration of centriole orientation are shown to the right of the interphase cell. Overview panel shows centrosome separation (Scale bar 5 µm, lower panel). (B) γ-Tubulin localization in interphase and mitosis is shown relative to Pericentrin, Cep215 and Cep192. Arrowheads indicate mutually exclusive staining.
We next compared the localization of γ-Tubulin with that of three prominent PCM proteins, notably Pericentrin, Cep215 and Cep192. In interphase, all three proteins also formed rings surrounding the mother centriole, but none of them localized to the centriole lumen, as indicated by the absence of central dot staining (Fig. 1B, left panel). Consistent with the notion that PCM undergoes a drastic expansion at the onset of mitosis, Pericentrin, Cep192 and Cep215 formed large extended networks in mitotic cells (Fig. 1B, right panels). Interestingly, γ-Tubulin staining did not show extensive co-localization with any of the three PCM proteins (arrowheads), suggesting that different PCM components cooperate to trap γTuRCs within a mitotic meshwork.
Centrosomal proteins localize in distinctive dot- and ring-like patterns
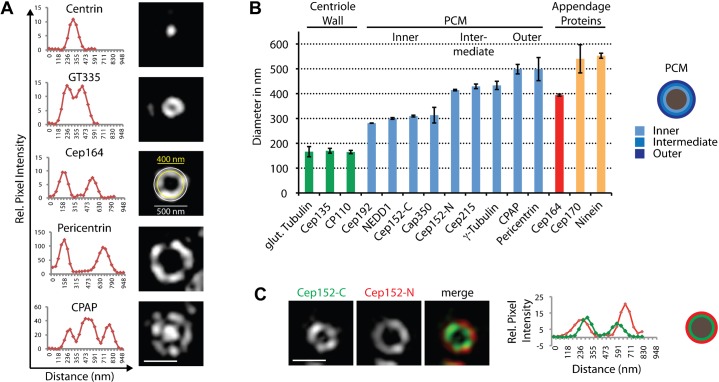
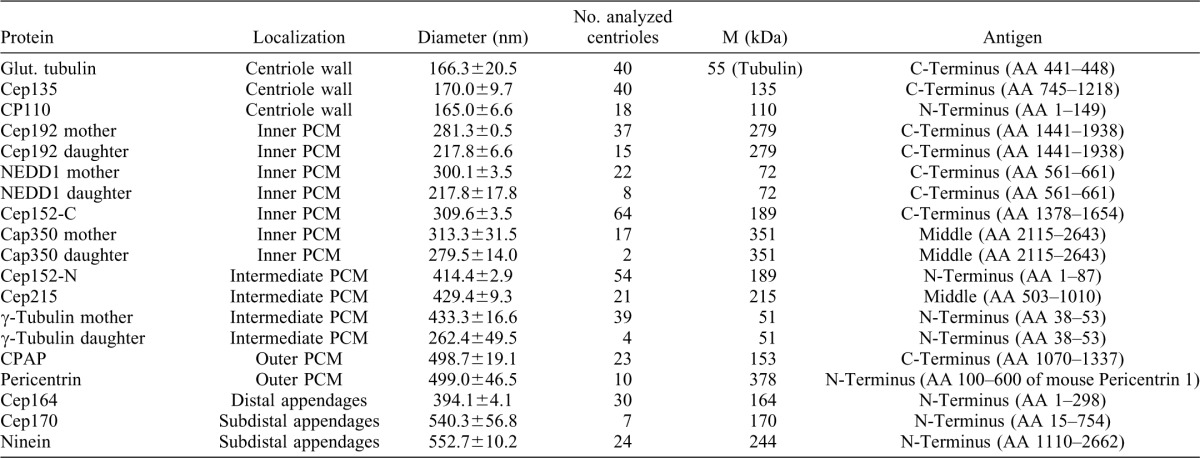
Having established the efficacy of 3D-SIM for examination of centrosome architecture, we proceeded to examine the disposition of 18 different centriolar/centrosomal proteins within the interphase centrosome. This survey made use of a battery of well-characterized antibodies and revealed two distinct patterns of protein localization. In top-views of centrioles, either dot- or ring-like patterns (or a combination of the two) could be observed (Fig. 2A). Of the 18 proteins analyzed in this study, 4 (Centrin, Sas-6, STIL and Plk4) revealed compact dots. All other proteins were disposed in ring-like patterns around the centriole and, as shown above for γ-Tubulin and NEDD1, CPAP showed both ring and central dot localization (Fig. 1A, Fig. 2A). Ring staining was also recently observed for Cep152 and Cep63 in an independent 3D-SIM study (Sir et al., 2011). To measure ring diameters, we scanned fluorescence intensities and determined the distances between intensity maxima (Fig. 2A,B; Table 1). The monoclonal antibody GT335 was used to stain glutamylated tubulin (Bobinnec et al., 1998; Wolff et al., 1992), thereby providing a reference for the width of the centriolar cylinder. This approach revealed a ring with a diameter of about 170 nm (Fig. 2A,B; Table 1). This is consistent with electron microscopy data (Paintrand et al., 1992) and validates the method employed in the present study. Interestingly, Cep135 and CP110 were arranged as rings of very similar sizes (Fig. 2B), in line with immuno-EM data suggesting that Cep135 and CP110 concentrate at the proximal and distal ends, respectively, of the centriolar cylinder (Kleylein-Sohn et al., 2007; Schmidt et al., 2009).
Fig. 2. Centrosomal proteins display distinct localizations and radial distances from centriole walls.
U2OS cells were fixed and stained with the indicated antibodies. Scale bar 0.5 µm. (A) Exemplary top-views of proteins localizing to centrioles in different patterns, along with intensity plots. In the middle panel (Cep164) rings corresponding to diameters of 400 and 500 nm are depicted. (B) For quantification, distances between intensity maxima were measured (mean ± sd) (see also Table 1). Proteins were grouped according to average ring diameter and centriole localization. Schematic of cross-section of mother centriole is given on the right to illustrate subdivision into inner, intermediate and outer PCM. (C) Staining of centrioles with antibodies against the N- or C-termini of Cep152 reveals distinct distances from the centriole wall. Representative images and the corresponding intensity plots along with a schematic illustration of centriolar cross-section are shown.
Table 1. Measurement of ring diameters of proteins localizing in ring-like patterns around centrioles.
Consideration of the size of IgG (about 8 nm) raises the potential of minor deviations (less than 32 nm) from the measured diameters.
