Summary
The assembly of a functional mitotic spindle is essential for cell reproduction and requires a precise coordination between the nuclear cycle and the centrosome. This coordination is particularly prominent in organisms that undergo closed mitosis where centrosomes must not only respond to temporal signals, but also to spatial considerations, e.g. switching from the production of cytoplasmic microtubule arrays to the generation of dynamic intra-nuclear microtubules required for spindle assembly. We utilize a gene knockout of Kif9, a Dictyostelium discoideum Kin-I kinesin, to destabilize the physical association between centrosomes and the nuclear envelope. This approach presents a unique opportunity to reveal temporal and spatial components in the regulation of centrosomal activities in a closed-mitosis organism. Here we report that centrosome–nuclear engagement is not required for the entry into mitosis. Although detached centrosomes can duplicate in the cytoplasm, neither they nor nuclei alone can produce spindle-like microtubule arrays. However, the physical association of centrosomes and the nuclear envelope is required to progress through mitosis beyond prometaphase.
Key words: Centrosome, Nucleus, Closed mitosis, Dictyostelium
Introduction
In many lower eukaryotes (e.g. fungi, protists, unicellular algae), cells divide via a mitotic mechanism that functions within a closed nuclear compartment (Heath, 1980; Kubai, 1976). The nuclear envelope remains mostly or completely intact and thus requires that mitotic components be specifically imported for spindle assembly and function. This closed mitosis mechanism differs from the open system characteristic of vertebrate cells whereby the nuclear envelope is disassembled at prophase and reassembled at telophase. Understanding the differences between these two types of mitoses provides an opportunity to develop reagents that specifically perturb closed systems, and offers a means to selectively target these types of organisms in complex environments. (De Souza and Osmani, 2007; Güttinger et al., 2009; Sazer, 2010).
A primary difference between open and closed systems lies in the management of their centrosome activities. Closed mitotic systems require microtubule-organizing centers to play spatially separate roles. Centrosomes here must nucleate microtubules in the cytosol during interphase but transition into nuclear-based spindle poles during mitosis. In organisms such as S. cerevisiae and A. nidulans, permanently embedding centrosomes into the nuclear envelope facilitates these two activities. The cytoplasmic and nuclear surfaces remain spatially segregated and therefore can be differentially regulated (Jaspersen and Winey, 2004; Oakley and Morris, 1983). In other organisms (e.g. Dictyostelium, S. pombe, Cryptococcus) the centrosome is only embedded in the nuclear envelope during mitosis (Ding et al., 1997; Moens, 1976; Roos, 1975; Yamaguchi et al., 2009). This transient arrangement complicates the picture and requires careful coordination between cytoplasmic and nuclear activities to ensure centrosomes duplicate and enter the nuclear envelope in a timely fashion to successfully complete division (e.g. Hagan, 2008). Although centrosome and nuclear growth cycles are intimately linked, little is known about the functional requirements of centrosome–nuclear engagement in a closed mitotic system. If one could separate the centrosome from the nucleus in a closed mitotic system, it would then be possible to differentiate processes that independently target the two organelles vs what events require the concerted actions of both.
Centrosomes in Dictyostelium consist of a trilaminar disc shaped core surrounded by an electron dense corona (Moens, 1976; Roos, 1975; Ueda et al., 1999). The core constitutes the replicative structure analogous to centrioles in vertebrate cells and the corona functions to nucleate and anchor microtubules. During interphase, the centrosome lies in the cytoplasm adjacent to the nucleus, and is recruited into the nuclear envelope for mitosis (Moens, 1976; Roos, 1975; Ueda et al., 1999). Centrosomes here begin to duplicate at the G2/M transition and the onset of mitosis is visually marked by the near complete loss of cytoplasmic microtubules. During prophase, the core doubles in length and width (Ueda et al., 1999), and at the prophase/prometaphase transition, this structure begins to split lengthwise down the middle to expose internal surfaces that nucleate spindle microtubules. At about the same time, the centrosome docks into an opening in the nuclear envelope. The twin daughter centrosomes separate a short distance forming a nascent spindle, and pause in prometaphase for 4–5 minutes. Once spindle elongation begins, it proceeds in a linear fashion from metaphase into telophase. Ultrastructure analyses of the poles demonstrate that centrosomes become thinner throughout elongation, and curl back on themselves such that the surface exposed to the nuclear volume begins to protrude out into the cytosol (Ueda et al., 1999). These protrusions are thought to nucleate the astral microtubules. In the later stages of telophase, the centrosome completely curls back on itself such that the surface previously exposed to the nuclear volume now surrounds the entire core, and is ejected from the nuclear envelope into the cytosol. Astral microtubules interact with the cell cortex to facilitate cleavage furrow positioning. In wild-type cells, the entire mitotic process takes on the order of 15 minutes.
Deletion of the kif9 kinesin in Dictyostelium disrupts a microtubule-based mechanism that maintains a centrosome–nuclear linkage (Tikhonenko et al., 2009). These cells exhibit growth defects and accumulate supernumerary centrosomes, but nonetheless proceed through division. Using this strain as a tool, we can examine cytoplasmic and nuclear-based centrosome activities during the transitions from G2 through M. As presented below, we find that centrosome–nuclear engagement is not required for mitotic entry, or for centrosome duplication and separation. But engagement is required to fully commit the cell to division, to assemble a spindle, and to restrict centrosome reduplication processes. Our work drafts a set of rules for centrosome–nuclear engagement in a closed mitotic system.
