Summary
The cullin-RING family of ubiquitin ligases regulates diverse cellular functions, such as cell cycle control, via ubiquitylation of specific substrates. CUL3 targets its substrates through BTB proteins. Here we show that depletion of CUL3 and the BTB protein KLHL18 causes a delay in mitotic entry. Centrosomal activation of Aurora-A, a kinase whose activity is required for entry into mitosis, is also delayed in depleted cells. Moreover, we identify Aurora-A as a KLHL18-interacting partner. Overexpression of KLHL18 and CUL3 promotes Aurora-A ubiquitylation in vivo, and the CUL3-KLHL18-ROC1 ligase ubiquitylates Aurora-A in vitro. Our study reveals that the CUL3-KLHL18 ligase is required for timely entry into mitosis, as well as for the activation of Aurora-A at centrosomes. We propose that the CUL3-KLHL18 ligase regulates mitotic entry through an Aurora-A-dependent pathway.
Keywords: ubiquitin, BTB, POZ, CUL3, Aurora-A, mitotic entry
Introduction
The regulation of mitotic progression relies predominantly on two post-translational mechanisms: protein phosphorylation and ubiquitylation. Ubiquitylation requires the sequential activities of three enzymes: E1 (ubiquitin activating), E2 (ubiquitin conjugating) and E3 (ubiquitin ligase) enzymes. E3 ligases regulate ubiquitylation through the specific binding of a substrate to an E2 (Hershko and Ciechanover, 1998). Since E3 ligases specifically interact with a substrate protein and catalyze ubiquitylation of the substrate, E3 ligases are primarily a regulatory factor for the ubiquitin signaling pathway. The cullin-RING ligases (CRLs) are the largest family of E3 ligases (Petroski and Deshaies, 2005). CRLs consist of a cullin scaffold protein, a small RING finger catalytic subunit ROC1 or ROC2 (also known Rbx1 or Rbx2), substrate targeting molecules such as SKP1 and the F-box protein, and regulatory molecules such as Nedd8, CAND1 and COP9 signalosome (Petroski and Deshaies, 2005). The human genome encodes eight cullin proteins (CUL1, 2, 3, 4A, 4B and 5, 7 and PARC/CUL9). All cullin scaffolds interact with the ROC protein to recruit the E2, and each cullin brings its unique substrate targeting machinery to ubiquitylate a large number of specific substrates. Therefore CRLs are thought to regulate many diverse cellular processes, yet most of their substrates and functions are still unknown (Petroski and Deshaies, 2005).
CUL3 targets its substrates through Bric-a-brac-Tramtrack-Broad complex (BTB) domain containing proteins (BTB proteins) (Furukawa et al., 2003; Geyer et al., 2003; Pintard et al., 2003; Xu et al., 2003; Kobayashi et al., 2004; Furukawa and Xiong, 2005). In humans, there are over 300 BTB proteins, each potentially targets a specific substrate for ubiquitylation, yet only a small number of CUL3-BTB protein ligases and their substrates have been identified. Related to cell cycle control, it has been shown that the CUL3-KLHL9/KLHL13, and CUL3-KLHL21 ligases ubiquitylate Aurora-B to control mitotic progression and cytokinesis (Sumara et al., 2007; Maerki et al., 2009), the CUL3-KLHDC5 ligase ubiquitylates p60Katanin for its degradation to regulate microtubules (Cummings et al., 2009), and Cyclin E has been proposed to be ubiquitylated for its degradation in a CUL3-dependent manner (Singer et al., 1999; McEvoy et al., 2007).
Entry and progression of mitosis is tightly regulated by multiple kinases including Aurora-A (Marumoto et al., 2005). Mitotic entry is controlled by the activation of cyclin-dependent kinase 1 (Cdk1), whose activity is regulated directly by mitotic cyclins and phosphatase Cdc25, (Murray, 2004) and indirectly by polo-like kinase 1 (Plk1) (Hansen et al., 2004; Lenart et al., 2007). Plk1, Cdc25, and Cdk1 form a feedback loop and positively regulate each other's activity (Murray, 2004). Aurora-A localizes at the centrosome and activates Plk1 with Bora, leading to the activation of Cdk1 and in turn mitotic entry (Hirota et al., 2003; Macurek et al., 2008; Seki et al., 2008). After initiation of mitosis, Aurora-A accumulates at spindle poles and the mitotic spindle to regulate the mitotic spindle through the phosphorylation of spindle assembly factors (Marumoto et al., 2005).
Here we show that depletion of CUL3 or a BTB protein, KLHL18, causes a delay in mitotic entry and delayed activation of Aurora-A at the centrosome. CUL3 and KLHL18 interact with Aurora-A, and KLHL18 directly binds to Aurora-A. CUL3 and KLHL18 promote Aurora-A ubiquitylation in vivo and the CUL3-KLHL18-ROC1 complex is sufficient to ubiquitylate Aurora-A in vitro. We propose that the CUL3-KLHL18 ligase regulates mitotic entry through Aurora-A ubiquitylation that leads to Aurora-A activation at the centrosome, and the initiation of mitotic entry.