All other proteins examined formed rings of larger diameter. Based on the measured diameters, PCM proteins were arbitrarily subdivided into three groups, termed inner (ca. 280–320 nm), intermediate (ca. 400–420 nm) and outer PCM (ca. 500 nm) (Fig. 2B; Table 1). According to this classification, NEDD1 belongs to the inner category, but γ-Tubulin to the intermediate group. This difference can be rationalized by considering that NEDD1 and γ-Tubulin localize at the proximal and distal ends, respectively, of γTuRCs. Whereas NEDD1 functions as a γTuRC-recruitment factor, γ-Tubulin makes contact with the minus ends of MTs and hence is expected to face outwards. Thus, in interphase centrosomes the bulk of γTuRCs is located within a region that spans from the inner to the intermediate PCM.
Whenever staining could be detected at both mother and nascent daughter centrioles (Cep192, NEDD1, CAP350 and γ-Tubulin), the diameter of the daughter ring was invariably smaller than that of the mother (Table 1). This observation is in line with the notion that PCM is more developed around mother than daughter centrioles (Piel et al., 2000; Wang et al., 2011).
Studying protein disposition and orientation
When interpreting the above results, it is important to bear in mind that large proteins may span considerable distances, particularly if they comprise extended coiled-coil domains as is the case for many centrosomal proteins (Andersen et al., 2003). In principle, this offers the interesting possibility to chart the localization of distinct protein domains through use of antibodies directed against appropriate epitopes. To illustrate this point, we focused on the PCM component and Plk4-interaction partner Cep152 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). Specifically, we raised antibodies against the N- and C-termini of Cep152 (for antibody specificity, see supplementary material Fig. S2) and used these reagents to explore the disposition of Cep152 in relation to the centriolar cylinder. Interestingly, we found that Cep152-C consistently stained a smaller ring than Cep152-N (diameters of ca. 310 and 415 nm, respectively) (Fig. 2B,C; Table 1). This implies that Cep152 spans the PCM from the inner to the intermediate region, potentially in an extended rod-like shape, with the C-terminus located close to the centriolar cylinder and the N-terminus facing outwards. These results demonstrate that the combination of 3D-SIM with site-specific antibodies represents a powerful approach for determining protein orientation and disposition.
Appendage proteins
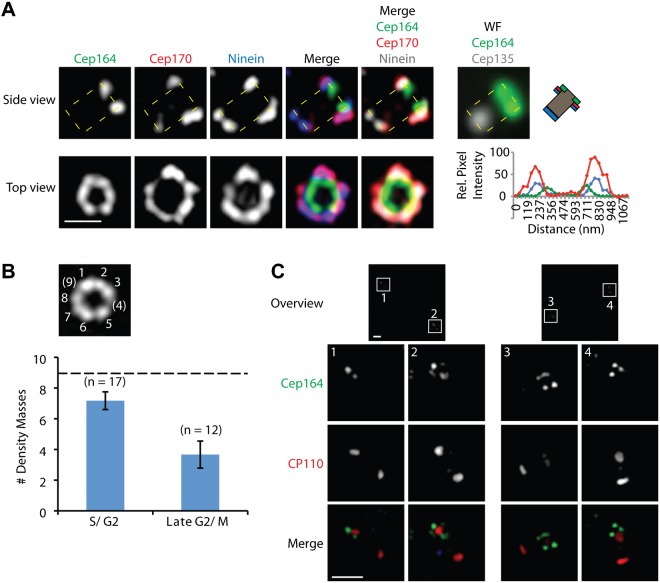
The rings with largest diameters (ca. 550 nm) were seen when staining centrosomes with antibodies against the subdistal appendage proteins Ninein (Mogensen et al., 2000) and Cep170 (Guarguaglini et al., 2005) (Fig. 2B, Fig. 3A; Table 1). In comparison, the distal appendage protein Cep164 (Graser et al., 2007a) was arranged in rings of about 400 nm (Fig. 2B; Fig. 3A, top-view; Table 1). In side-views, Ninein and Cep170 localized to very similar positions but slightly more proximally than the distal appendage protein Cep164 (Fig. 3A, side-view). Our measurements are in good agreement with previous estimates of appendage size, as determined by electron microscopy (Paintrand et al., 1992) and other super-resolution microscopy techniques, notably photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) (Sillibourne et al., 2011). Minor differences in the reported estimates are readily explained by differences in data representation (Fig. 2A, panel Cep164).
Fig. 3. Localization of centriolar appendage proteins.
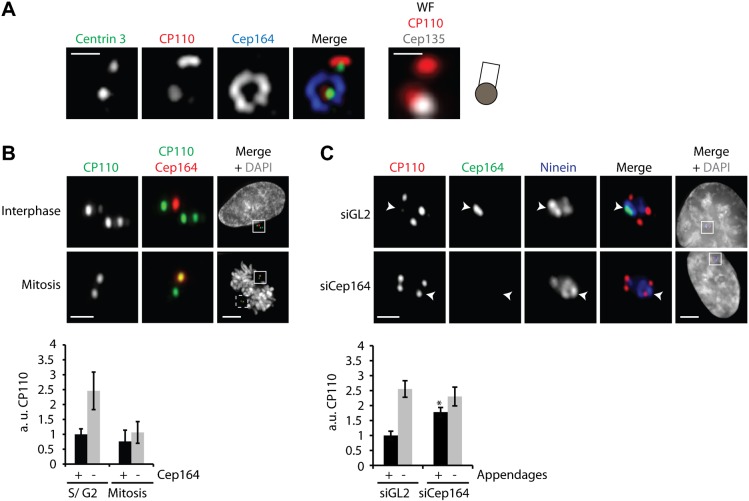
U2OS cells were fixed and stained with the indicated antibodies and images were acquired with 3D-SIM. (A) Relative localization of distal and subdistal appendage proteins. Shown are side-view (a single stack, upper panel, boxed area indicates centriole) and top-view images (lower panel) of a representative centriole. Also shown are correlative wide-field (WF) images of the green (Cep164) and far-red (Cep135) channels of the same centriole that is visualized in side-view, along with a schematic representation to illustrate the orientation of the centriole. Intensity plots of Cep164, Cep170 and Ninein are given for the top-view. Scale bar 0.5 µm. (B,C) Staining for Cep164 reveals distinct density masses. The number of these masses is lower in G2/M cells (after centrosome separation) than in S/G2 cells (before centrosome separation). Panel B shows a representative image of a centriole in S/G2 phase; panel C shows two examples of centrioles in late G2/M phase. Scale bars 1 µm.