Results
As in wild-type (WT), centrosomes that are located adjacent to nuclei upon mitotic entry in kif9 null cells are integrated into the nuclear envelope and proceed through division (Fig. 1). Nuclear engagement is visible in the light microscope (LM) as an increase in the intensity of GFP-α-tubulin labeling of the centrosome. The mitotic stages at the LM level are indistinguishable from WT cells; however, timing of the early mitotic events is significantly more variable in the kif9 mutant (supplementary material Fig. S1). For example, after centrosome docking and before spindle elongation, WT cells spend an average of 3.9 min in prometaphase (± 2.0 min s.d., n = 12). Although 7 out of 20 kif9 null cells roughly duplicate this timing, the average length of prometaphase (15.0 min) and standard deviation (± 14.0 min, n = 20) in the mutant is much greater. These results suggest that while centrosome–nuclear docking occurs in the absence of Kif9, the insertion process or an immediate downstream event is subject to error and is actively monitored by a checkpoint component.
Fig. 1. Mitosis in Dictyostelium.
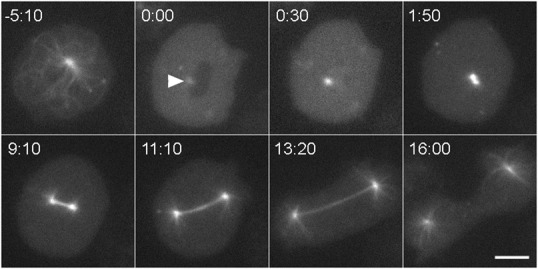
Still frames from a live cell recording, showing GFP-tubulin distribution as a cell transitions from interphase (−5:10) into mitosis and continues into cytokinesis. Although this cell lacks the Kif9 protein, it shows the process and timing characteristic of wild-type cells. The interphase microtubule array rapidly disassembles at the G2/M transition, leaving a dim spot to mark centrosome position (arrowhead, 0:00) adjacent to the dark area of the nucleus. At 30 s, the nucleus has flooded with tubulin. The centrosome is noticeably brighter indicating that it has docked into the nuclear envelope and begun to incorporate tubulin into a nascent spindle. At 1:50, the replicated centrosomes have separated into a bipolar, prometaphase configuration. The subsequent three frames show spindle elongation. In the final frame, the spindle has disassembled, astral microtubules have grown, and the cleavage furrow is constricting the cell into two. Time in min:sec, bar = 5 µm.
Centrosomes in kif9 null cells that do not integrate into the nuclear envelope at the onset of mitosis undergo duplication and separation events in the cytosol (Fig. 2). Cytosolic daughters separate after their nuclear-engaged counterparts establish spindle bipolarity during prometaphase (Fig. 2). As evidenced by their independent movements, cytosolic daughters do not remain connected nor do they form a visible spindle apparatus. Therefore centrosome duplication does not require nuclear envelope insertion, but the nuclear environment during mitosis is necessary for spindle assembly. During prometaphase, cytosolic daughters incorporate less GFP-α-tubulin, and are only 53% as bright (± 11% s.d., n = 20) as single daughter centrosomes docked into the nuclear envelope (e.g. frame 22:00 in Fig. 2). As cells proceed into telophase, the cytosolic daughters begin to nucleate astral microtubules at the same time as nuclear engaged centrosomes, and both centrosome types are indistinguishable from one another by late telophase (e.g. frame 47:30 in Fig. 2). All centrosomes, whether integrated in the nuclear envelope or free in the cytosol, trigger cytokinetic furrow activity following telophase (supplementary material Fig. S2). Thus cytosolic and nuclear-engaged centrosomes undergo similar if not identical maturation processes near the end of mitosis to restore interphase microtubule arrays.
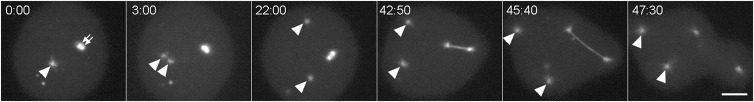
Fig. 2. Cytosolic centrosome dynamics.
Panel shows mitotic sequence of a mononucleated kif9 null cell with two centrosomes, labeled with GFP-tubulin. One centrosome has docked into the nucleus and separated into daughter units in a prometaphase configuration (small arrows in frame 0:00). These daughters progress on to anchor the spindle apparatus. The second centrosome is free in the cytosol and marked with an arrowhead. In frame 3:00, the cytosolic centrosome has split into two daughters, which move around in the cytosol during mitosis (arrowheads). Note that this cell takes about four times longer to progress through mitosis than a normal cell (e.g. Fig. 1). The other GFP-dots visible in the cytosol are not centrosomes. These inclusions are commonly seen in WT cells. Their variations in size and their absence of nucleated tubulin make them visually distinct from centrosomes. Time in min:sec, bar = 2 µm.