Results
CUL3 functions in mitotic entry
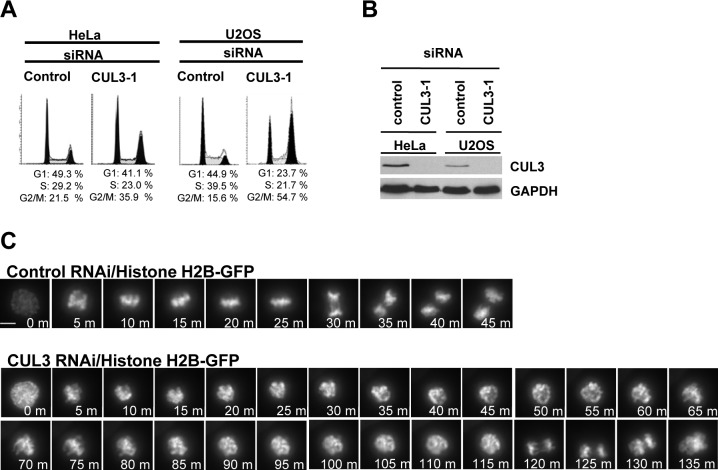
To examine possible cell cycle functions of CUL3 in human cultured cells, we analyzed cell cycle distribution of CUL3 knockdown cells by flow cytometry. The accumulation of cells with 4N DNA content was observed in both HeLa and U2OS cells after knockdown of CUL3 by small interfering RNA (siRNA) transfection, suggesting that CUL3 has mitotic functions (Fig. 1A). We also observed increased number of multinucleated cells and mitotic progression delay in CUL3 depleted cells compared to control cells (data not shown, Fig. 1C) suggesting CUL3 has functions in mitotic progression and cytokinesis, consistent with previous reports (Sumara et al., 2007; Maerki et al., 2009). In addition to the mitotic progression delay and the cytokinesis defect, an accumulation of cells with 4N DNA can also be caused by a delay or defect in mitotic entry. To determine whether CUL3 is involved in mitotic entry, we combined cell synchronization with RNA interference (RNAi). U2OS cells were transfected with siRNAs in the interval between two thymidine blocks and examined after release from the second thymidine block (Fig. 2A). CUL3 protein was almost completely depleted at the release from the 2nd thymidine block (Fig. 2C) and the number of mitotic cells was decreased in CUL3 depleted cells as compared to control cells at 8 hours after release (Fig. 2B). This result indicates that depletion of CUL3 may delay mitotic entry. To further confirm the function of CUL3 in mitotic entry, we monitored the mitotic index of control and CUL3 depleted cells after release from double thymidine block (Fig. 2D). Mitotic entry was significantly delayed in CUL3 depleted cells compared to control cells. These results suggest that CUL3 may regulate mitotic entry.
Fig. 1. CUL3 functions in mitotic progression.
(A) Asynchronous HeLa and U2OS cells were transfected with indicated siRNAs. 72 hours after transfection, cell cycle profile was analyzed by flow cytometry, and (B) cell lysate was analyzed by western blotting (WB) with anti-CUL3 or anti-GAPDH (control). (C) Histone H2B-GFP stably expressing HeLa cells were transfected with either control or CUL3 siRNAs. 48h after transfection, cells were analyzed by time-lapse antibodies fluorescence microscopy. One representative cell for control and CUL3 RNAi is shown from chromosome condensation (0 min). Scale bars, 10 μm.
Fig. 2. CUL3 functions in mitotic entry.
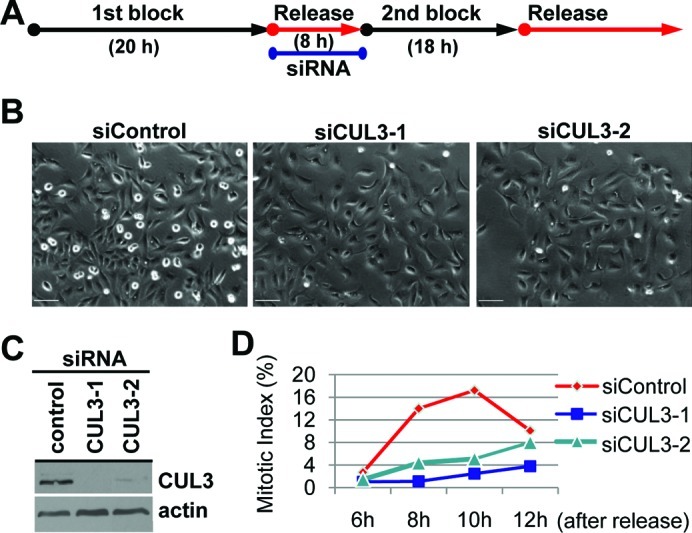
(A) A schematic protocol for cell synchronization (by double thymidine block) and RNAi. (B, C, D) U2OS cells were synchronized and transfected as indicated and subjected to the protocol shown in (A) with indicated siRNAs. (B) Phase pictures of indicated siRNA transfected cells at 8 hours after release. Scale bars, 100 μm. (C) When released from 2nd thymidine block, cells were lysed and subjected to WB with indicated antibodies. (D) Cells were fixed and stained with DAPI at the indicated times after release. Over 500 cells were analyzed and mitotic index was determined.