In top-views of Cep164 staining different density maxima could readily be discerned (Fig. 3B). In interphase centrioles, we most commonly observed only 6–8 such masses, even though the centriolar cylinder clearly offers space for 9 density masses (Fig. 3B, numbers in brackets), as expected from the 9-fold symmetry of centrioles. Even more striking was the result obtained when analyzing Cep164 staining in mitotic cells (Fig. 3B,C). In this case, only 3–4 Cep164-positive density masses could be observed. When taken at face value, these observations suggest that individual centrioles carry fewer than 9 appendages and/or that not all appendages share the same complement of proteins. A caveat to this interpretation is that antibody inaccessibility could also explain the absence of staining at some of the 9 expected positions.
CP110 levels are reduced on centrioles that bear Cep164
Since CP110 caps the distal end of centrioles (Kleylein-Sohn et al., 2007; Schmidt et al., 2009), anti-CP110 antibodies are expected to stain 4 dots in G2 phase cells (e.g. Arquint et al., 2012; Guderian et al., 2010). However, we noticed that one of the four centrioles often showed much weaker CP110 staining. Most interestingly, we found that the centriole showing weak CP110 staining was invariably the one carrying Cep164-positive appendages (Fig. 4A). In contrast, the distally located protein Centrin (Paoletti et al., 1996) was not affected by the presence of Cep164 (Fig. 4A), suggesting that Cep164 exerts a specific effect on CP110. To examine the relationship between CP110 and Cep164 in more detail, wide-field microscopy was used to analyze and quantify the dependency of centriolar CP110 levels on Cep164 during the cell cycle. In interphase S/G2 cells, Cep164-positive centrioles showed 2.5-fold reduced levels of CP110 signal as compared to Cep164-negative centrioles (Fig. 4B). This reduction cannot be attributed to antibody competition, as one centriole showed reduced CP110 staining even when staining for Cep164 was omitted (data not shown). Furthermore, near-identical CP110 staining was seen in Cep164-positive and -negative centrioles in mitotic cells (Fig. 4B). Our analysis also showed that CP110 staining was 2–2.5-fold lower in mitotic cells than interphase cells (Fig. 4B, graph), consistent with cell cycle-dependent CP110 degradation (D'Angiolella et al., 2010). In contrast, Cep164 staining was higher during interphase than mitosis (Fig. 3B,C) (Graser et al., 2007a). Similar results were also observed in RPE-1 cells (supplementary material Fig. S3A).
Fig. 4. CP110 levels are reduced at Cep164-bearing centriole.
U2OS cells were fixed and stained with the indicated antibodies. Scale bars 0.5 µm. (A) 3D-SIM image of a centrosome stained for Centrin, CP110, and Cep164 alongside a wide-field image (WF) of the same centrosome stained for CP110 (red channel) and Cep135 (far-red channel). The schematic indicates the orientation of the centriole pair. (B) Centriolar CP110 levels at Cep164-negative and -positive centrioles vary during the cell cycle. The upper panels show representative wide-field images (scale bars 1 µm and 5 µm for the overview images), the histogram below a quantification of CP110 levels in arbitrary units (a.u.; three independent experiments, 10 cells each, mean ± sd). (C) U2OS were transfected for 48 h with the indicated siRNA oligonucleotides before being stained with antibodies against CP110, Cep164 and Ninein and counterstained with DAPI. The upper panels show representative wide-field images (scale bars 1 µm and 5 µm for the overview images), the histogram below a quantification of CP110 levels in arbitrary units (a.u.; three independent experiments, 10 cells each, mean ± sd, student's t test: * P<0.05).
To corroborate a causal relationship between the presence of Cep164 and reduced CP110 levels, Cep164 was depleted from U2OS cells and CP110 levels were quantified at each centriole. Co-staining for the subdistal appendage marker Ninein was used to identify the oldest centriole. Under these conditions, CP110 levels were comparable at all four centrioles, including the appendage-bearing but Cep164-depleted centriole (Fig. 4C). This restoration of CP110 staining upon Cep164 depletion argues that the presence of Cep164-positive distal appendages antagonizes CP110 association with distal ends of fully mature centrioles.
Centriole biogenesis
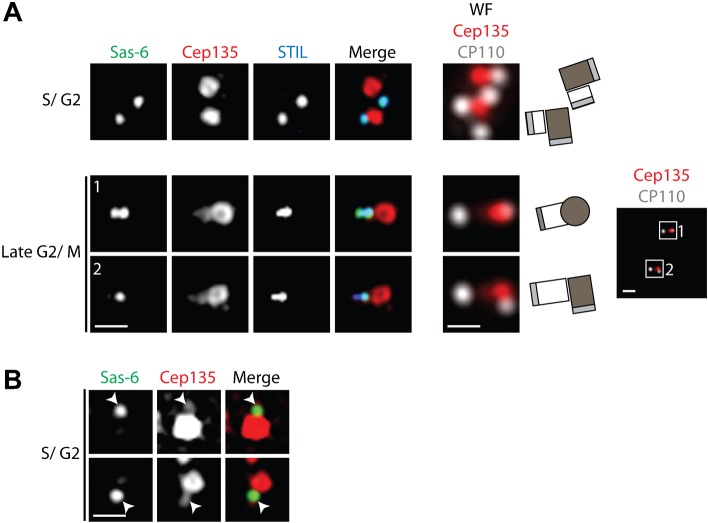
In a next series of experiments we examined the localization of Sas-6, Cep135 and STIL, three proteins implicated in the early stages of centriole biogenesis. Sas-6 and Cep135 are putative homologs, respectively, of Chlamydomonas Bld12 and Bld10 (Hiraki et al., 2007; Matsuura et al., 2004; Nakazawa et al., 2007), whereas STIL, a putative homolog of Caenorhabditis elegans Sas-5 and Drosophila Ana2 (Stevens et al., 2010), is a potential partner of Sas-6 (Arquint et al., 2012; Tang et al., 2011; Vulprecht et al., 2012). Recent structural studies identified Sas-6 as a major component of the cartwheel that confers 9-fold symmetry to the nascent centriole (Kitagawa et al., 2011; van Breugel et al., 2011). Using wide-field microscopy for detection of CP110, we identified S/G2 phase cells with four centrioles and then examined the localization of STIL, Sas-6 and Cep135 by 3D-SIM. Whereas STIL and Sas-6 co-localized precisely, in line with recent results (Arquint et al., 2012; Tang et al., 2011; Vulprecht et al., 2012), the bulk of Cep135 was clearly distant from Sas-6/STIL (Fig. 5A, upper panel). Similar results were obtained in RPE-1 cells (supplementary material Fig. S3B). The localization of Cep135 coincided with that of C-Nap1 (data not shown), a marker for the proximal ends of mother centrioles (Fry et al., 1998). In late G2 or M phase cells, Cep135 staining could be seen to extend from mother centrioles to the area occupied by Sas-6 and STIL (Fig. 5A, lower panels). This indicates that Cep135 progressively associates with the proximal ends of daughter centrioles, concomitant with their growth during the cell cycle. In support of this view, post-acquisition adjustment of fluorescence intensity for S/G2 phase centrioles allowed the visualization of a faint but reproducible Cep135 signal overlapping Sas-6 (Fig. 5B). This overlap does not reflect bleed-through between channels, as these were acquired sequentially and the microscope filter setup allowed clear separation of the utilized fluorophores (supplementary material Fig. S1C). Because Cep135 and Sas-6 are considered likely homologs of the Chlamydomonas cartwheel components Bld10 and Bld12, respectively (Hiraki et al., 2007; Matsuura et al., 2004; Nakazawa et al., 2007), the distinct localizations of Cep135 and Sas-6 came as a surprise. Our data do not exclude that Cep135 is a genuine cartwheel component of nascent centrioles in human cells, but they indicate that the bulk of Cep135 localizes to the proximal end of the mother centriolar cylinder (see also Kleylein-Sohn et al., 2007).
