Abstract
Amino acid analysis of internal sequences of purified NADH-hexacyanoferrate(III) oxidoreductase (NFORase), obtained from highly purified plasma membranes (PM) of spinach (Spinacia oleracea L.) leaves, showed 90 to 100% homology to internal amino acid sequences of monodehydroascorbate (MDA) reductases (EC 1.6.5.4) from three different plant species. Specificity, kinetics, inhibitor sensitivity, and cross-reactivity with anti-MDA reductase antibodies were all consistent with this identification. The right-side-out PM vesicles were subjected to consecutive salt washing and detergent (polyoxyethylene 20 dodecylether and 3-[(3-cholamido-propyl)-dimethylammonio]-1-propane sulfonate [CHAPS]) treatments, and the fractions were analyzed for NFORase and MDA reductase activities. Similar results were obtained when the 300 mm sucrose in the homogenization buffer and in all steps of the salt-washing and detergent treatments had been replaced by 150 mm KCl to mimic the conditions in the cytoplasm. We conclude that (a) MDA reductase is strongly associated with the inner (cytoplasmic) surface of the PM under in vivo conditions and requires washing with 1.0 m KCl or CHAPS treatment for removal, (b) the PM-bound MDA reductase activity is responsible for the majority of PM NFORase activity, and (c) there is another redox enzyme(s) in the spinach leaf PM that cannot be released from the PM by salt-washing and/or CHAPS treatment. The PM-associated MDA reductase may have a role in reduction of ascorbate in both the cytosol and the apoplast.
The plant PM contains intrinsic redox activities that have been implicated in numerous biological processes such as ion uptake, Fe3+ reduction, hormonal growth control, perception of blue light, and defense against pathogens (Møller and Crane, 1990; Rubinstein and Luster, 1993; Lüthje et al., 1997). These redox activities have been characterized with regard to their substrates and are known as oxidoreductases (Bérczi and Asard, 1995). The highest activity can be measured with NADH as electron donor and HCF(III) as electron acceptor, although redox activities can also be detected with NADPH as electron donor and quinones (e.g. duroquinone and ubiquinone-0) or other Fe(III) complexes (e.g. Fe-EDTA and Cyt c) as electron acceptors. In addition to the NAD(P)H-dependent oxidoreductases, a high-potential b-type Cyt has recently been shown to be involved in trans-PM electron transport using ascorbic acid as the electron donor (Asard et al., 1995).
Three different kinds of PM redox activity, classified by their acceptor specificity, were separated 10 years ago (Luster and Buckhout, 1988). However, in spite of this and in spite of the physiological importance of these enzymes, to date only a few of them have been purified (mostly only partially) and characterized in plants (Bérczi and Asard, 1995). Redox enzymes that have been purified to homogeneity and characterized are a 27-kD redox protein from maize root PM (Luster and Buckhout, 1989), two distinct NAD(P)H dehydrogenases from onion root PM (Serrano et al., 1994), and a 45-kD, FAD-containing NFORase from spinach (Spinacia oleracea) leaf PM (Bérczi et al., 1995). A PM-bound thioredoxin has recently been identified by immunoscreening in soybean (Shi and Bhattacharyya, 1996). This protein may play a key role in the regulation of the thiol-disulfide balance of PM proteins.
The production of reactive oxygen species is a rapid response of plant cells to pathogens and can be considered to be the most general mechanism in the plant defense system (Mehdy et al., 1996). Even under optimal conditions, photooxidative stress in photosynthetic cells and many metabolic processes produce reactive oxygen species (Foyer et al., 1994). Ascorbate plays a central role in protecting plant cells against the action of reactive oxygen species in excess. When ascorbate scavenges reactive oxygen species (or other free radicals), it is univalently oxidized to the MDA radical (Heber et al., 1996). Plants have many systems for reducing MDA to regenerate the ascorbate pool, and one of them is MDA reductase (Heber et al., 1996; Smirnoff, 1996). The localization of an ascorbate-regenerating system in association with the PM would be as useful to the plant cell as it is for MDA reductase in chloroplasts (Hossain et al., 1984), glyoxysomes/peroxisomes (Bowditch and Donaldson, 1990; Mullen and Trelease, 1996), and mitochondria (De Leonardis et al., 1995).
In the present paper we show that the physiological electron acceptor for the previously purified NFORase is MDA and that the enzyme is located on the inner (cytoplasmic) surface of the PM. The importance of the localization of an MDA reductase on the cytoplasmic surface of the PM is also discussed.
MATERIALS AND METHODS
Plant Material and PM Purification
Spinach (Spinacia oleracea L.) was grown under controlled conditions as described by Askerlund et al. (1991). PM vesicles were isolated from 4-week-old leaves by aqueous polymer two-phase partitioning using the batch procedure (Larsson et al., 1987). PM vesicles were isolated both in the presence of 300 mm Suc without added monovalent salt (physiological osmotic conditions) and in the presence of 150 mm KCl without added Suc (physiological osmotic and ionic strength conditions) in the homogenization medium. Purified PM vesicles were stored at −80°C in 25 mm Mops-KOH buffer, pH 7.0, supplemented with either 300 mm Suc or with 100 mm KCl, 100 mm Suc, and 1% (v/v) glycerol.
Enzyme Purification
Frozen PM vesicles, purified after homogenization with 300 mm Suc-containing homogenization buffer, were thawed, diluted 10-fold with 10 mm His-HCl buffer, pH 5.8, and then pelleted by high-speed centrifugation (45-Ti rotor, Beckman) at 100,000g at 4°C for 30 min to change the storage buffer to the proper solubilization buffer. NFORase from the pellet was purified as described by Bérczi et al. (1995) and will be referred to as enzyme 1 (the CHAPS-solubilized enzyme). The supernatant was concentrated using a pressure cell concentrator (Amicon, Beverly, MA) with a membrane filter (YM10, Amicon), and the chromatographic steps given by Bérczi et al. (1995) to purify NFORase were applied to the supernatant, except that CHAPS was omitted from the solutions used. This enzyme will be referred to as enzyme 2 (the osmotically released enzyme).