The loss of Kif9 does not lead to an increase in multinucleated cells, but does result in greater variability from the normal 1:1 ratio of centrosomes to nuclei (Tikhonenko et al., 2009). How might the loss of centrosome–nuclear connectivity lead to such variability? One possibility is that the centrosome remains detached from the nucleus, duplicates in the cytosol, and cells exit mitosis in the absence of karyokinesis. We examined 209 mitotic kif9 null cells and could only find four examples where the centrosomes were clearly detached from the nucleus upon mitotic entry. In these cases, cells remained in a prophase-like state for an extended period of time (66 min ± 40), until one or both daughter centrosomes docked into the nuclear envelope and initiated mono or bipolar spindle assembly (e.g. Fig. 7). In these few examples, cells did not exit mitosis in the absence of centrosome–nuclear engagement. However, a larger scale population assay revealed that while mononucleated WT and kif9 null cells with a single centrosome contain approximately the same size nucleus, mononucleated kif9 null cells containing two centrosomes are more variable, with a nuclear size peak approximately double WT (Fig. 3). This result supports the idea that cells can exit mitosis in the absence of karyokinesis. We postulate that this mechanism is one of the primary means of supernumerary centrosome accumulation.
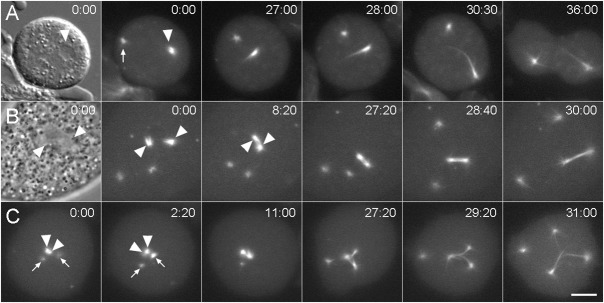
Fig. 7. Daughter centrosome dynamics.
Three sequences that demonstrate spindle arrangements resulting from daughter centrosome–nuclear interactions. (A) Mononucleated kif9 null cell with a single centrosome. Daughter centrosomes have already separated at 0:00: the significantly brighter daughter (arrowhead) has docked into the nuclear envelope while other daughter (arrow) remains in the cytosol. The first frame shows a DIC image where the nucleus is clearly visible; the subsequent frames show GFP-tubulin fluorescence at the times indicated. The single nuclear daughter forms a monopolar spindle as this cell progresses through mitosis. (B) Mononucleated kif9 null cell with two centrosomes. Both centrosomes have separated forming four daughters; two of which have independently docked into the nuclear envelope (0:00) (arrowheads). The first frame shows a phase contrast image where the nuclear daughter centrosome positions are indicated around the relatively clear area of the nucleus. The two nuclear daughter centrosomes coalesce to form a functional bipolar spindle apparatus, demonstrating plasticity of the spindle assembly process. (C) Mononucleated kif9 null cell with two centrosomes. One centrosome has docked into the nucleus and has split into a prometaphase spindle arrangement (arrowheads in 0:00), while the other centrosome has separated into daughters in the cytosol (arrows). By 2:10, one of the cytosolic daughter centrosomes has docked into the nucleus (right arrow) and has begun to incorporate GFP-tubulin. The three nuclear daughter centrosomes then proceed to form a tripolar spindle apparatus, while the unengaged daughter remains in the cytosol. Time in min:sec, bar = 2 µm.
Fig. 3. Centrosome-number nuclear-size relationship.
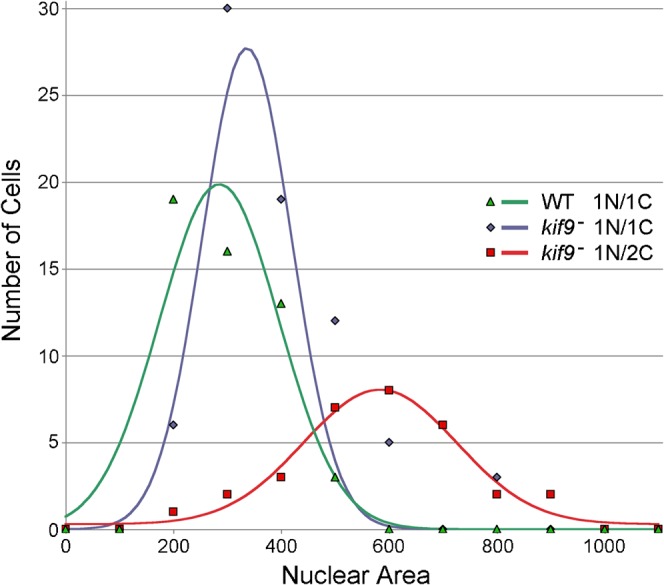
Mononucleated wild-type and kif9 null cells with single centrosomes both contain nuclei of similar size (255 ± 94 pixels, n = 50; 343 ± 133 pixels, n = 74 respectively; area calculated from 2D projections of Z-stacks). However, mononucleated kif9− cells with 2 centrosomes contain an average nucleus nearly twice this size (531 ± 58 pixels, n = 30). This result correlates centrosome accumulation with an increased DNA content, suggesting a population of cells that entered mitosis, duplicated centrosomes, failed to segregate nuclear content, but continued on in the cell cycle.