Fig. 5. The bulk of Cep135 localizes distantly from Sas-6 and STIL.
U2OS cells were stained with the indicated antibodies. Representative 3D-SIM and wide-field (WF) images are shown. Scale bars 0.5 µm or 1 µm (overview image). (A) Localization of Sas-6, Cep135 and STIL in cells in S/G2 phase (before centrosome separation, upper panel) or late G2/M phase (after centrosome separation, lower panel). Wide-field images of the red (Cep135) and far-red (CP110) channels of the same centrosome were acquired to determine the orientation of centrioles (schematics illustrate centriole orientations and overview panel shows centrosome separation). (B) Brightness/contrast-enhanced image to indicate co-localization of weak Cep135 signal with Sas-6 (arrowheads).
Plk4 constitutes an early marker for the location of nascent centrioles
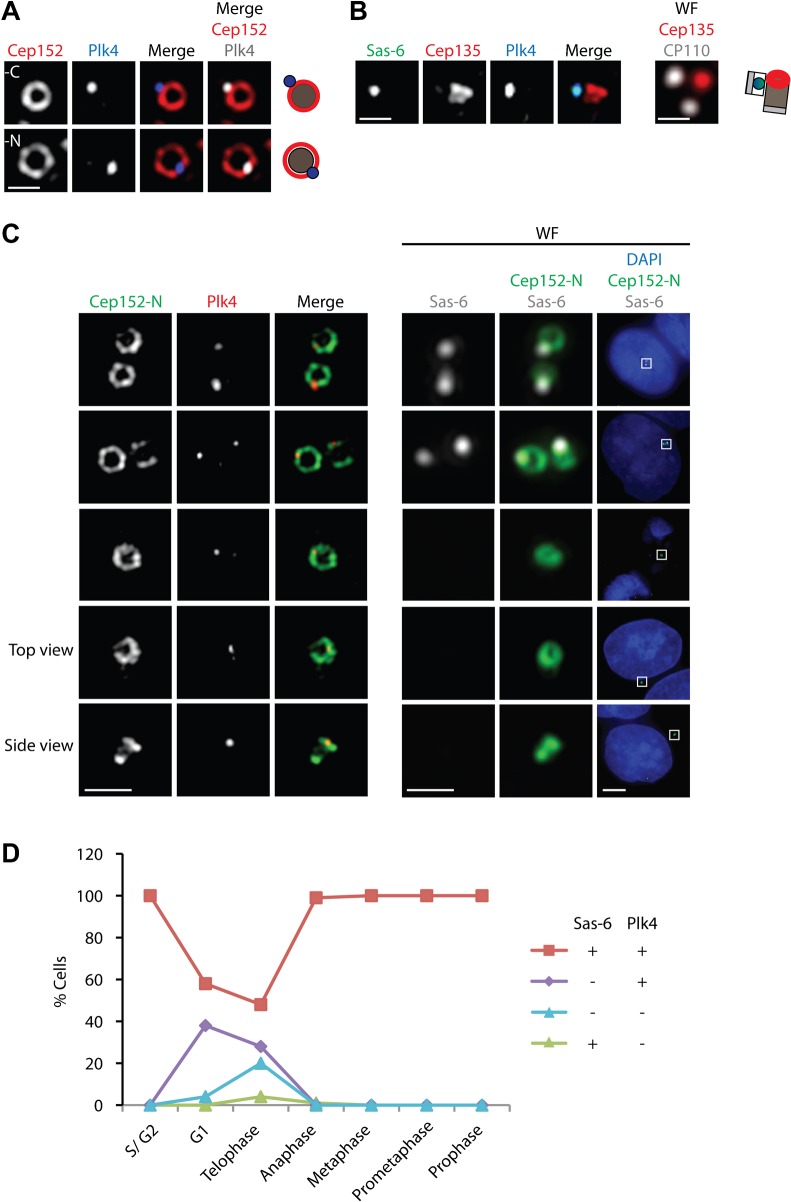
In view of the key regulatory role of Plk4 in centriole biogenesis (Bettencourt-Dias et al., 2005; Habedanck et al., 2005), we carefully examined the localization of this protein kinase. Co-staining for Plk4 and its interaction partner Cep152 revealed that Plk4 localized to a spot that fell onto the ring stained by Cep152-N antibodies but outside of the ring defined by Cep152-C (Fig. 6A), consistent with the reported interaction between Plk4 and the N-terminus of Cep152 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). Dot staining was also produced when using a monoclonal anti-Plk4 antibody (supplementary material Fig. S5B). Moreover, Plk4 co-localized with Sas-6 (Fig. 6B), raising the intriguing question of which of these proteins would constitute the earlier marker for the precise site of centriole biogenesis. Both Sas-6 and STIL are degraded upon exit from mitosis and, as a consequence, the centrioles of early G1 cells lack detectable levels of these proteins (Arquint et al., 2012; Strnad et al., 2007; Tang et al., 2011; Vulprecht et al., 2012) (see also Fig. 6C,D). Plk4 levels also peak in mitosis (Sillibourne et al., 2010), but, in contrast to Sas-6, Plk4 can be seen at centrioles at all cell cycle stages, albeit at variable levels (supplementary material Fig. S4A,B).