Fractionation of the MDA Reductase Activity
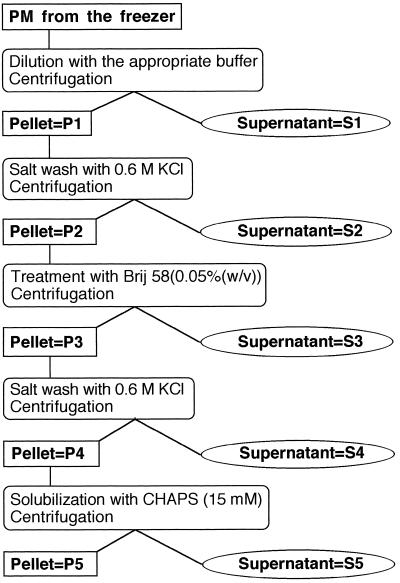
A fractionation procedure was performed to localize the MDA reductase activity in the PM preparation (Fig. 1). The storage buffer used throughout the fractionation procedure contained either 25 mm Mops-KOH buffer, pH 7.0, 300 mm Suc, and 1% (v/v) glycerol, or 25 mm Mops-KOH buffer, pH 7.0, 100 mm KCl, 100 mm Suc, and 1% (v/v) glycerol.
Figure 1.
Scheme of the fractionation protocol.
Frozen PM vesicles were thawed, diluted 10-fold with their storage buffer, and pelleted by high-speed centrifugation at 200,000g at 4°C for 45 min to separate membrane vesicles (both sealed and unsealed) from soluble proteins released from leaky vesicles during thawing. The supernatant was concentrated using the pressure cell with the membrane filter (S1 fraction). The pellet was resuspended in the proper storage buffer (P1 fraction) and then diluted 10-fold with 25 mm Mops-KOH buffer, pH 7.0, supplemented with 0.6 m KCl. After a 5-min incubation, membrane vesicles were pelleted as above. The pellet was resuspended in the proper storage buffer (P2 fraction) and supplemented with Brij 58 to obtain a final detergent concentration of 0.05% (w/v) and a detergent-to-lipid ratio of about 1:1 (w/w) in the suspension. The supernatant (S2 fraction) was concentrated as above.
The Brij-58 treatment converts right-side-out PM vesicles into inside-out vesicles (Johansson et al., 1995). After a 5-min incubation, the Brij 58-treated vesicles were pelleted and the supernatant (S3 fraction) was concentrated as above. The pellet was resuspended in the proper storage buffer (P3 fraction) and then diluted 10-fold with 25 mm Mops-KOH buffer, pH 7.0, supplemented with 0.6 m KCl. After a 5-min incubation, the salt-washed membrane vesicles were pelleted and the supernatant (S4 fraction) was concentrated as described above. The pellet was resuspended in the proper storage buffer supplemented with Brij 58 to a detergent concentration of 0.05% (w/v) (P4 fraction) and then diluted 5-fold with 10 mm His-HCl buffer, pH 5.8. This suspension was combined with the same volume of 10 mm His-HCl, pH 5.8, 2 mm EDTA, 2 mm EGTA, and 30 mm CHAPS (the final CHAPS-to-protein ratio was about 15:1 [w/w]). After a 15-min incubation at room temperature, the unsolubilized membrane proteins were pelleted as described above, the supernatant was again concentrated (S5 fraction), and the pellet was resuspended in 25 mm Mops-KOH buffer, pH 7.0, supplemented with 1% (v/v) glycerol (P5 fraction). Redox activities were determined as soon as the fractions were obtained (i.e. before freezing and storing them at −80°C), and the protein content of the fractions was determined on the next day.
Enzyme Assays and Protein Determination
NAD(P)H-utilizing redox activity with HCF(III) and MDA was measured as given by Bérczi et al. (1991) and Hossain et al. (1984), respectively. MDA was generated by enzymatic oxidation of ascorbic acid as given by Hossain et al. (1984). The MDA concentration was determined by measuring the A360 according to the method of Bielski et al. (1971). Protein concentration was estimated according to the method of Markwell et al. (1978) using BSA as a standard.
Electrophoresis and Immunoblotting
Samples for SDS-PAGE separation and enhanced chemiluminescence visualization of MDA reductase were prepared by the method of Wessel and Flügge (1984), which uses an organic solvent method for precipitating soluble as well as hydrophobic proteins. This was done to remove lipids, detergents, and salts from the different samples and to permit the loading of equal MDA reductase activity from each fraction.
SDS-PAGE was run on a 10 to 15% gradient gel in the buffer system of Laemmli (1970) with a Protean TM II apparatus (Bio-Rad). The proteins were electroblotted onto cellulose nitrate membrane (0.45 μm, BA85, Schleicher & Schuell), reacted with polyclonal antibody raised against the MDA reductase, which was purified from cucumber fruits (a kind gift from Prof. K. Asada, Kyoto University, Uji, Kyoto, Japan), and visualized by the enhanced chemiluminescence method according to the manufacturer's instructions (Amersham). The primary and secondary antibodies were diluted 1:3000 and 1:5000, respectively.
Amino Acid Sequencing
The purified NFORase from both the CHAPS-solubilized (enzyme 1) and the osmotically released (enzyme 2) protein mixture was separated on gradient SDS-polyacrylamide gel. The 45-kD bands were cut out and sequenced by Dr. Bo Ek (Department of Cell Research, Swedish University of Agricultural Sciences, Uppsala, Sweden). For internal sequencing, the gel pieces were incubated with LysC from Achromobacter lyticus (Waco, Osaka, Japan) essentially as described by Rosenfeld et al. (1992). The eluted peptides were separated using a SMART chromatography station equipped with a μRPC SC C2/C18 2.1/10 column (Pharmacia). Amino acid sequencing was performed on a sequencer according to the manufacturer's instructions (model 476A, Applied Biosystems/Perkin-Elmer).