In multinucleated Dictyostelium cells, mitosis proceeds in a synchronous fashion (Neujahr et al., 1998). The kif9 null mutant further provides an opportunity to examine the contribution of centrosome–nuclear engagement to the synchrony machinery. We observed 14 examples of multinucleated kif9 null cells entering mitosis that contained a heterogeneous mixture of nuclei with and without engaged centrosomes (e.g. Figs 4, 5). Nuclei lacking centrosomes correlated with a significant prometaphase delay in nuclei in the same cytosol that contained integrated centrosomes (Fig. 4). This delay (76 min ± 33 min, n = 14) is even longer than the average prometaphase period observed in mononucleated kif9 null cells. Moreover, we observed two outcomes that appeared coupled to mitotic progression. In the first, the delay provides a time window for cytosolic centrosomes to find and engage unoccupied nuclei through Brownian motion (6 out of 14 cells); once docked, mitosis then proceeds. In the second (8 out of 14 cells) the delay triggers centrosome endoduplication or fragmentation events that also lead to mitotic exit (Figs 5, 8). These latter events were observed as a splitting of fluorescent tubulin centers, followed by a burst of microtubule polymerization and cleavage furrow formation. No such delays are seen in multinucleated cells containing the Kif9 protein. These results indicate that mitotic nuclei lacking integrated centrosomes exert a global negative control over mitotic progression, and that there exists an endoduplication pathway that can be triggered to generate additional centrosomes.
Fig. 4. Unengaged nuclei trigger mitotic arrest.
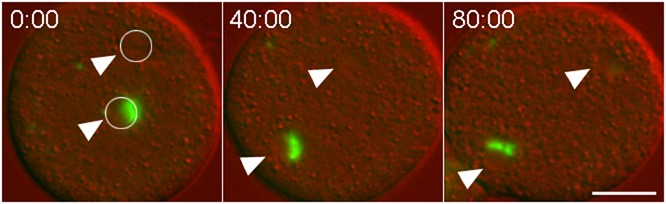
Merged DIC (red) and GFP-tubulin (green) images of a binucleate kif9 null cell containing a single centrosome. Nuclei are marked with arrowheads and encircled in the first frame. A single centrosome has docked into the lower nucleus and separated into a prometaphase spindle arrangement, but this cell remains arrested in this configuration for at least 80 min. Time, min:sec, bar, 5 µm.
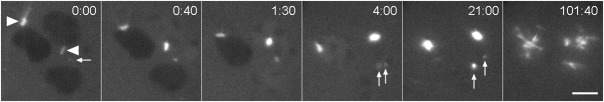
Fig. 5. Centrosome reduplication.
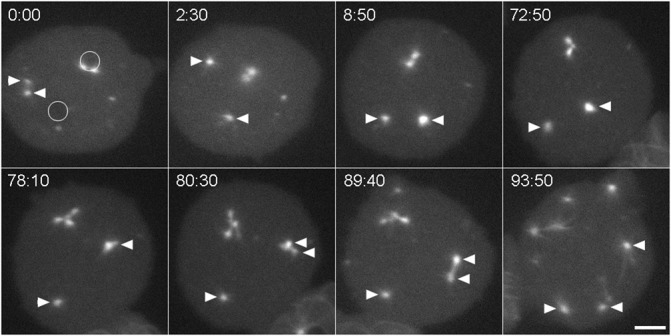
A GFP-tubulin labeled binucleate kif9 null cell with a centrosome pair in one nucleus (upper circle in panel 0:00), one unoccupied nucleus (lower circle in panel 0:00), and a cytosolic centrosome that has separated into daughters (arrowheads). Over the next 8 min, one of the cytosolic daughter centrosomes (right arrowhead) moves to and into the unoccupied nucleus (visible at 8:50 as enhanced fluorescence). The cell remains in this configuration for about one hour, at which time both nuclear centrosomal complexes appear to reduplicate. The top pair forms an aberrant spindle complex with four poles, the bottom daughter centrosome forms an abbreviated spindle with two poles. This cell eventually cleaved into six parts. Time, min:sec, bar, 5 µm.
Fig. 8. Tubulin incorporation does not require centrosome integration.
GFP-tubulin labeled trinucleate kif9 null cell. Three centrosomes are marked, two with arrowheads, one with a small arrow. The two brighter centrosomes (arrowheads) engage nuclei and incorporate tubulin. The third centrosome remains in the cytosol and splits into two daughters at 4:00. Only well after this split does one of the daughters engage the third nucleus and begin to brighten (left arrow at 21:00). However, the nearby nucleus has already flooded with tubulin. As in Fig. 6, the delayed integration event triggers a mitotic arrest and centrosomes appear to attempt reduplication. By 101:40, the nuclear centrosomes fragment into multiple pieces. This cell eventually goes on to form multiple cleavage furrows. Time, min:sec, bar, 2 µm.
Mother or daughter centrosomes that are present in the cytosol after cells enter mitosis remain competent to integrate into nuclei and contribute to spindle assemblies (Figs 6, 7). More than one centrosome can bind to the same nucleus (Fig. 6), and combinations of daughter centrosomes engage nuclei to form mono- bi- and multi-polar spindles (Fig. 7). These results indicate that the nuclear envelope is promiscuous to multiple centrosome binding and that the spindle assembly process is dynamic and flexible. The docking of a single daughter or of multiple centrosomes leads to abnormal spindle arrangements.
Fig. 6. Multiple centrosomes can bind to the same nucleus.