Fig. 6. Comparison of Plk4 with Sas-6 localization.
(A) U2OS cells were stained with antibodies against Plk4 and either Cep152-C (upper panel) or Cep152-N (lower panel); representative 3D-SIM images are shown. Scale bar 0.5 µm. Schematic illustrates relative localizations. (B) Representative 3D-SIM and wide-field (WF) images of cells stained with antibodies against Sas-6, Cep135, Plk4 and CP110 (far-red channel); schematic illustrates orientation of centrioles. Scale bar 0.5 µm. (C) U2OS cells were stained with antibodies against Sas-6, Cep152 and Plk4 and counterstained with DAPI. Representative 3D-SIM and correlative wide-field (WF) images are shown. Scale bars 1 µm or 5 µm (overview). (D) U2OS cells were stained as in C and wide-field microscopy was used to count cells showing the indicated combinations of Plk4 and Sas-6 staining at each cell cycle stage (n = 100 for each cell cycle stage).
Importantly, Sas-6 is the earliest protein known to localize to the newly forming daughter centriole (Strnad et al., 2007). Thus, we examined Plk4 localization in relation to Sas-6 staining. We used coupled wide-field microscopy to search for cells in late mitosis or early G1 in which Sas-6 was either present or absent from nascent centrioles and then analyzed Plk4 localization with 3D-SIM. Remarkably, Plk4 staining showed a dot staining pattern, regardless of whether or not Sas-6 was detectable at centrioles (Fig. 6C). Quantitative analyses of these data showed that Plk4 localized to 38% of G1 cells that lacked detectable Sas-6 staining (Fig. 6D). Similar results were obtained for RPE-1 cells (supplementary material Fig. S5A) and staining of Plk4 with a monoclonal antibody also revealed a centriolar dot pattern at different cell cycle stages (supplementary material Fig. S5B). Taken together, these data suggest that the site for centriole duplication is determined on the mother centriole prior to detectable recruitment of Sas-6 to nascent centrioles and that Plk4 represents an early marker for this site.
Discussion
We have used super-resolution 3D-structured illumination microscopy to get a better understanding of the architecture of human centrosomes. The dimensions of this organelle and its constituent centrioles are barely above the Abbe-Rayleigh diffraction limit, making it difficult to study protein disposition by conventional light microscopy. Our study encompasses a total of 18 different proteins and illustrates the power of 3D-SIM for detailed structural analysis of centrosomal PCM as well as centrioles and their substructures, notably appendages, at different cell cycle stages.
Our analysis reveals a wealth of detail on the localization and disposition of several proteins. To the extent that prior information from immuno-EM is available, our data are generally in excellent agreement. However, whereas 3D-SIM clearly revealed a ring-like disposition of CP110 in top-views of centrioles, the side-views afforded by previous immuno-EM had suggested a uniform distribution of gold particles at the distal tips of centrioles (Kleylein-Sohn et al., 2007). This discrepancy can be explained by considering that a sample thickness of about 70 nm will not readily allow the visualization of a ring when a centriolar cylinder with a diameter of about 170 nm is cut in longitudinal sections. Thus, we conclude that CP110 is likely to form a ring on the cylinder's wall rather than a disk-like cap at the distal end of centrioles. In support of this interpretation we note that discontinuous staining, consistent with ring-like arrangements, can be seen by immuno-EM in longitudinal sections of centrioles when proteins are arranged in rings of wider diameters, such as Ninein (e.g. Schmidt et al., 2009).
Interestingly, in interphase cells γ-Tubulin was detected not only in the PCM that surrounds the centrioles but also within the centriolar lumen (see also Fuller et al., 1995). Whereas the former population is expected to nucleate microtubules, the latter likely reflects the requirement for γ-Tubulin in the biogenesis of centrioles and cilia. During M phase, γ-Tubulin showed the expected distribution throughout a massively enlarged PCM, but we have not observed extensive co-localization with any of the 3 major PCM components analyzed in this study. Thus, it seems plausible that γTuRCs associates with a mitotic meshwork formed cooperatively by multiple PCM components rather than binding predominantly to any one particular PCM component.
At the distal end of centrioles, we have analyzed the disposition of both subdistal and distal appendage proteins. Both classes of proteins showed ring-like arrangements and distinct density masses could be discerned. Interestingly, however, the number of readily resolved density masses was generally below 9, even though the rings clearly offered enough space to accommodate 9 masses. For example, Cep164 most commonly showed 6–8 pronounced density masses on interphase centrioles, but only 3–4 during mitosis. It is conceivable that antibody accessibility is limited for some of the 9 distal appendages, perhaps due to steric hindrance caused by anchored MTs (Delgehyr et al., 2005). If so, this shielding would be particularly prominent during mitosis. Alternatively, it is possible that not all centrioles carry the full complement of nine distal appendages and/or that Cep164 protein does not accumulate to the same extent on all appendages. If the latter interpretation was correct, it would indicate that the formation or protein composition of distal appendages is regulated during the cell cycle.
Another interesting observation concerns the relationship between the distal appendage protein Cep164 and the distal end capping protein CP110. Specifically, we have found that the one centriole that carries Cep164 commonly shows substantially lower levels of CP110. As control experiments showed that antibody competition cannot account for this observation, we conclude that the presence of Cep164-bearing appendages interferes with CP110 binding to the distal end of the mature centriole. This observation is interesting from the perspective of ciliogenesis. Removal of CP110 from the tip of the basal body is in fact required for the outgrowth of the ciliary axoneme (Kohlmaier et al., 2009; Schmidt et al., 2009; Spektor et al., 2007; Tang et al., 2009). This prompts the question of how, in quiescent cells, CP110 can be selectively removed from the mature centriole during ciliogenesis, while persisting on the daughter centriole. Our data indicate that CP110 levels are reduced at the Cep164-positive centriole already in proliferating cells. This suggests that the formation of distal appendages may represent an early step in the preparation of the mature centriole for future function as a basal body. Thus, Cep164-bearing appendages appear to play an important role not only in the anchoring of the basal body to the plasma membrane (Graser et al., 2007a), but also in the removal of CP110 and the consequent uncapping of the basal body's distal end.