RESULTS
Identification of the NFORase as MDA Reductase
The NFORase purified and characterized from spinach leaf PM by Bérczi et al. (1995) was obtained from frozen/thawed PM that were diluted in a low-salt medium and pelleted before the solubilization and purification of the membrane-associated NFORase. About one-half of the NFORase activity found in the frozen PM was released into the supernatant by this procedure and was discarded. However, we decided to continue the search for the natural substrate for this enzyme and therefore purified the NFORase both from the pellet (enzyme 1) and from the supernatant (enzyme 2) using essentially the same purification procedure, except that CHAPS was omitted in all steps involving enzyme 2. No significant difference was observed between the two enzymes either during the purification or in their catalytic properties. They had similar substrate specificity, specific activity, inhibitor sensitivity (not shown), and apparent molecular size on SDS-PAGE.
N-Terminal sequencing of the two enzymes failed, but after LysC digestion and HPLC separation, two internal sequences were analyzed for each of the enzymes. One of the sequences was identical in both preparations (Fig. 2). Both this common sequence and the two other sequences were identical or very similar (more than 90% identical) to amino acid sequences of MDA reductase obtained from pea (Pisum sativum) (Murthy and Zilinskas, 1994), cucumber (Cucumis sativus) (Sano and Asada, 1994), or tomato (Lycopersicon esculentum) (Grantz et al., 1995). MDA reductase is an FAD-containing enzyme of 47 kD (Sano and Asada, 1994), which is consistent with the results of Bérczi et al. (1995). Thus, it is highly probable that both enzymes 1 and 2 are MDA reductase and that the physiological substrate of the PM NFORase purified by Bérczi et al. (1995) is MDA.
Figure 2.
Internal amino acid (aa) sequences of the NFORase purified from spinach leaf PM vesicles. Frozen PM vesicles were thawed and diluted with buffer with low ionic strength and low osmotic potential and then pelleted by centrifugation. Enzyme 1 was purified from the pellet after CHAPS solubilization, and enzyme 2 was purified from the supernatant containing proteins released by the freezing-thawing procedure and hypo-osmotic shock. Search for homologous internal sequences in the Swiss-Prot databank identified highly homologous sequences in monodehydroascorbate radical reductases in three different species. Numbers on the right are the amino acid positions of the highly homologous sequences in the known MDA reductases. Bold letters indicate identity.
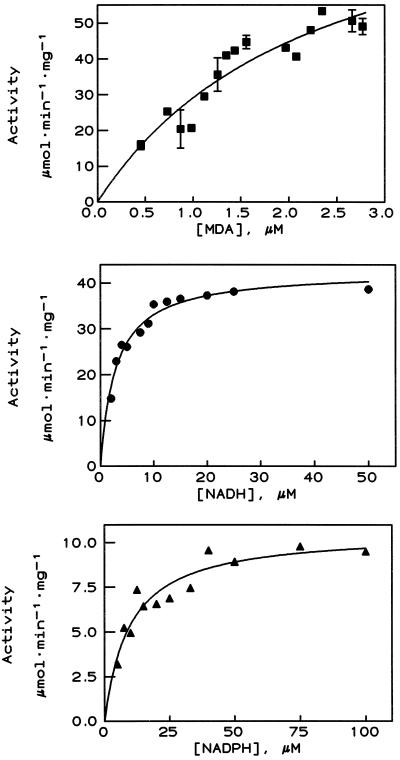
When the MDA reductase activity of the PM and purified enzyme 1 was tested, it showed the same donor specificity and almost the same degree of purification as the NFORase activity (Table I). The effect of three inhibitors of NFORase activity, mersalyl, PCMB, and dicumarol, was very similar with MDA as electron acceptor when both the PM and the purified enzyme (enzyme 1) were tested (Table II; also in Bérczi et al., 1995). The following Km values were obtained by varying the MDA, NADH, or NADPH concentrations (Fig. 3): Km(MDA) = 2.2 ± 0.3 μm (with 50 μm NADH, n = 3); Km(NADH) = 2.5 ± 0.8 μm; and Km(NADPH) = 13.2 ± 3.6 μm (with 2.0 ± 0.2 μm MDA and n = 3 in both cases). These values are in good agreement with earlier results published for other MDA reductases (Hossain et al., 1984; Hossain and Asada, 1985; Borraccino et al., 1986, 1989; Bowditch and Donaldson, 1990).
Table I.
Substrate specificity of HCF(III) and MDA reduction of the PM and the purified NFORase (enzyme 1)
| Fraction | Specific Activity
|
|||
|---|---|---|---|---|
| MDA
|
HCF(III)
|
|||
| + NADH | + NADPH | + NADH | + NADPH | |
| μmol NAD(P)H oxidized min−1mg−1 protein | ||||
| PM | 0.082 | n.m.a | 0.644 | n.m. |
| Enzyme 1 | 61.3 | 11.4 | 251 | 42.2 |
n.m., Not measured.
Table II.
Inhibition of NADH oxidation of PM and NFORase (enzyme 1) with HCF(III) and MDA as electron acceptors
| Inhibitor | Inhibition
|
|||
|---|---|---|---|---|
| PM
|
NFORase (enzyme 1)
|
|||
| HCF(III) | MDA | HCF(III) | MDA | |
| % | ||||
| Mersalyl (100 μm) | 71 | 100 | 99 | 97 |
| PCMB (100 μm) | 93 | 100 | 96 | 100 |
| Dicumarol (400 μm) | n.m.a | n.m. | 93 | 100 |
n.m., Not measured.
Figure 3.
Redox activity of the NFORase (enzyme 1) when the concentration of MDA (▪), NADH (•), and NADPH (▴) was varied and the concentrations of NADH and MDA, respectively, were held constant. Results presented are from one enzyme preparation; two other preparations gave similar results (for Km values, see Results).