CUL3 localizes at centrosomes and the mitotic spindle
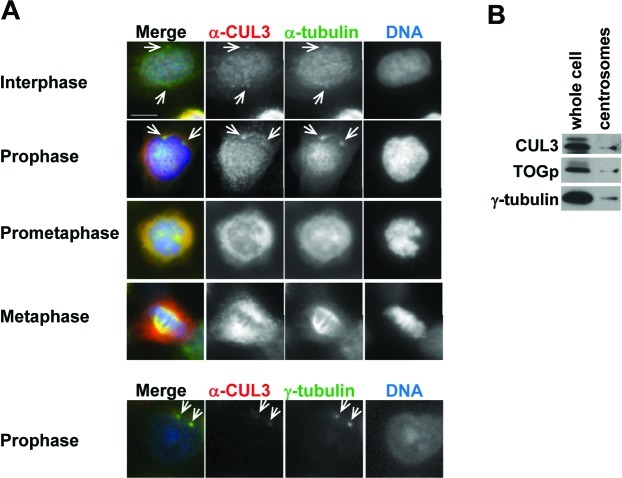
To understand the early mitotic function of CUL3, we determined endogenous CUL3 localization during mitosis in human cultured cells by indirect immunofluorescence microscopy (Fig. 3A). CUL3 localized predominantly in the nucleus in interphase cells. CUL3 was observed at the centrosome at late G2 or prophase, started accumulating at the spindle poles in prometaphase, and stayed on the spindle poles and the mitotic spindle at metaphase (Fig. 3A). To further confirm our observation from the immunofluorescence staining, centrosomes were purified from nocodazole/cytochalasin D-treated U2OS cells by sucrose gradient ultracentrifugation, and subjected to western blotting (Fig. 3B). In agreement with the immunofluorescence data, CUL3 was observed in the centrosomal fraction along with two other known centrosomal proteins: γ-tubulin and TOGp. These results demonstrate that CUL3 localizes and may function at the centrosome, mitotic spindle, and the spindle poles.
Fig. 3. CUL3 localizes at centrosomes and the mitotic spindle.
(A) U2OS cells were fixed with paraformaldehyde (top panels) or methanol (bottom panels) and immunostained with indicated antibodies. DNA was counter-stained by DAPI. Representative pictures of cells in late G2 phase (Interphase), prophase, prometaphase, and metaphase are shown. White arrowheads indicate colocalization of α-tubulin or γ-tubulin and CUL3 in interphase and propahse. Scale bars, 10 μm. (B) Whole cell extract and purified centrosomes from U2OS cells were subjected to WB with indicated antibodies.
Identification of KLHL18 as a mitotic BTB protein
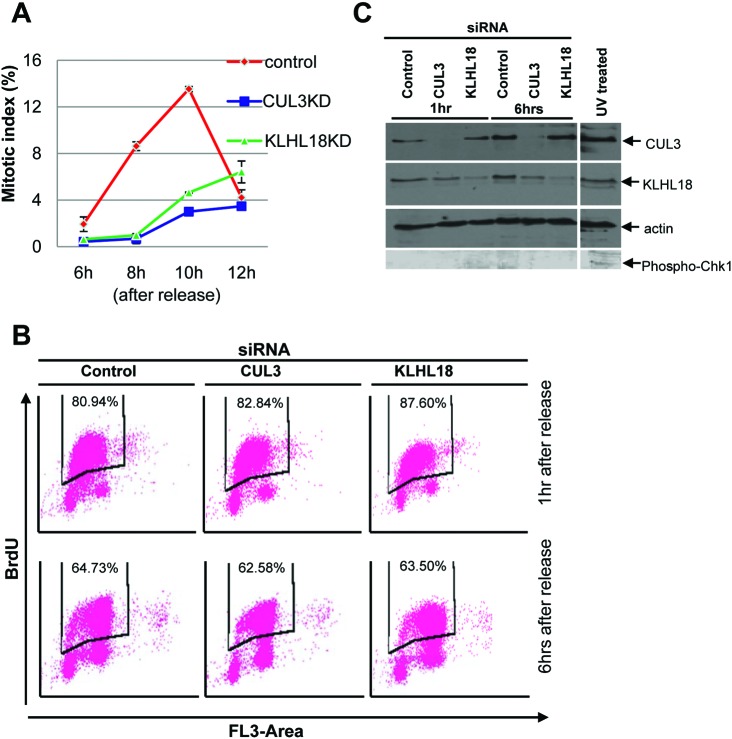
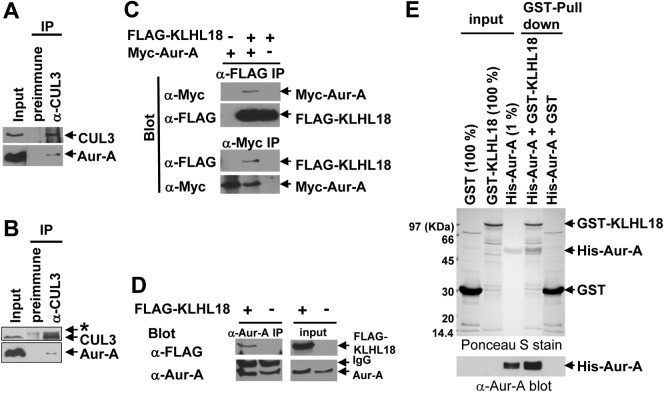
To investigate the role of the CUL3 ligase in mitotic entry, we analyzed the CUL3 complex, especially looking for BTB proteins which could be potential substrate adaptors of CUL3 E3 ligases in early mitosis. We utilized cytostatic factor (CSF)-arrested Xenopus egg extract, which contains all the proteins and complexes present during mitosis in abundance, making it ideal for the study of mitosis. Using a Xenopus CUL3 antibody, we immunoprecipitated CUL3 complexes from the egg extracts. The CUL3 and control pre-immune serum precipitated complexes were resolved by SDS-PAGE and stained with Coomassie blue. The specific protein bands in the CUL3 immunocomplex and corresponding bands in pre-immune serum precipitated complex were analyzed by tandem mass-spectrometry and we found two uncharacterized