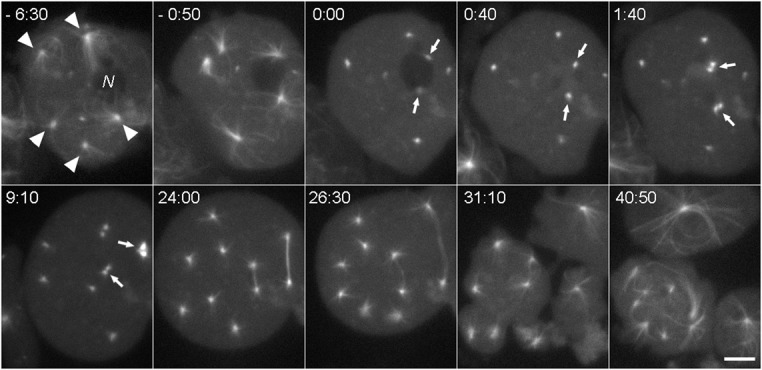
A GFP-tubulin labeled mononucleated kif9 null cell containing five centrosomes. At time −6:30, all five centrosomes (arrowheads) are separate from the nucleus (N). At −0:50, two centrosomes are adjacent to the nucleus as the cell begins to enter mitosis. At 0:00, the interphase MT arrays have disassembled, and two centrosomes (arrows) remain adjacent to the same nucleus. These centrosomes integrate into the nucleus and duplicate by the 1:40 frame; the three cytosolic centrosomes duplicate and separate just prior to the 9:10 frame. This cell continues through division, forming two unequal spindles that segregate chromosomes, and ultimately cleaves into three parts. Time, min:sec, bar, 5 µm.
During prophase, the dark nuclear volume floods with fluorescent GFP-labeled tubulin from the cytosol (Fig. 1). The fluorescent intensity of nuclei in cells due to the GFP-tubulin influx increases by 71% ± 18%, and peaks for 10–30 sec at 27% ± 11% (n = 6) above cytosolic levels (supplementary material Fig. S3). This result indicates that tubulin import is tightly regulated and appears to be driven by an active process. Detailed observation of multi-nucleated cells indicates that tubulin import is not directly triggered by centrosome integration into the nuclear envelope. In Fig. 8, three nuclei show slightly asymmetric tubulin import timing, but only two of the nuclei contain adjacent centrosomes. The third nucleus integrates a daughter centrosome well after tubulin import.
Discussion
The Kif9 kinesin provides a microtubule-based, mechanical linkage between the nucleus and centrosome to ensure that the centrosome remains close to (within 1 µm) the outer nuclear envelope (Tikhonenko et al., 2012; Tikhonenko et al., 2009). Electron microscopy suggests that physical contact between these two organelles also occurs through non-microtubule structures, indicating that other components of the centrosome corona are likely involved with docking and integration into the nuclear envelope during division (Moens, 1976; Roos, 1975; Ueda et al., 1999). In the absence of Kif9, centrosomes can be pulled away from the nucleus through cortical or cytosolic forces acting through the microtubule array. Since Dictyostelium centrosomes duplicate during prophase and must integrate into the nuclear envelope for spindle assembly, this action uncouples the centrosome replication cycle from nuclear engagement during mitosis and provides an opportunity to examine the individual activities of these two organelles in chromosome partitioning. To our knowledge, the ability to uncouple these two organelles in a closed mitotic system is unique and can be used to address a long-standing question of how they coordinate their activities to effect a successful division. Our initial analyses presented here establish several “rules of engagement” that serve as a guideline for future dissection of regulatory pathways.
A centrosome in the nuclear envelope is required for spindle assembly
Our results demonstrate that centrosomes are able to duplicate and separate in the cytosol during prometaphase, and do not require nuclear integration for this process. Short microtubules can be seen protruding from cytosolic daughter centrosomes in the kif9 null cells but there is no visible evidence for a microtubule-based spindle. Cytosolic centrosome splitting was previously observed in Dictyostelium cells overexpressing the coronal protein, CP224 (Gräf et al., 2003). However, in this case, supernumerary centrosomes were seen to separate later during anaphase, and initially contained some interconnecting CP224-GFP material. It is possible that the overexpression defect produces centrosomes that duplicate earlier but retain some degree of affinity, and remain together until astral microtubule formation in anaphase drives the two daughters apart. The absence of cytoplasmic spindle formation in the kif9 null cells is consistent with centrosome activities in mitotic extracts from cell models such as Xenopus, where DNA or at least manipulations to the Ran GTPase and Aurora kinase components are required to stabilize higher-ordered microtubule assemblies (Desai et al., 1998; Heald et al., 1997; Ohba et al., 1999; Sardon et al., 2008; Wilde and Zheng, 1999). Since the nuclear envelope remains intact during Dictyostelium division, the stabilizing effects of chromatin are unavailable to cytosolic centrosomes. And unlike the situation in higher plants, the nuclear envelope does not participate in the spindle assembly process (Wadsworth et al., 2011).