Fractionation of the Redox Activity in the PM Vesicles
A very detailed fractionation procedure was carried out for two reasons: (a) Since MDA reductase is found in the cytoplasm, we wanted to ascertain whether the activity in our PM vesicles could be due to an artifactual association of cytoplasmic MDA reductase with the vesicles. (b) If MDA reductase is strongly associated with the PM, we wanted to know its orientation, i.e. whether it is associated with the outer, apoplastic surface or with the inner, cytoplasmic surface.
Traditionally, plant tissues are homogenized in the presence of a 0.3 to 0.5 m concentration of an osmoticum such as Suc or mannitol to avoid rupturing organelles such as mitochondria and chloroplasts. Therefore, our standard PM preparation included 0.3 m Suc in all media. However, the cytosol contains salts rather than sugars as osmotica, and at a much lower ionic strength in the homogenization medium there is a risk that soluble proteins adhere electrostatically and unspecifically to membranes, thereby creating artifactual associations. To avoid this, we also purified our PM vesicles in the presence of 150 mm KCl instead of 300 mm Suc.
The frozen/thawed PM vesicles, purified with Suc or KCl, were both right-side-out and tightly sealed, since the latency of the NFORase was more than 80% (Table III). The two kinds of PM preparation contained the same specific activity of both NFORase and MDA reductase (Table IV).
Table III.
Latency of NFORase and MDA reductase activities in the pellets of the fractionation experiments
| Fraction | Activity
|
|||
|---|---|---|---|---|
| NFORase
|
MDA reductase
|
|||
| Suc | KCl | Suc | KCl | |
| % | ||||
| PM | 82 ± 1 | 88 ± 4 | 72 ± 2 | 81 ± 3 |
| P1 | 91 ± 1 | 93 ± 2 | 86 ± 3 | 84 ± 3 |
| P2 | 86 ± 1 | 89 ± 6 | 72 ± 7 | 86 ± 3 |
| P3 | 33 ± 4 | 44 ± 8 | 27 ± 5 | 28 ± 8 |
| P4 | 19 ± 11 | 28 ± 6 | 5 ± 4 | 13 ± 18 |
| P5 | 24 ± 7 | 35 ± 29 | 35 ± 23 | 11 ± 16 |
PM vesicles were purified in the presence of either 300 mm Suc or 150 mm KCl (see Methods). The origin of the fractions is explained in Figure 1. Latency of enzyme activity was measured with Triton X-100 (± 0.015% [w/v]) and calculated as a percentage (Larsson et al., 1984). Values are averages of three independent series of PM preparations both with Suc and with KCl.
Table IV.
Fractionation of the NFORase and MDA reductase activity of spinach leaf PM vesicles
| Parameter | Protein
|
NFORase
|
MDA
Reductase
|
|||
|---|---|---|---|---|---|---|
| Suc | KCl | Suc | KCl | Suc | KCl | |
| nmol min−1mg−1 protein | ||||||
| PM-specific activity | N/A | N/A | 671 ± 135 | 600 ± 42 | 54 ± 22 | 62 ± 5 |
| % | ||||||
| Relative values | ||||||
| PM | 100 | 100 | 100 | 100 | 100 | 100 |
| S1 | 5 | 4 | 6 | 6 | 14 | 11 |
| S2 | 2 | 1 | 1 | 1 | 2 | 2 |
| S3 | 13 | 16 | 17 | 31 | 30 | 54 |
| S4 | 14 | 10 | 22 | 18 | 31 | 20 |
| S5 | 14 | 15 | 7 | 10 | 12 | 21 |
| P5 | 44 | 59 | 13 | 12 | 6a | 5a |
| Recovery | 92 | 105 | 66 | 78 | 95 | 113 |
PM vesicles were purified in the presence of either 300 mm Suc or 150 mm KCl (see Methods). The origin of the fractions is explained in Figure 1. Values are the averages of three independent series of PM preparations both with Suc and with KCl and are given as percentages of the PM (100%).
Values are not significantly different from 0.
Very little protein, NFORase, and some MDA reductase activity were released into the supernatant (S1; Table IV) when these PM vesicles were diluted with an iso-osmotic medium. This is different from results reported previously (Bérczi et al., 1995), in which the dilution was with a hypo-osmotic medium known to cause rupture and resealing of the vesicles (Bérczi and Møller, 1986) and to lead to the loss of some enzymatic activity from the vesicle lumen (S1; Table IV).
When the right-side-out vesicles were washed with 0.6 m KCl, which will release loosely attached proteins, little protein or redox activity was released (S2; Table IV). The majority of the NFORase activity and MDA activity was inside the permeability barrier of the right-side-out vesicles at this point.
The addition of Brij 58 converts sealed right-side-out PM vesicles into sealed inside-out vesicles (Johansson et al., 1995). This would release any proteins merely enclosed within the right-side-out vesicles but not proteins firmly attached to their inner surface. With our vesicles, Brij-58 treatment caused the release of 17 to 31% of the NFORase and 30 to 54% of the MDA activity (S3; Table IV). The percentage released from the KCl-prepared PM was higher, showing that more enzyme activity was bound to the cytoplasmic side of the PM in the Suc-prepared vesicles and was not released by the inversion. Consistent with the results of Johansson et al. (1995), the latency in the pellet after Brij treatment (P3; Table III) was very low for both NFORase and MDA reductase. This indicates that the residual enzyme activity (about one-third to one-half) was bound to the cytoplasmic surface of the PM vesicles now exposed to the medium.
Washing these inside-out PM vesicles with 0.6 m KCl released a substantial part of the residual activity (S4; Table IV), and washing them with 1.0 m KCl released virtually all of the MDA activity (results not shown). After the 0.6 m KCl treatment, a further gentle solubilization with CHAPS removed the remaining MDA reductase activity, and left 10 to 15% of the NFORase activity and about one-half of the protein in the final pellet (S5, P5; Table IV).