BTB proteins, KLHL18 and KBTBD2, exclusively in the CUL3 immunocomplex (supplementary material Table S1). Since CSF arrests Xenopus embryos at metaphase, KLHL18 and KBTBD2 are major, if not the only, CUL3 interacting BTB proteins in early mitosis. Therefore, we chose these two proteins for further analysis in early mitotic regulation. We tested mitotic entrance delay in KLHL18 or KBTBD2 depleted synchronized cells and found KLHL18 depletion caused mitotic entrance delay similar to CUL3 depleted cells (Fig. 4A), but KBTBD2 depleted cells did not (data not shown).
Fig. 4. CUL3 and KLHL18 knockdown cells have delayed mitotic entry but progress through S phase normally.
(A) U2OS cells were synchronized and transfected with indicated siRNAs as shown in Fig. 2A. Cells were fixed and stained with DAPI at the indicated times after release. Over 500 cells were analyzed and the mitotic index was determined, data are represented as mean ± SD. (B, C) U2OS cells were synchronized and transfected as indicated in Fig. 2A. (B) At the indicated times following release cells were pulse labeled with 10uM BrdU for 2 hours and stained with anti-BrdU-FITC and 7-AAD, and analyzed by flow cytometry. The percentages of incorporated BrdU are indicated. (C) Lysates from the indicated time points after release were analyzed by WB with indicated antibodies. Lysates from U2OS cells treated with UV were used as a positive control for phospho-Chk1 blot.
To ensure that the mitotic delay that we were observing in CUL3 and KLHL18 knockdown cells was not a consequence of an earlier defect occurring in S phase, we performed a BrdU assay to monitor if S phase was progressing and completing normally in these cells. We BrdU pulsed our synchronized control, CUL3 and KLHL18 knockdown cells at 1 hour and 6 hours after release, followed by flow cytometry analysis to assess BrdU incorporation. At both the early and late time points: control, CUL3 and KLHL18 knockdown cells showed almost the same pattern of BrdU incorporation, indicating that S phase was occurring normally in all the cells (Fig. 4B). Additionally to assess if the reduced mitotic index was due to any DNA damage being caused earlier, we performed a western blot for phospho-Chk1 protein, a marker for DNA damage. Control, CUL3 and KLHL18 knockdown cells showed no signal for the phospho-Chk1 protein, whereas U2OS cells treated with UV showed a phospho-Chk1 signal (Fig. 4C). These results illustrate that CUL3 and KLH18 knockdown cells progress through S phase normally and do not display any DNA damage.