Interestingly, mitotic nuclei that lack integrated centrosomes do not support microtubule assembly, even though the nuclear volume contains tubulin. This result contrasts spindle assemblies in open systems where chromatin and specifically kinetochores can nucleate microtubules. In vertebrates, mitotic spindles can form through a centrosome-mediated microtubule search and capture mechanism, or through a centrosome independent chromosome-based pathway (O'Connell and Khodjakov, 2007; Wadsworth et al., 2011). In Dictyostelium, centrosomes appear strictly required and must be docked into the nuclear envelope for spindle assembly to occur.
Tubulin import is an active process independent of centrosome docking
In closed mitotic systems, all of the protein components required for intranuclear spindle assembly must be imported from the cytosol. The nuclear pore complex (NPC) is the primary gateway in the nuclear envelope and there are complex series of transport factors known to shuttle components in and out (Aitchison and Rout, 2012). The rapid increase in nuclear tubulin fluorescence during prophase highlights a regulation point for this transport machinery. Only after the interphase microtubule network is disassembled and the cell is visually primed for division do nuclei become permeable to tubulin. The influx occurs rapidly and reaches transient nuclear levels that exceed cytosolic tubulin concentration, implying that the nuclear envelope remains intact and there is a dedicated carrier-mediated mechanism that is activated to drive tubulin through the NPC. A similar tubulin influx has been reported for A. nidulans nuclei (Ovechkina et al., 2003). The increased tubulin concentration may facilitate microtubule nucleation on the centrosome surfaces exposed to the nuclear environment and may partially explain the increased GFP-tubulin incorporation over cytosolic centrosomes. We further show that the tubulin transport machinery is not linked to centrosome–nuclear integration, indicating that the mitotic entry machinery independently regulates nuclear and centrosomal activities to prepare a cell for division.
Nuclei can integrate multiple centrosomes
Our results also demonstrate that multiple centrosomes can integrate into the same nucleus, to produce independent bipolar spindles or coalesce to form multipolar arrangements. Both of these scenarios produce spindles that elongate through telophase and proceed into cytokinesis. However, the resulting daughter cells are likely aneuploid and unable to survive long term. This work indicates that the spindle assembly process here is dynamic and malleable, and that there is little or no error control that monitors overall spindle morphology. Similar results have been obtained in yeasts, where it is possible to drive the production of additional spindle pole bodies through mutation (Flory et al., 2002; McGrew et al., 1992; Shirk et al., 2011), and these arrangements lead to aberrant spindles and DNA mis-segregation. However, our work also underscores the importance of maintaining a firm linkage between centrosomes and nuclei during interphase, to insure that only one centrosome is available per nucleus for division. For example, Dictyostelids spontaneously form multinucleated cells in culture and can be induced to form extensive syncytial arrangements by impacting cytokinetic machinery (De Lozanne and Spudich, 1987; Knecht and Loomis, 1987; Neujahr et al., 1998). Yet despite multiple nuclei and centrosomes in a common cytoplasm, mitosis proceeds in an orderly fashion because of the linkage machinery to ensure a 1:1 paring between these two organelles.
Prometaphase arrest
Perhaps the most striking defect in the kif9 null cells is the variability in the length of prometaphase. Even centrosomes that appear properly docked into the nuclear envelope by light microscopy (i.e. enhanced tubulin fluorescence and daughter separation) can pause in this stage before the spindle initiates elongation. The most likely trigger for the delay is the spindle assembly checkpoint (SAC) (Musacchio and Salmon, 2007). Although this checkpoint formally monitors chromosome attachment to the spindle, there are at least two components addressed here that potentially impact the SAC, centrosome anchorage and nuclear occupancy.
Anchorage
In previous work, we showed that the Sun1 protein distribution in the nuclear envelope is influenced by Kif9 activity (Tikhonenko et al., 2012). SUN-domain containing proteins are well conserved in eukaryotes and participate in linkages that couple the outer and inner nuclear envelopes (Starr and Fridolfsson, 2010). These linkages provide NE membrane stability and provide a physical conduit to interconnect cytoplasmic and nuclear activities. In wild-type interphase Dictyostelium cells, Sun1 distribution is enriched in the NE region underlying the centrosome (Schulz et al., 2009; Tikhonenko et al., 2012); in the absence of Kif9, Sun1 is more evenly distributed around the nuclear envelope indicating that Kif9 and Sun1 interact. In S. cerevisiae, mutations to the SUN protein Mps3, directly affect the insertion of newly formed spindle pole bodies into the nuclear envelope, indicating a critical role for SUN in the nuclear anchorage of these organelles (Friederichs et al., 2011). Although the replication of Dictyostelium centrosomes is structurally different than in S. cerevisiae (Jaspersen and Winey, 2004), Sun1 could play a conserved role in integration and anchorage. The observed redistribution of Sun1 may be sufficient to alter how the Dictyostelium centrosome is inserted or anchored into the nuclear envelope in kif9 null cells and this in turn could trigger a delay in proper spindle assembly mechanics.
Occupancy
In the complete absence of spindle assembly, there are no microtubules available to connect chromosomes and thus the checkpoint stalls mitotic progression. This condition can be experimentally induced in closed mitotic systems through mutation or by treatment with microtubule poisons (De Souza et al., 2011; Hagan, 2008; Lim et al., 2009). However, these methods evenly impact all nuclei. The Dictyostelium kif9 null model presents a unique condition; in multinucleated cells there can be a mixture of centrosome occupied and unoccupied nuclei. Since syncytial nuclei divide synchronously in Dictyostelium, we can directly ask what impact centrosome–nuclear heterogeneity has on mitotic progression. In our results, unoccupied nuclei appear to dominate and delay the mitotic progression of other nuclei that do contain centrosomes. The simplest explanation of this effect is that the wait signal produced by the SAC is freely diffusible out of an unoccupied nucleus and it globally controls mitotic progression of other nuclei in the cytoplasm.