We have also examined the localization of centriole duplication proteins at the interface between mother and nascent daughter centrioles. These analyses have yielded two remarkable observations. First, whilst Sas-6 staining showed a tight dot at the interface between mother and daughter centrioles, consistent with the association of Sas-6 with the hub and spokes of the cartwheel (Kitagawa et al., 2011; Nakazawa et al., 2007; van Breugel et al., 2011), the bulk of Cep135 was seen primarily as a ring decorating the proximal end of the mother centriole. This contrasts with the localization of Bld10, the putative Cep135 homolog of Chlamydomonas, which in basal bodies clearly localizes to the spoke tips of the cartwheel (Hiraki et al., 2007; Matsuura et al., 2004). If Cep135 were to localize prominently to the tips of cartwheel spokes also during early biogenesis of human centrioles, we would have expected to see this protein in close proximity to Sas-6. We emphasize that our data do not exclude that Cep135 is a genuine cartwheel component in human cells, as absence of staining can always be attributed to impaired antibody-accessibility. However, taken at face value our data suggest that the number of Cep135 proteins at cartwheel structures is likely to be low in comparison to the amount of Cep135 that can be detected by 3D-SIM at mother centrioles. It is tempting to speculate that the latter Cep135 population is important for centriole stabilization (see also Hiraki et al., 2007).
Finally, we have examined the localization of Plk4, a key regulator of centriole biogenesis (Bettencourt-Dias et al., 2005; Habedanck et al., 2005). Using two independently raised anti-Plk4 antibodies, we found that Plk4 localized to a distinct dot on the centriolar cylinder. This dot always lay on a circle defined by the antibody directed against the N-terminus of Cep152, consistent with direct binding between the two proteins (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). The observed dot-staining pattern for Plk4 contrasts with previous immunofluorescence and immuno-EM data from our laboratory, showing a ring-like disposition of Plk4 around the circumference of the mother centriole (Habedanck et al., 2005; Kleylein-Sohn et al., 2007). However, we emphasize that our previous data reported on localizations observed after overexpression of Plk4, whilst the present 3D-SIM study examines Plk4 localization at physiological expression levels. Under these latter conditions, both anti-Plk4 antibodies utilized concurred to reveal a striking dot staining.
It is interesting to compare the localization of Plk4 and Sas-6 from a spatio-temporal perspective. In space, Plk4 always co-localized with Sas-6, which, together with STIL, defines the site of emergence of the nascent centriole (Arquint et al., 2012; Kleylein-Sohn et al., 2007). On the time axis, however, we were intrigued to find that Plk4 dot staining was visible throughout the cell cycle, even in early G1 cells before any Sas-6 staining was detectable. As Sas-6 staining of most interphase centrioles was generally stronger than Plk4 staining, this is unlikely to reflect differences in antibody quality. Instead, our data suggest that Plk4 localizes to a distinct site on the surface of the mother centriolar cylinder before Sas-6 contributes to form the cartwheel of the nascent daughter centriole. This makes it tempting to speculate that a high local concentration of Plk4 on the mother centriole surface contributes to define the location of future centriole assembly. The confined Plk4 localization inbetween mother and daughter centrioles throughout the cell cycle could also explain why mother centrioles can only re-duplicate after centriole disengagement at the M-G1 transition, when Plk4 becomes accessible for another round of duplication.
Collectively, our observations raise a number of intriguing questions for future studies. In particular, it is not clear what mechanism leads to the concentration of Plk4 at a specific site on the mother centriole surface and whether the choice of this site reflects a stochastic event or the action of upstream regulatory proteins. Furthermore, considering that Plk4 abundance is regulated by βTrCP-mediated proteolytic degradation (Cunha-Ferreira et al., 2009; Rogers et al., 2009) and βTrCP-binding to Plk4 is in turn triggered by trans-autophosphorylation (Guderian et al., 2010; Holland et al., 2010; Sillibourne et al., 2010), the question arises of what mechanisms allow the local concentration of Plk4 without triggering auto-destruction. Finally, and most importantly, it remains to be elucidated how localized Plk4 is able to promote the assembly of a cartwheel structure.
Materials and Methods
Antibodies
Polyclonal rabbit antibodies against His-tagged Cep152 (aa1–87 (-N) and aa1378–1654 (-C)) were raised at Charles River Laboratories (Elevages Scientifique des Dombes, Charles River Laboratories, Romans, France) and purified according to standard protocols. Antibodies against CPAP, Sas-6, Cep135 (Kleylein-Sohn et al., 2007), CP110, Cep192 (Schmidt et al., 2009), Plk4 (Guderian et al., 2010), centrin-3 (Thein et al., 2007), Cep164, Ninein (Graser et al., 2007a), Cep215 (Graser et al., 2007b), CAP350 (Yan et al., 2006), Cep170 (Guarguaglini et al., 2005), and STIL (Arquint et al., 2012) have been described previously. Anti-NEDD1, γ-Tubulin, Pericentrin, Flag and GT335 antibodies were purchased from Sigma (Taufkirchen, Germany). Alexa 488-, Alexa 594-, Alexa 647-labeled secondary anti-mouse and anti-rabbit antibodies were purchased from Life technologies (Carlsbad, CA, USA) and CF405S-labelled secondary anti-mouse and anti-rabbit antibodies were purchased from Biotium (Hayward, CA, USA). To simultaneously visualize different polyclonal rabbit antibodies, these were directly labeled by Alexa 488, Alexa 594 and Alexa 647 fluorophores, using the corresponding Antibody Labeling Kits (Life technologies). Whenever required secondary antibodies were blocked using anti-Flag antibodies prior to the incubation with directly coupled antibodies of the same species.
Cell culture and siRNA-mediated protein depletion
U2OS and RPE-1 cells were cultured as described previously (Graser et al., 2007a; Habedanck et al., 2005). Cep152 and Cep164 were depleted using the previously described siRNA duplex oligonucleotides (Graser et al., 2007a) and the luciferase duplex GL2 was used for control (Elbashir et al., 2001). Transfections were performed using Oligofectamin (Life technologies) according to the manufacturer's protocol.