KLHL18 interacts with CUL3
KLHL18 has a BTB domain at its N-terminus, six kelch repeats at its C-terminus, and a BTB and C-terminal Kelch (BACK) domain in between (Fig. 5A). It has been reported that the Drosophila homolog of human KLHL18 forms a complex with CUL3 to target GC5808 and GC10324 for ubiquitylation, and is required for normal thorax development (Fujiyama-Nakamura et al., 2009). Functions of human KLHL18 in cell cycle control are not known. We first determined whether human KLHL18 forms a complex with CUL3 in mitosis. Ectopically expressed FLAG-tagged KLHL18 interacted with endogenous CUL3 in nocodazole arrested mitotic HeLa cells (Fig. 5B).The N-terminal of CUL3 interacts with the BTB domain in a large number of BTB proteins (Furukawa et al., 2003; Xu et al., 2003). We determined the residues important for interaction between CUL3 and KLHL18 (Fig. 5C). As expected, the deletion of N-terminus 41 amino acids of CUL3 (CUL3ΔN41) disrupts interaction with KLHL18, and the deletion of N-terminus 65 amino acids within the BTB domain of KLHL18 (KLHL18ΔBTB) disrupts interaction with CUL3 (Fig. 5C). These data demonstrate that the BTB domain in KLHL18 interacts with the N-terminus of CUL3, similar to known BTB proteins in other CUL3 ligases (Furukawa et al., 2003; Geyer et al., 2003; Furukawa and Xiong, 2005). We also confirmed endogenous CUL3 and KLHL18 interaction in U2OS cells (Fig. 5D). These findings support that KLHL18 forms an E3 ligase with CUL3 and the CUL3-KLHL18 ligase may regulate mitotic entry.
Fig. 5. KLHL18 interacts with CUL3.
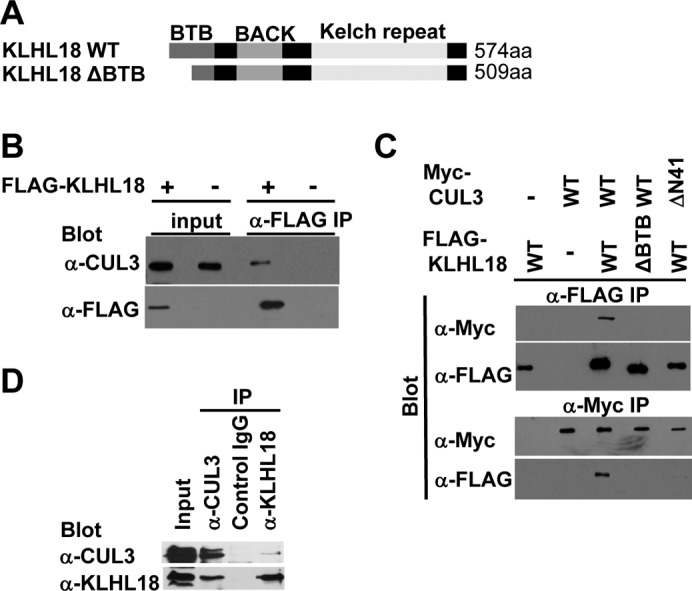
(A) Schematic figure of wild type and mutant of KLHL18. (B) FLAG-tagged KLHL18 expressing plasmids were transfected to HeLa cells and treated with 100 ng/mL Nocodazole for 12 hours before lysis. FLAG KLHL18 and endogenous CUL3 interaction was examined by immunoprecipitation (IP) and WB with indicated antibodies. (C) Wild type and mutant KLHL18 and CUL3 expressing plasmids were transfected to HeLa cells. Interactions were examined by IP-WB analysis with indicated antibodies. (D) Endogenous CUL3 and KLHL18 interaction in U2OS cells was examined by IP-WB with indicated antibodies
The CUL3-KLHL18 ligase is involved in Aurora-A activation at the centrosome
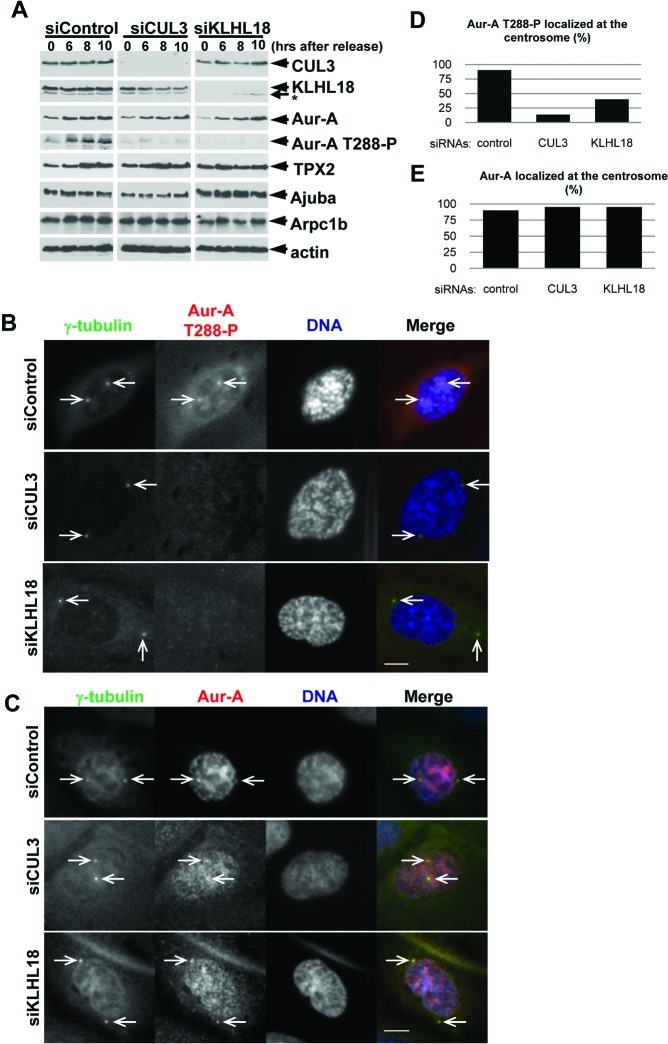
Next, we investigated how the CUL3-KLHL18 ligase controls mitotic entry. Since CUL3 or KLHL18 depletion delays mitotic entry (Fig. 4A) and CUL3 localizes at centrosomes and spindle poles (Fig. 3), the CUL3-KLHL18 ligase may target a centrosomal protein that controls mitotic entry. Aurora-A kinase localizes at the centrosome in interphase and is activated for initiating mitotic entry (Hirota et al., 2003; Macurek et al., 2008; Seki et al., 2008). The phosphorylation of Aurora-A kinase on Thr288, located in the activation T loop of the kinase, markedly increases its enzymatic