The recovery of protein and MDA reductase was 90 to 110%, and the recovery of NFORase was approximately 70% in this multistep fractionation procedure (Table IV), indicating that all of the enzyme activities were accounted for.
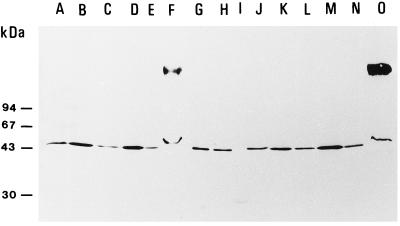
Samples from the various subfractions containing the same amount of MDA reductase activity were separated by SDS-PAGE, blotted, and immunoreacted with antibodies raised against cytosolic MDA reductase from cucumber (Fig. 4). Only a band at 45 kD reacted with the antibodies and was seen in all fractions. The broad band at high molecular mass in fractions S5 (Fig. 4, lanes F and O) is artifactual, since (a) it was not seen in the preceding fractions and (b) hardly any protein was seen at that position after silver or Coomassie blue staining. The band at 45 kD, and, thus, the antigenicity per activity unit, was stronger in fractions S1 and S3 (Fig. 4, lanes B, D, K, and M), where enzyme 2 is recovered, than in fractions S4 and S5 (Fig. 4, lanes E, F, N, and O), which contain enzyme 1, the bound form.
Figure 4.
Immunoblotting of the PM and supernatant fractions of the MDA reductase activity fractionation experiment using anti-MDA reductase antibodies. Lanes A to F, Fractions from PM preparation with Suc in the homogenization buffer; lanes J to O, fractions from PM preparation with KCl in the homogenization buffer. A and J, PM; B and K, S1 fraction; C and L, S2 fraction; D and M, S3 fraction; E and N, S4 fraction; F and O, S5 fraction; G and H, Mono-Q fraction Q27 and affinity fraction A40 (Bérczi et al., 1995); and I, standard proteins. About the same total MDA reductase activity (approximately 5 nmol min−1) was applied in each lane (except lane I). The positions of standard proteins of known molecular mass are shown on the left. All fractions were from the same experiment. Three independent fractionation experiments gave similar results. The high-molecular-mass band in lanes F and O is an artifact probably caused by CHAPS.
DISCUSSION
The 45-kD FAD-containing NFORase from spinach leaf PM (Bérczi et al., 1995) is here identified as MDA reductase based on a high degree of sequence identity (Fig. 1), substrate specificity (Table I), inhibitor sensitivity (Table II), kinetic constants (Fig. 3), and immunological cross-reactivity (Fig. 4).
Very little MDA reductase was associated with the outer, apoplastic surface of the PM, as shown by the high latency of MDA reductase in P1 and the low MDA activity in the first two supernatants (S1 and S2, Tables III and IV). Between one-third and one-half of the MDA activity seemed to be soluble inside the vesicles or weakly bound to the inner, cytoplasmic surface, since it was released by Brij-58 treatment (S3, Table IV). This would be equivalent to enzyme 2 as described in Methods.
However, even when the PM vesicles had been isolated under high-salt conditions (150 mm KCl), approximately 40% of the MDA reductase activity remained bound to the PM vesicles after Brij-58 treatment (P3 = S4 plus S5; Table IV). A wash with 0.6 m KCl removed only part of this activity (Table IV); a wash with 1.0 m KCl (not shown) or CHAPS treatment was necessary to remove the last 20% (S5, Table IV). These observations indicate that a large part of the MDA reductase, copurifying with spinach leaf PM (enzyme 1; see Methods), was tightly bound by mainly electrostatic forces to the inner cytoplasmic surface. It is unlikely that the enzyme has membrane-spanning helices or is anchored to the PM via a fatty acid, as was reported for a subpopulation of nitrate reductase (Stöhr et al., 1995), but there may well be a docking protein in the PM. The presence of MDA reductase (often called ascorbate free radical reductase in the literature) activity in the plant PM preparations confirms previous reports (Morré et al., 1986; Luster and Buckhout, 1988; Gonzalez-Reyes et al., 1992), but in none of these studies was it shown how or where MDA reductase was associated with the membrane.
Apart from the difference in the binding to the PM, we have detected only one difference between enzyme 2, the soluble form of MDA reductase released from the lumen of the PM vesicles by hypo-osmotic rupture, and enzyme 1, the form bound to the PM surface. This difference relates to the immuno-cross-reactivity (Fig. 4): enzyme 2 (S1 and S3) reacted stronger than enzyme 1 (fractions S4 and S5), although the same MDA reductase activity had been loaded into all lanes. However, this difference could be an artifact caused by the sample preparation method (see Methods) and caution should be shown in the interpretation. With our present knowledge it is not possible to determine whether enzymes 1 and 2 are encoded by different genes or whether the two populations are formed by posttranslational modification of one MDA reductase gene product.
The Km(NADH) for the purified MDA reductase with MDA as electron acceptor was 2.5 μm (Fig. 3; Results), whereas it has been reported to be 25 to 77 μm for the enzyme in PM vesicles from various sources with ferricyanide as acceptor (Askerlund et al., 1988; Møller and Crane, 1990, and refs. therein). This large difference may partly be due to the difference in electron acceptor or in species. However, it may also be a product of electrostatic interactions: Both the outer and inner surface of the plant PM has a pI below pH 4.0 (Larsson et al., 1990, and refs. therein). This means that the net charge is negative at neutral pH, giving rise to a negative surface potential. This will repel the likewise negatively charged NADH (Edman et al., 1985, and refs. therein). Thus, the concentration of NADH at the active site would be less than that in the bulk solution, leading to an overestimation of the Km when the enzyme is attached to the PM surface. On the other hand, Askerlund et al. (1988) determined Km(NADH) in the presence of 7.5 mm Mg2+, which will reduce the size of the surface potential to near 0, so at least in that case the electrostatic component was probably small.