Even in the absence of spindle assembly, cells can slip past a SAC-mediated arrest and exit mitosis (De Souza et al., 2011; Rieder and Maiato, 2004). In Dictyostelium, we observe two manifestations of that slippage. First, if all nuclei eventually acquire at least one daughter centrosome integrated into the NE, the cell enters anaphase and progresses through cytokinesis. Regardless of the spindle arrangement, microtubule assembly is stimulated to drive elongation. Interestingly, in monopolar spindles (e.g. Fig. 7A), the spindle retains a compact rod-like shape even in the absence of interdigitating microtubules from the opposite pole. This result implies microtubule bundling factors are active to maintain order in the half spindles. Second, we have also observed centrosomes undergo a reduplication cycle or fragmentation (Fig. 5), as if there is a pathway present to rescue bipolar spindle formation.
At the end of mitosis, all cells proceed through cytokinesis. The cytosolic centrosomes in the kif9 null cells, as well as CP224 mutant cells and closely spaced centrosomes in multinucleated spindle arrangements trigger cleavage furrow activity (Gräf et al., 2003; Neujahr et al., 1998). This process sequesters extra centrosomes into cytoplasts and has been described as one mechanism to restore the normal 1:1 centrosome–nucleus cell configuration (Gräf et al., 2003). In contrast to CP224 mutant cells and vertebrate tumor cells, we see little evidence of extra centrosome coalescence or clustering (Godinho et al., 2009). In fact, the cytosolic centrosomes even appear to repel one another, an activity that could play into a mechanism to support a syncytial environment, by maintaining distance between nuclei.
The kif9 null cell model presents an opportunity to spatially segregate mitotic centrosomes and nuclei in a closed mitotic organism. Centrosome duplication is not dependent on nuclear engagement; however, the microtubule-nucleation capability of the centrosome is strictly required for spindle assembly. Multiple centrosomes can bind to the same nucleus; therefore syncytial cells must have a robust process to ensure that only one centrosome is available per nucleus. The SAC monitored in one nucleus can influence activities in other closed nuclei in the same cytosol, indicating a global cytoplasmic control over mitotic progression. Altogether, these initial observations lay the groundwork for examining pathways that independently regulate centrosome and nuclear activities during mitotic entry, mitotic exit, and spindle assembly.
Materials and Methods
Live cell imaging
Generation of the kif9 null strain is described by Tikhonenko et al. (Tikhonenko et al., 2009). GFP-α-tubulin labeled WT and kif9 null cells were attached to acid-cleaned glass coverslips, washed in phosphate buffer (20 mM KCl, 2.5 mM Na2HPO4, 2.5 mM Na2HPO4, 0.24 mM MgCl2, pH 6.4), and placed under agarose in humidified Rose chambers (Brito et al., 2005). Paired image frames (fluorescent and DIC or phase contrast) were recorded at 10 sec intervals on a Nikon TE2000 inverted microscope, using a 60× objective and an Orca-ER camera (Hamamatsu) controlled by IP Lab software (BD Biosciences). Distance, intensity, and area measurements were performed using ImageJ (NIH) and analyzed with Microsoft Excel. Image sequences were assembled with Adobe Photoshop.
Supplementary Material
Acknowledgments
Work in our laboratories is supported in part by grants from the NSF (MCB-1051612 to M.P.K.) and the NIH (GM59363 to A.K.).