activity (Walter et al., 2000). Knockdown of Aurora-A in human cultured cells results in mitotic entrance delay that is similar to CUL3 or KLHL18 knockdown (Hirota et al., 2003) (Fig. 4A). To determine the possibility of CUL3-KLHL18 regulating mitotic entry by regulation of Aurora-A, we monitored Aurora-A and Thr288 phosphorylated Aurora-A (Aurora-A T288-P) in control, CUL3, or KLHL18 depleted U2OS cells after release from double thymidine block, by western blotting. In control cells, both total Aurora-A and Aurora-A T288-P expression increased in 6 to 8 hours after release from double thymidine block (Fig. 6A), consistent with previous reports (Hirota et al., 2003; Seki et al., 2008). In CUL3 or KLHL18 depleted cells, Aurora-A protein expression also increased in 6 to 8 hours after release, similar to control cells. However, Aurora-A T288-P expression did not increase by this time (Fig. 6A), suggesting Aurora-A activation was delayed in CUL3 or KLHL18 depleted cells. We also monitored several Aurora-A activators including TPX2, Ajuba, and Arpc1b but the kinetics of the expression of these proteins was not significantly different between control, CUL3 or KLHL18 siRNA transfected cells (Fig. 6A).
Fig. 6. The CUL3-KLHL18 ligase is involved in Aurora-A activation at the centrosomes.
U2OS cells were synchronized and transfected with indicated siRNAs according to the protocol in Fig. 2A. (A) Cells were lysed at the indicated times after release and examined by WB with indicated antibodies. (B, C) Cells were fixed at 10 hours after release and immunostained with indicated antibodies. DNA was counter-stained by DAPI. Representative pictures of cells in prophase are shown. White arrowheads indicate colocalization of γ-tubulin and Aurora-A T288-P or γ-tubulin and Aurora-A. Scale bars, 10 μm. (D, E) Colocalization of γ-tubulin and Aurora-A T288-P (D) and γ-tubulin and Aurora-A (E) in more than 50 of each siRNA transfected prophase cells were quantified.
To further determine the spatiotemporal regulation of Aurora-A by the CUL3-KLHL18 ligase, we analyzed localizations of Aurora-A and Aurora-A T288-P at mitotic entry in control, CUL3 or KLHL18 depleted U2OS cells by immunofluorescence microscopy. Synchronized and siRNA transfected U2OS cells were fixed 10 hours after release form double thymidine block, and Aurora-A T288-P was detected at centrosomes in 90% of control siRNA transfected prophase cells but only in 13% of CUL3 siRNA and 40% of KLHL18 siRNA transfected prophase cells (Fig. 6B,D), suggesting that Aurora-A activation at the centrosome at mitotic entry is delayed in CUL3 or KLHL18 depleted cells. In the same conditions, Aurora-A localized at centrosomes in most of control, CUL3, or KLHL18 siRNA transfected prophase cells (Fig. 6C,E). These results reveal that the CUL3-KLHL18 ligase is required for activation of Aurora-A at centrosomes at mitotic entry.