The majority of the NFORase activity in spinach leaf PM is caused by MDA reductase. However, there was always a significant part (10–15%) of the NFORase activity left in the PM after the CHAPS treatment (P5), which removed the last MDA reductase (Table IV). This indicates that the spinach leaf PM contains at least one more redox enzyme for which the natural substrates are still unknown, in addition to one or two b-type Cyts (Askerlund et al., 1989; Møller et al., 1991).
Dicotyledons and nongrass monocotyledons reduce Fe3+ to Fe2+ before the uptake of Fe2+. This reduction is probably carried out by a transmembrane reductase in the PM of root cells (Møller and Crane, 1990; Lüthje et al., 1997). Once Fe2+ has been taken up, it is reoxidized to Fe3+, which is transported up to the shoot, where it is thought to require another reduction to Fe2+ before it can be taken up into the leaf cells. The PM of leaf cells would therefore also be expected to contain an Fe-reducing enzyme, although perhaps at a fairly low level. It is possible that the residual NFORase activity in spinach leaf PM after salt-washing and mild-detergent treatment is the Fe-reducing enzyme. Experiments are in progress to investigate this possibility.
MDA reductase has been found in both particulate and soluble (cytosolic?) fractions (Arrigoni et al., 1981; Borraccino et al., 1986), in spinach chloroplasts (Hossain et al., 1984), potato tubers (Borraccino et al., 1986), cucumber fruit (Hossain and Asada, 1985; Sano et al., 1995), leaves of various plants (Heber et al., 1996), soybean root nodules (Dalton et al., 1992), and castor bean endosperm glyoxysomes (Bowditch and Donaldson, 1990; Mullen and Trelease, 1996). In glyoxysomes the enzyme was located in the outer, cytoplasmic surface of the membrane, where it was suggested to be part of a transmembrane transfer of reducing equivalents very similar to that depicted in Figure 5 (Bowditch and Donaldson, 1990; Mullen and Trelease, 1996). It is quite possible that MDA reductase is associated with the surface of other intracellular membranes. This would explain why NFORase activity can be measured in many purified membrane fractions, including the ER and the tonoplast (Fredlund et al., 1994). It would also explain why antibodies raised against a 44-kD NFORase from potato tuber microsomes (Galle et al., 1984) cross-react with one or several polypeptides of 45 to 55 kD in the same membrane fractions (Fredlund et al., 1994). Perhaps Galle et al. (1984) isolated MDA reductase from these membranes rather than the NADH-Cyt b5 reductase they reported. The latter is smaller: 36 kD in erythrocyte membranes (Kitajima et al., 1981), 34.5 kD in pea microsomes (Jollie et al., 1987), and 32 kD in rat liver PM (Kim et al., 1995).
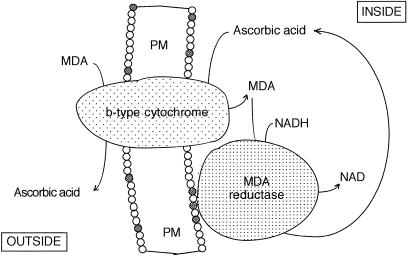
Figure 5.
Schematic representation of the location of PM-bound MDA reductase and of the functional interaction between the trans-PM b-type Cyt and the PM-bound MDA reductase.
There are at least two known membrane processes in which MDA reductase placed on the inner surface of the PM can be useful. Ascorbate has a key role in scavenging oxidative radicals because of its ability to reduce tocopheryl free radicals appearing when tocopherol quenches lipid hydroperoxyl radicals (Packer et al., 1979; Scarpa et al., 1984). Also, there is a high-potential b-type Cyt present in the plant PM (Askerlund et al., 1989; Møller et al., 1991) that transports electrons from ascorbate in the cytoplasm to MDA in the apoplast (Horemans et al., 1994; Asard et al., 1995). In both cases, ascorbate is oxidized to MDA and the presence of an MDA reductase in situ would facilitate the recovery of the ascorbate pool in the vicinity of the membrane surface. In the latter case, apoplastic ascorbate, which has important functions in cell wall synthesis and in host-pathogen interactions (Smirnoff, 1996), can be kept reduced as illustrated in Figure 5. It would be interesting to see whether pathogen infection or oxidative stress changes the degree to which MDA reductase associates with the PM. The study plants with a changed expression of MDA reductase (e.g. by transformation with antisense RNA) with respect to resistance to pathogens and oxidative stress would also be illustrative.
ACKNOWLEDGMENTS
We are grateful to Prof. Christer Larsson, Dr. Kenneth M. Fredlund, and Dr. Per Askerlund for practical advice and many helpful discussions, to Dr. Miguel A. Quinones for comments about the manuscript, and to Mrs. Lena Carlsson and Mrs. Christina Nilsson for excellent technical assistance.
Abbreviations:
- Brij 58
polyoxyethylene 20 dodecylether or C12E20
- CHAPS
3-[(3-cholamido-propyl)-dimethylammonio]-1-propane sulfonate
- dicumarol
3,3′-methylene-bis-(4-hydroxy-coumarin)
- HCF(III)
potassium hexacyanoferrate (III) or ferricyanide
- MDA
monodehydroascorbate radical
- NFORase
NADH-HCF(III) oxidoreductase
- PCMB
p-chloromercurobenzoate
- PM
plasma membrane(s)
Footnotes
This work was supported by grants to A.B. from the Hungarian National Science Foundation (nos. OTKA T012747 and T019863), the Hungarian Academy of Sciences, the Swedish Royal Academy of Sciences, and Kungliga Fysiografiska Saellskapet and to I.M.M. from the Swedish Natural Science Research Council and the Wenner-Gren Stiftelserna.