Wide-field and structured illumination microscopy
Cells were fixed in methanol for 5 min at −20°C. Antibody incubations and washings were performed as described previously (Meraldi et al., 1999). Super-resolution 3D-SIM imaging was performed on a DeltaVision OMX V3 system (Applied Precision) equipped with a 100×/1.40 NA PlanApo oil immersion objective (Olympus), and 405, 488 and 592 nm diode lasers and Cascade II:512 EMCCD cameras (Photometrics). Exciting light was directed through a movable optical grating to generate a fine-striped interference pattern on the sample plane. Emitted fluorescence light was directed through a set of appropriate dichroic beam splitter and emission filter according to its wavelength range onto three cameras. Image stacks of typically 4 µm height with 15 images per plane (5 phases, 3 angles) and a z-distance of 0.125 µm were acquired and computationally reconstructed to generate super-resolution optical serial sections with two-fold extended resolution in all three axes. Color channels were carefully aligned using alignment parameter from control measurements with 0.5 µm diameter multi-spectral fluorescent beads (Life technologies). Images were acquired in the sequential imaging mode to rule out bleed-through (supplementary material Fig. S1C). SI reconstruction and image processing was performed with the SoftWoRx 3.7 imaging software package (Applied Precision).
Wide-field imaging was typically performed with a PersonalDV microscope (Applied Precision) equipped with a 60×/1.42 PlanApo oil objective (Olympus), CoolSNAP ES2 interline CCD camera (Photometrics), Xenon illumination and appropriate filter sets for DAPI, FITC, TRITC and Cy5. Image stacks were recorded with a z-distance of 0.2 µm and subjected to a constrained iterative deconvolution (conservative ratio, 4 cycles, medium noise filtering, SoftWoRX, 3.7. imaging software package, Applied Precision). Motorized stage positions could be mapped between the two systems, allowing to re-locate cells for consecutive imaging on both systems. This coupled setup allowed us to image cells with conventional wide-field deconvolution microscopy prior to super-resolution imaging in order to visualize Alexa 647-stained centrioles as reference to determine their orientation and age within a centriole pair whenever required. By this approach we could notably distinguish between the mature mother centriole and the orthogonally growing daughter centriole. Of note, the 592 nm laser line of the 3D-SI microscope excites Alexa 647 with only low efficiency of which only a minor fraction of the emitted signal transmits through the bandpass filter used for the red channel. Accordingly, bleed-through of the 647 nm signal can be neglected when detecting the Alexa 594 fluorophore signal with 3D-SIM (supplementary material Fig. S1B).
For fluorescence intensity quantifications stainings were analyzed using a DeltaVision wide-field imaging system (Applied Precision) on a TE200 base equipped with an APOPLAN 100×/1.4 NA oil immersion objective (Nikon). Wide-field image stacks were recorded and deconvolved as described above using SoftWoRX. For quantitation of centrosomal CP110 and Plk4 levels at the centrosome with ImageJ, z-stacks were acquired with the same exposure and maximum intensity projections were carried out. Background signal intensity was subtracted from CP110 and Plk4 signal intensity.
Supplementary Material
Acknowledgments
We thank Jürgen Neumann for technical help with the OMX, and Oliver Biehlmaier and all members of the Nigg laboratory for helpful discussions. This work was supported by an EMBO Short-term fellowship (to K.F.S.), the Swiss National Science Foundation [31003A_132428/1] to E.A.N., and the Bioimaging Network Munich to H.L. In addition, K.F.S. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds and the International Max Planck Research School for Molecular and Cellular Life Sciences (Martinsried, Germany).
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Andersen J. S., Wilkinson C. J., Mayor T., Mortensen P., Nigg E. A., Mann M. (2003). Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Arquint C., Sonnen K. F., Stierhof Y. D., Nigg E. A. (2012). Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125, 1342–1352 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- Bahtz R., Seidler J., Arnold M., Haselmann-Weiss U., Antony C., Lehmann W. D., Hoffmann I. (2012). GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125, 486–496 10.1242/jcs.093930 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D. M. (2007). Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. (2005). SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15, 2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G., Godinho S. A. (2011). Centrosomes and cilia in human disease. Trends Genet. 27, 307–315 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden S. P., Glover D. M. (2003). Polar expeditions–provisioning the centrosome for mitosis. Nat. Cell Biol. 5, 505–511 10.1038/ncb0603-505 [DOI] [PubMed] [Google Scholar]
- Bobinnec Y., Moudjou M., Fouquet J. P., Desbruyères E., Eddé B., Bornens M. (1998). Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton 39, 223–232 [DOI] [PubMed] [Google Scholar]
- Bornens M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- Bornens M. (2012). The centrosome in cells and organisms. Science 335, 422–426 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B., Bettencourt-Dias M. (2011). Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casenghi M., Meraldi P., Weinhart U., Duncan P. I., Körner R., Nigg E. A. (2003). Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5, 113–125 10.1016/S1534-5807(03)00193-X [DOI] [PubMed] [Google Scholar]
- Chrétien D., Buendia B., Fuller S. D., Karsenti E. (1997). Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 120, 117–133 10.1006/jsbi.1997.3928 [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., Hoffmann I. (2010). Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191, 731–739 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D. M., Bettencourt-Dias M. (2009). The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19, 43–49 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- D'Angiolella V., Donato V., Vijayakumar S., Saraf A., Florens L., Washburn M. P., Dynlacht B., Pagano M. (2010). SCFCyclin F controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature 466, 138–142 10.1038/nature09140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., Bornens M. (2005). Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118, 1565–1575 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Dictenberg J. B., Zimmerman W., Sparks C. A., Young A., Vidair C., Zheng Y., Carrington W., Fay F. S., Doxsey S. J. (1998). Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141, 163–174 10.1083/jcb.141.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., Raff J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., Theurkauf W. (2005a). Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21, 411–434 10.1146/annurev.cellbio.21.122303.120418 [DOI] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. (2005b). Centrosome control of the cell cycle. Trends Cell Biol. 15, 303–311 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N. S., Yu Q. D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D. M. (2010). Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Elbashir S. M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 10.1038/35078107 [DOI] [PubMed] [Google Scholar]
- Fry A. M., Mayor T., Meraldi P., Stierhof Y. D., Tanaka K., Nigg E. A. (1998). C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S. D., Gowen B. E., Reinsch S., Sawyer A., Buendia B., Wepf R., Karsenti E. (1995). The core of the mammalian centriole contains γ-tubulin. Curr. Biol. 5, 1384–1393 10.1016/S0960-9822(95)00276-4 [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Lavoie S. B., Gassner O. S., Lamla S., Le Clech M., Nigg E. A. (2007a). Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Nigg E. A. (2007b). Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120, 4321–4331 10.1242/jcs.020248 [DOI] [PubMed] [Google Scholar]