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Aitchison J. D., Rout M. P. (2012). The yeast nuclear pore complex and transport through it. Genetics 190, 855–883 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito D. A., Strauss J., Magidson V., Tikhonenko I., Khodjakov A., Koonce M. P. (2005). Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium. Mol. Biol. Cell 16, 3334–3340 10.1091/mbc.E05-01-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J. A. (1987). Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236, 1086–1091 10.1126/science.3576222 [DOI] [PubMed] [Google Scholar]
- De Souza C. P. C., Osmani S. A. (2007). Mitosis, not just open or closed. Eukaryot. Cell 6, 1521–1527 10.1128/EC.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Hashmi S. B., Yang X., Osmani S. A. (2011). Regulated inactivation of the spindle assembly checkpoint without functional mitotic spindles. EMBO J. 30, 2648–2661 10.1038/emboj.2011.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T. J., Walczak C. E. (1998). The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61, 385–412 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Ding R., West R. R., Morphew D. M., Oakley B. R., McIntosh J. R. (1997). The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 8, 1461–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory M. R., Morphew M., Joseph J. D., Means A. R., Davis T. N. (2002). Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13, 47–58. [PubMed] [Google Scholar]
- Friederichs J. M., Ghosh S., Smoyer C. J., McCroskey S., Miller B. D., Weaver K. J., Delventhal K. M., Unruh J., Slaughter B. D., Jaspersen S. L. (2011). The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 7, e1002365 10.1371/journal.pgen.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho S. A., Kwon M., Pellman D. (2009). Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 28, 85–98 10.1007/s10555-008-9163-6 [DOI] [PubMed] [Google Scholar]
- Gräf R., Euteneuer U., Ho T.-H., Rehberg M. (2003). Regulated expression of the centrosomal protein DdCP224 affects microtubule dynamics and reveals mechanisms for the control of supernumerary centrosome number. Mol. Biol. Cell 14, 4067–4074 10.1091/mbc.E03-04-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger S., Laurell E., Kutay U. (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178–191 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Hagan I. M. (2008). The spindle pole body plays a key role in controlling mitotic commitment in the fission yeast Schizosaccharomyces pombe. Biochem. Soc. Trans. 36, 1097–1101 10.1042/BST0361097 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Habermann A., Karsenti E., Hyman A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615–628 10.1083/jcb.138.3.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath I. B. (1980). Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis. Int. Rev. Cytol. 64, 1–80 10.1016/S0074-7696(08)60235-1 [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. (2004). The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20, 1–28 10.1146/annurev.cellbio.20.022003.114106 [DOI] [PubMed] [Google Scholar]
- Knecht D. A., Loomis W. F. (1987). Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science 236, 1081–1086 10.1126/science.3576221 [DOI] [PubMed] [Google Scholar]
- Kubai D. F. (1976). The evolution of the mitotic spindle. Int. Rev. Cytol. 43, 167–227 10.1016/S0074-7696(08)60069-8 [DOI] [PubMed] [Google Scholar]
- Lim H. H., Zhang T., Surana U. (2009). Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 19, 325–333 10.1016/j.tcb.2009.03.008 [DOI] [PubMed] [Google Scholar]
- McGrew J. T., Goetsch L., Byers B., Baum P. (1992). Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae. Mol. Biol. Cell 3, 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P. B. (1976). Spindle and kinetochore morphology of Dictyostelium discoideum. J. Cell Biol. 68, 113–122 10.1083/jcb.68.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E. D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Neujahr R., Albrecht R., Köhler J., Matzner M., Schwartz J. M., Westphal M., Gerisch G. (1998). Microtubule-mediated centrosome motility and the positioning of cleavage furrows in multinucleate myosin II-null cells. J. Cell Sci. 111, 1227–1240. [DOI] [PubMed] [Google Scholar]
- O'Connell C. B., Khodjakov A. L. (2007). Cooperative mechanisms of mitotic spindle formation. J. Cell Sci. 120, 1717–1722 10.1242/jcs.03442 [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Morris N. R. (1983). A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 96, 1155–1158 10.1083/jcb.96.4.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. (1999). Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356–1358 10.1126/science.284.5418.1356 [DOI] [PubMed] [Google Scholar]
- Ovechkina Y., Maddox P., Oakley C. E., Xiang X., Osmani S. A., Salmon E. D., Oakley B. R. (2003). Spindle formation in Aspergillus is coupled to tubulin movement into the nucleus. Mol. Biol. Cell 14, 2192–2200 10.1091/mbc.E02-10-0641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C. L., Maiato H. (2004). Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7, 637–651 10.1016/j.devcel.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Roos U. P. (1975). Mitosis in the cellular slime mold Polysphondylium violaceum. J. Cell Biol. 64, 480–491 10.1083/jcb.64.2.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T., Peset I., Petrova B., Vernos I. (2008). Dissecting the role of Aurora A during spindle assembly. EMBO J. 27, 2567–2579 10.1038/emboj.2008.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S. (2010). Nuclear membrane: nuclear envelope PORosity in fission yeast meiosis. Curr. Biol. 20, R923–R925 10.1016/j.cub.2010.10.005 [DOI] [PubMed] [Google Scholar]
- Schulz I., Baumann O., Samereier M., Zoglmeier C., Gräf R. (2009). Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur. J. Cell Biol. 88, 621–638 10.1016/j.ejcb.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Shirk K., Jin H., Giddings T. H., Jr, Winey M., Yu H. G. (2011). The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J. Cell Sci. 124, 2891–2896 10.1242/jcs.086652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Fridolfsson H. N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26, 421–444 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenko I., Nag D. K., Robinson D. N., Koonce M. P. (2009). Microtubule-nucleus interactions in Dictyostelium discoideum mediated by central motor kinesins. Eukaryot. Cell 8, 723–731 10.1128/EC.00018-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenko I., Magidson V., Gräf R., Khodjakov A., Koonce M. P. (2012). A kinesin-mediated mechanism that couples centrosomes to nuclei. (in press).</othinfo> [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Schliwa M., Euteneuer U. (1999). Unusual centrosome cycle in Dictyostelium: correlation of dynamic behavior and structural changes. Mol. Biol. Cell 10, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P., Lee W. L., Murata T., Baskin T. I. (2011). Variations on theme: spindle assembly in diverse cells. Protoplasma 248, 439–446 10.1007/s00709-010-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359–1362 10.1126/science.284.5418.1359 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Biswas S. K., Ohkusu M., Takeo K. (2009). Dynamics of the spindle pole body of the pathogenic yeast Cryptococcus neoformans examined by freeze-substitution electron microscopy. FEMS Microbiol. Lett. 296, 257–265 10.1111/j.1574-6968.2009.01643.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.