LITERATURE CITED
- Arrigoni O, Dipierro S, Borraccino G. Ascorbate free radical reductase, a key enzyme of the ascorbate acid system. FEBS Lett. 1981;125:242–245. [Google Scholar]
- Asard H, Horemans N, Caubergs RJ. Involvement of ascorbic acid and a b-type cytochrome in plant plasma membrane redox reactions. Protoplasma. 1995;184:36–41. [Google Scholar]
- Askerlund P, Larsson C, Widell S. Localization of donor and acceptor sites of NADH dehydrogenase activities using inside-out and right-side-out plasma membrane vesicles from plants. FEBS Lett. 1988;239:23–28. [Google Scholar]
- Askerlund P, Larsson C, Widell S. Cytochromes of plant plasma membranes. Characterization by absorbance difference spectrophotometry and redox titration. Physiol Plant. 1989;76:123–134. [Google Scholar]
- Askerlund P, Laurent P, Nakagawa H, Kader J-C. NADH-ferricyanide reductase of leaf plasma membrane. Plant Physiol. 1991;95:6–13. doi: 10.1104/pp.95.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérczi A, Asard H. NAD(P)H-utilizing oxidoreductases of the plasma membrane. An overview of presently purified proteins. Protoplasma. 1995;184:140–144. [Google Scholar]
- Bérczi A, Fredlund KM, Møller IM. Purification and characterization of an NADH-hexacyanoferrate(III) reductase from spinach leaf plasma membrane. Arch Biochem Biophys. 1995;320:65–72. doi: 10.1006/abbi.1995.1343. [DOI] [PubMed] [Google Scholar]
- Bérczi A, Møller IM. Comparison of the properties of plasmalemma vesicles purified from wheat roots by phase partitioning and by discontinuous sucrose gradient centrifugation. Physiol Plant. 1986;68:59–66. [Google Scholar]
- Bérczi A, Sizensky JA, Crane FL, Faulk WP. Diferric transferrin reduction by K562 cells. A critical study. Biochim Biophys Acta. 1991;1073:562–570. doi: 10.1016/0304-4165(91)90231-5. [DOI] [PubMed] [Google Scholar]
- Bielski BHJ, Comstock DA, Bowen RA. Ascorbic acid free radicals. I. Pulse radiolysis study of optical absorption and kinetic processes. J Am Chem Soc. 1971;93:5624–5629. [Google Scholar]
- Borraccino G, Dipierro S, Arrigoni O. Purification and properties of ascorbate free-radical reductase from potato tubers. Planta. 1986;167:521–526. doi: 10.1007/BF00391228. [DOI] [PubMed] [Google Scholar]
- Borraccino G, Dipierro S, Arrigoni O. Interaction of ascorbate free radical reductase with sulphhydryl reagents. Phytochemistry. 1989;28:715–717. [Google Scholar]
- Bowditch MI, Donaldson RP. Asorbate free-radical reduction by glyoxysomal membranes. Plant Physiol. 1990;94:531–537. doi: 10.1104/pp.94.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton DA, Langeberg L, Robbins M. Purification and characterization of monodehydroascorbate reductase from soybean root nodules. Arch Biochem Biophys. 1992;292:281–286. doi: 10.1016/0003-9861(92)90080-g. [DOI] [PubMed] [Google Scholar]
- De Leonardis S, De Lorenzo G, Barraccino G, Dipierro S. A specific ascorbate free radical reductase isoenzyme participates in the regeneration of ascorbate for scavenging toxic oxygen species in potato tuber mitochondria. Plant Physiol. 1995;109:847–851. doi: 10.1104/pp.109.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman K, Ericson I, Møller IM. The regulation of exogenous NAD(P)H oxidation in spinach (Spinaciaoleracea) leaf mitochondria by pH and cations. Biochem J. 1985;232:471–477. doi: 10.1042/bj2320471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Lelandais M, Kunert KJ. Photooxidative stress in plants. Physiol Plant. 1994;92:696–717. [Google Scholar]
- Fredlund KM, Struglics A, Widell S, Askerlund P, Kader J-C, Møller IM. Comparison of the stereospecificity and immunoreactivity of NADH-ferricyanide reductases in plant membranes. Plant Physiol. 1994;106:1103–1106. doi: 10.1104/pp.106.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle A-M, Bonnerot C, Jolliot A, Kader J-C. Purification of a NADH-ferricyanide reductase from plant microsomal membranes with a zwitterionic detergent. Biochem Biophys Res Commun. 1984;122:1201–1205. doi: 10.1016/0006-291x(84)91219-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes JA, Döring O, Navas P, Obst G, Böttger M. The effect of ascorbate free radical on the energy state of the plasma membrane of onion (Allium cepa L.) root cells: alteration of K+ efflux by ascorbate? Biochim Biophys Acta. 1992;1098:177–183. [Google Scholar]
- Grantz AA, Brummell DA, Bennett AB. Ascorbate free radical reductase mRNA levels are induced by wounding. Plant Physiol. 1995;108:411–418. doi: 10.1104/pp.108.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber U, Miyake C, Mano J, Ohno C, Asada K. Monodehydroascorbate radical detected by electron papamagnetic resonance spectrometry is a sensitive probe of oxidative stress in intact leaves. Plant Cell Physiol. 1996;37:1066–1072. [Google Scholar]
- Horemans N, Asard H, Caubergs RJ. The role of ascorbate free radical as an electron acceptor to cytochrome b-mediated trans-plasma membrane electron transport in higher plants. Plant Physiol. 1994;104:1455–1458. doi: 10.1104/pp.104.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MA, Asada K. Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. J Biol Chem. 1985;260:12920–12926. [PubMed] [Google Scholar]
- Hossain MA, Nakano Y, Asada K. Monodehydroascorbate reductase in spinach chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen peroxide. Plant Cell Physiol. 1984;25:385–395. [Google Scholar]
- Johansson F, Olbe M, Sommarin M, Larsson C. Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness. A new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J. 1995;7:165–173. doi: 10.1046/j.1365-313x.1995.07010165.x. [DOI] [PubMed] [Google Scholar]