CUL3 and KLHL18 physically interact with Aurora-A
Since CUL3 or KLHL18 depletion results in the delay of Aurora-A activation at the centrosomes and we observed that CUL3 localized at the centrosomes, we wanted to determine the possibility that CUL3-KLHL18 targets Aurora-A for ubiquitylation. For this we checked if Aurora-A physically interacts with the CUL3-KLHL18 ligase. We immunoprecipitated CUL3 from nocodazole arrested U2OS cells and found CUL3 specifically interacted with Aurora-A (Fig. 7A). We also found CUL3 specifically interacted with Aurora-A in Xenopus as well (Fig. 7B). These results demonstrated that CUL3 physically interacts with Aurora-A in early mitosis in human cells, and these interactions are conserved in Xenopus. Next we examined KLHL18 interaction with Aurora-A. Ectopically expressed FLAG-tagged KLHL18 and Myc-tagged Aurora-A interaction was detected reciprocally (Fig. 7C). Also, endogenous Aurora-A interacted with ectopically expressed FLAG-tagged KLHL18 in 293T cells (Fig. 7D).The direct interaction of KLHL18 and Aurora-A was determined in vitro. Bacterially expressed and purified histidine-tagged Aurora-A and GST-tagged KLHL18 specifically and directly interacted in vitro (Fig. 7E). These results demonstrate that the CUL3-KLHL18 ligase physically interacts with Aurora-A.
Fig. 7. KLHL18 interacts with Aurora-A directly.
(A) U2OS cells treated with 100 ng/mL Nocodazole for 12 hours were lysed and examined by IP-WB with indicated antibodies. (B) CSF-arrested Xenopus egg extracts were examined by IP-WB analysis with indicated antibodies. Asterisk shows non-specific band. (C) FLAG-tagged KLHL18 and Myc-tagged Aurora-A (Myc-Aur-A) expressing plasmids were co-transfected to 293T cells and their interactions were determined by IP-WB analysis with indicated antibodies. (D) FLAG-tagged KLHL18 expressing plasmid was transfected to 293T cells and the FLAG-KLHL18 and endogenous Aurora-A interaction was determined by IP-WB with indicated antibodies. (E) Bacterially expressed and purified GST fused KLHL18 or GST and 6x Histidine fused (His-) Aurora-A were incubated in vitro as indicated and their interactions were determined by GST-pull down assay. The input and GST-pull down reactions were separated by SDS-PAGE, transferred to a nitrocellulose membrane and examined by Ponceau S staining (upper panel). His-Aurora-A was examined by WB with anti-Aurora-A antibody as well (bottom panel).
CUL3-KLHL18 ligase ubiquitylates Aurora-A
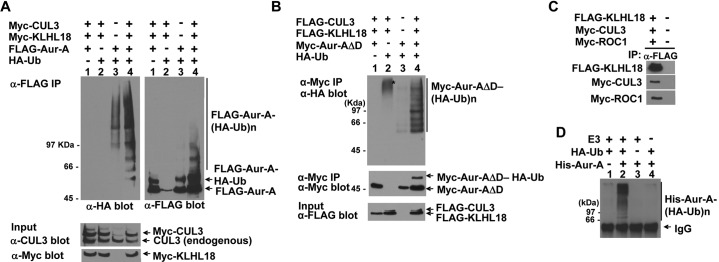
To determine if Aurora-A not only interacts with, but also is a substrate of the CUL3-KLHL18 ligase, we performed an in vivo ubiquitylation assay with ectopically expressed CUL3 and KLHL18. FLAG-tagged Aurora-A and HA-tagged ubiquitin were transfected to 293T cells with or without CUL3 and KLHL18, ubiquitylated Aurora-A was then immunoprecipitated by anti-FLAG antibody, and detected by anti-HA antibody. Overexpression of CUL3 and KLHL18 caused an increase in Aurora-A ubiquitylation (Fig. 8A top panels). Although Aurora-A was ubiquitylated by CUL3-KLHL18, Aurora-A was not stabilized upon the knockdown of CUL3 or KLHL18 indicating that Aurora-A ubiquitylation was not leading to degradation (Fig. 6A). Thus the ubiquitylation of Aurora-A by CUL3-KLHL18 we were observing was not for targeting Aurora-A to the proteasome. Aurora-A is ubiquitylated for degradation by the anaphase promoting complex/Cyclosome (APC/C) at mitotic exit in an N-terminal A box, and C-terminal D box dependent manner (Littlepage and Ruderman, 2002; Crane et al., 2004). To examine Aurora-A ubiquitylation specifically by the CUL3-KLHL18 ligase, and not by APC/C, we constructed a C-terminal D box deletion mutant of Aurora-A (Aurora-A ΔD) for in vivo ligase assays. We confirmed Aurora-A ΔD interacts with KLHL18 by transfection and IP-western blot assay (data not shown). Indeed Aurora-A ΔD ubiquitylation was also stimulated in the presence of CUL3 and/or KLHL18 (Fig. 8B). We also noticed that overexpression of both CUL3 and KLHL18 stimulated mono-ubiquitylation of Aurora-A as well (Fig. 8A,B). To determine if the CUL3-KLHL18-ROC1 complex is sufficient for Aurora-A ubiquitylation, we performed an in vitro ubiquitylation assay with recombinant Aurora-A as a substrate and an immunoprecipitated CUL3-KLHL18-ROC1 complex from 293T cells as a source of E3 ligase (Fig. 8C). Aurora-A was ubiquitylated in vitro when the recombinant Aurora-A was incubated with the CUL3-KLHL18-ROC1 E3 complex in addition to HA-tagged ubiquitin, E1, E2, and ATP (Fig. 8D). This ubiquitylation is CUL3-KLHL18-ROC1 complex dependent (Fig. 8D). Taken together, these results suggest that the CUL3-KLHL18 ligase ubiquitylates Aurora-A, which appears to be important for Aurora-A activation at the centrosome, and proper mitotic entry.