- Guarguaglini G., Duncan P. I., Stierhof Y. D., Holmström T., Duensing S., Nigg E. A. (2005). The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell 16, 1095–1107 10.1091/mbc.E04-10-0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian G., Westendorf J., Uldschmid A., Nigg E. A. (2010). Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J. Cell Sci. 123, 2163–2169 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Guichard P., Chrétien D., Marco S., Tassin A. M. (2010). Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29, 1565–1572 10.1038/emboj.2010.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., Agard D. A., Sedat J. W. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A. (2005). The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7, 1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Haren L., Stearns T., Lüders J. (2009). Plk1-dependent recruitment of γ-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE 4, e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E. M., Kulukian A., Holland A. J., Cleveland D. W., Stearns T. (2010). Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191, 721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S. W. (2007). Far-field optical nanoscopy. Science 316, 1153–1158 10.1126/science.1137395 [DOI] [PubMed] [Google Scholar]
- Hiraki M., Nakazawa Y., Kamiya R., Hirono M. (2007). Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 17, 1778–1783 10.1016/j.cub.2007.09.021 [DOI] [PubMed] [Google Scholar]
- Holland A. J., Lan W., Niessen S., Hoover H., Cleveland D. W. (2010). Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188, 191–198 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Babcock H., Zhuang X. (2010). Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143, 1047–1058 10.1016/j.cell.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W. F. (2011). Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L. G., Bennetzen M., Westendorf J., Nigg E. A.et al. (2011). Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30, 1520–1535 10.1038/emboj.2011.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M. C., Flückiger I., Gönczy P.et al. (2011). Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.-D., Nigg E. A. (2007). Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G., Lončarek J., Meng X., McEwen B. F., Mogensen M. M., Spektor A., Dynlacht B. D., Khodjakov A., Gönczy P. (2009). Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19, 1012–1018 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Fernandez J. J., Marshall W. F., Agard D. A. (2012). Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 31, 552–562 10.1038/emboj.2011.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. (2007). Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U. K., Stearns T. (2006). GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P. A., Kamiya R., Hirono M. (2004). Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 165, 663–671 10.1083/jcb.200402022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A. M., Bartek J., Nigg E. A. (1999). Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1, 88–93 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- Mogensen M. M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. (2000). Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113, 3013–3023. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M. (2007). SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17, 2169–2174 10.1016/j.cub.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Nigg E. A. (2002). Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer 2, 815–825 10.1038/nrc924 [DOI] [PubMed] [Google Scholar]
- Nigg E. A., Raff J. W. (2009). Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Nigg E. A., Stearns T. (2011). The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13, 1154–1160 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., Bornens M. (1992). Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108, 107–128 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Palazzo R. E., Vogel J. M., Schnackenberg B. J., Hull D. R., Wu X. (1999). Centrosome maturation. Curr. Top. Dev. Biol. 49, 449–470 10.1016/S0070-2153(99)49021-0 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J. L., Bornens M. (1996). Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C. L., Bornens M. (2000). The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149, 317–330 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Roberts D. M., Peifer M., Rogers S. L. (2009). The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184, 225–239 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G.et al. (2008). Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336 10.1126/science.1156947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Heintzmann R., Leonhardt H. (2010). A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190, 165–175 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T. I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S. B., Stierhof Y.-D., Nigg E. A. (2009). Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005–1011 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Sillibourne J. E., Tack F., Vloemans N., Boeckx A., Thambirajah S., Bonnet P., Ramaekers F. C., Bornens M., Grand-Perret T. (2010). Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol. Biol. Cell 21, 547–561 10.1091/mbc.E09-06-0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne J. E., Specht C. G., Izeddin I., Hurbain I., Tran P., Triller A., Darzacq X., Dahan M., Bornens M. (2011). Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton 68, 619–627 10.1002/cm.20536 [DOI] [PubMed] [Google Scholar]
- Sir J. H., Barr A. R., Nicholas A. K., Carvalho O. P., Khurshid M., Sossick A., Reichelt S., D'Santos C., Woods C. G., Gergely F. (2011). A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43, 1147–1153 10.1038/ng.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor A., Tsang W. Y., Khoo D., Dynlacht B. D. (2007). Cep97 and CP110 suppress a cilia assembly program. Cell 130, 678–690 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Stevens N. R., Dobbelaere J., Brunk K., Franz A., Raff J. W. (2010). Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 188, 313–323 10.1083/jcb.200910016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Gönczy P. (2008). Mechanisms of procentriole formation. Trends Cell Biol. 18, 389–396 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gönczy P. (2007). Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell 13, 203–213 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. J., Fu R. H., Wu K. S., Hsu W. B., Tang T. K. (2009). CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11, 825–831 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- Tang C. J., Lin S. Y., Hsu W. B., Lin Y. N., Wu C. T., Lin Y. C., Chang C. W., Wu K. S., Tang T. K. (2011). The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30, 4790–4804 10.1038/emboj.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein K. H., Kleylein-Sohn J., Nigg E. A., Gruneberg U. (2007). Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178, 345–354 10.1083/jcb.200701163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomre D., Bewersdorf J. (2010). A new wave of cellular imaging. Annu. Rev. Cell Dev. Biol. 26, 285–314 10.1146/annurev-cellbio-100109-104048 [DOI] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H. A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I. O., Robinson C. V.et al. (2011). Structures of SAS-6 suggest its organization in centrioles. Science 331, 1196–1199 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- Vorobjev I. A., Chentsov Y. S. (1980). The ultrastructure of centriole in mammalian tissue culture cells. Cell Biol. Int. Rep. 4, 1037–1044 10.1016/0309-1651(80)90177-0 [DOI] [PubMed] [Google Scholar]
- Vulprecht J., David A., Tibelius A., Castiel A., Konotop G., Liu F., Bestvater F., Raab M. S., Zentgraf H., Izraeli S.et al. (2012). STIL is required for centriole duplication in human cells. J. Cell Sci. 125, 1353–1362 10.1242/jcs.104109 [DOI] [PubMed] [Google Scholar]
- Wang W. J., Soni R. K., Uryu K., Tsou M. F. (2011). The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., Denoulet P. (1992). Distribution of glutamylated α and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59, 425–432. [PubMed] [Google Scholar]
- Yan X., Habedanck R., Nigg E. A. (2006). A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol. Biol. Cell 17, 634–644 10.1091/mbc.E05-08-0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.