- Jollie DR, Sligar SG, Schuler M. Purification and characterization of microsomal cytochrome b5 and NADH cytochrome b5 reductase from Pisum sativum. Plant Physiol. 1987;85:457–462. doi: 10.1104/pp.85.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Crane FL, Becker GW, Morré DJ. Purification of NADH-cytochrome b5 reductase from rat liver plasma membranes. Protoplasma. 1995;184:111–117. [Google Scholar]
- Kitajima S, Yasukochi Y, Minakami S. Purification and properties of human erythrocyte membrane NADH-cytochrome b5 reductase. Arch Biochem Biophys. 1981;210:330–339. doi: 10.1016/0003-9861(81)90196-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson C, Kjellbom P, Widell S, Lundborg T. Sidedness of plant plasma membrane vesicles purified by partition in aqueous two-phase systems. FEBS Lett. 1984;171:271–276. [Google Scholar]
- Larsson C, Møller IM, Widell S. Introduction to the plant plasma membrane—Its molecular composition and organization. In: Larsson C, Møller IM, editors. The Plant Plasma Membrane. Berlin: Springer-Verlag; 1990. pp. 1–15. [Google Scholar]
- Larsson C, Widell S, Kjellbom P. Preparation of high-purity plasma membranes. Methods Enzymol. 1987;148:558–568. [Google Scholar]
- Luster DG, Buckhout TJ. Characterization and partial purification of multiple electron transport activities in plasma membranes from maize (Zea mays) roots. Physiol Plant. 1988;73:339–347. doi: 10.1104/pp.91.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster DG, Buckhout TJ. Purification and identification of a plasma membrane associated electron transport protein from maize (Zea mays) roots. Plant Physiol. 1989;91:1014–1019. doi: 10.1104/pp.91.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje S, Döring O, Heuer S, Lüthen H, Böttger M. Oxidoreductases in plant plasma membranes. Biochim Biophys Acta. 1997;1331:81–102. doi: 10.1016/s0304-4157(96)00016-0. [DOI] [PubMed] [Google Scholar]
- Markwell MAK, Haas SB, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- Mehdy MC, Sharma YK, Sathasivan K, Bays NW. The role of activated oxygen species in plant disease resistance. Physiol Plant. 1996;98:365–374. [Google Scholar]
- Møller IM, Askerlund P, Widell S. Electron transport constituents in the plant plasma membrane. In: Crane FL, Morré DJ, Löw H, editors. Oxidoreduction at the Plasma Membrane: Relation to Growth and Transport, Vol II: Plants. Boca Raton, FL: CRC Press; 1991. pp. 35–59. [Google Scholar]
- Møller IM, Crane FL. Redox processes in the plasma membrane. In: Larsson C, Møller IM, editors. The Plant Plasma Membrane. Structure, Function, and Molecular Biology. Berlin: Springer-Verlag; 1990. pp. 93–126. [Google Scholar]
- Morré DJ, Navas P, Penel C, Castillo FJ. Auxin-stimulated NADH oxidase (semidehydroascorbate reductase) of soybean plasma membrane: role in acidification of cytoplasm? Protoplasma. 1986;133:195–197. [Google Scholar]
- Mullen RT, Trelease RN. Biogenesis and membrane properties of peroxisomes: does the boundary membrane serve and protect? Trends Plant Sci. 1996;1:389–394. [Google Scholar]
- Murthy SS, Zilinskas BA. Molecular cloning and characterization of a cDNA encoding pea monodehydroascorbate reductase. J Biol Chem. 1994;269:31129–31133. [PubMed] [Google Scholar]
- Packer JE, Slater TF, Willson RL. Direct observation of a free radical interaction between vitamin E and vitamin C. Nature. 1979;278:737–738. doi: 10.1038/278737a0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In gel digestion of proteins for internal sequencing analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Rubinstein B, Luster DG. Plasma membrane redox activity: components and role in plant processes. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:131–155. [Google Scholar]
- Sano S, Asada K. cDNA cloning of monodehydroascorbate radical reductase from cucumber: a high degree of homology in terms of amino acid sequence between this enzyme and bacterial flavoenzymes. Plant Cell Physiol. 1994;35:425–437. [PubMed] [Google Scholar]
- Sano S, Miyake C, Mikami B, Asada K. Molecular characterization of monodehydroascorbate radical reductase from cucumber highly expressed in Escherichia coli. J Biol Chem. 1995;270:21354–21361. doi: 10.1074/jbc.270.36.21354. [DOI] [PubMed] [Google Scholar]
- Scarpa M, Rigo A, Maiorino M, Ursini F, Gregolin C. Formation of α-tocopherol radical and recycling of α-tocopherol by ascorbate during peroxidation of phosphatidylcholine liposomes. An electron paramagnetic resonance study. Biochim Biophys Acta. 1984;801:215–220. doi: 10.1016/0304-4165(84)90070-9. [DOI] [PubMed] [Google Scholar]
- Serrano A, Cordoba F, Gonzales-Reyes JA, Navas P, Villalba JM. Purification and characterization of two distinct NAD(P)H dehydrogenases from onion (Allium cepa L.) root plasma membrane. Plant Physiol. 1994;106:87–96. doi: 10.1104/pp.106.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Bhattacharyya MK. A novel plasma membrane-bound thioredoxin from soybean. Plant Mol Biol. 1996;32:653–662. doi: 10.1007/BF00020206. [DOI] [PubMed] [Google Scholar]
- Smirnoff N. The function and metabolism of ascorbic acid in plants. Ann Bot. 1996;78:661–669. [Google Scholar]
- Stöhr C, Schuler F, Tischner R. Glycosyl-phosphatidyl-inositol-anchored proteins exist in the plasma membrane of Chlorella saccharophila (Krueger) Nadson: plasma-membrane-bound nitrate reductase as an example. Planta. 1995;196:284–287. [Google Scholar]
- Wessel D, Flügge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]