Fig. 8. The CUL3-KLHL18 ligase ubiquitylates Aurora-A in vivo and in vitro.
(A, B) FLAG-tagged Aurora-A (A) or Myc-tagged Aurora-A deleted C-terminal D-box (Aur-AΔD) (B) were cotransfected with Myc-tagged or FLAG-tagged CUL3, Myc-tagged or FLAG-tagged KLHL18, and HA-tagged Ubiquitin expressing plasmids to 293T cells as indicated. Twenty four hours after transfection, cells were lysed with a 1% SDS containing lysis buffer and boiled for 15 min. Denatured lysates were then diluted with 0.5% NP-40 lysis buffer and IP with anti-FLAG (A) or anti-Myc (B) antibody. The washed immunoprecipitates and the total cell lysate (input) were resolved by SDS-PAGE, followed by WB with indicated antibodies. (C) CUL3-KLHL18-ROC1 complexes were prepared from triply transfected 293T cells by IP using anti-FLAG antibody and used as the source of E3 ligase. (D) Bacterially expressed and purified His-tagged Aurora-A (His-Aur-A) (Fig. 6E) was incubated with the KLHL18-CUL3-ROC1 E3 complex (C), E1, E2, HA-tagged ubiquitin (HA-Ub), and ATP. The reactions were immunoprecipitated with anti-HA antibody and blotted with anti-Aurora-A antibody to examine Aurora-A ubiquitylation.
Discussion
In this study, we provide four lines of evidence indicating that the CUL3-KLHL18 ligase regulates mitotic entry by directly targeting Aurora-A for ubiquitylation. First, CUL3 localizes at the centrosome and forms a complex with KLHL18 in mitosis. Second, the depletion of CUL3 or KLHL18 caused a delay in mitotic entry as well as delay in Aurora-A activation at the centrosome. Third, Aurora-A interacts with the CUL3-KLHL18 ligase through direct interaction with KLHL18. Finally, overexpression of CUL3 and KLHL18 promotes Aurora-A ubiquitylation in vivo and the CUL3-KLHL18 ligase can ubiquitylate Aurora-A in vitro. Our data indicate that the CUL3-KLHL18 ligase interacts with, and ubiquitylates Aurora-A prior to the onset of mitosis. This ubiquitylation precedes Aurora-A activation, which also occurs before mitosis, and is required for mediating mitotic entry. Thus, CUL3-KLHL18 may regulate mitotic entry through an Aurora-A pathway.
CUL3 localization at the centrosome and interaction with KLHL18
We have defined a novel CUL3 based E3 ligase, CUL3-KLHL18. The temporal specificity of this E3 ligase at the onset of mitosis, as seen in the CSF arrested egg extract, is indicative of its importance for the regulation of mitotic entry. We also observed that CUL3 localizes at spindle poles and mitotic spindle during mitosis (Fig. 3). Does the CUL3-KLHL18 ligase function after entering mitosis? Aurora-A localizes at spindle poles and the mitotic spindle throughout mitosis and organizes mitotic spindle formation. Depletion or inactivation of Aurora-A causes chromosome misalignment and mitotic spindle disorganization (Marumoto et al., 2003; Marumoto et al., 2005; De Luca et al., 2006).The CUL3-KLHL18 ligase may regulate Aurora-A activity throughout mitosis. Interestingly, we observed chromosome misalignment in KLHL18 or CUL3 depleted asynchronous cells (S. Moghe and M. Furukawa, unpublished observations) suggesting that the CUL3-KLHL18 ligase also functions during mitotic spindle organization. Additionally time lapse studies monitoring the progression of cells throughout mitosis in KLHL18 knockdown cells will be valuable in addressing the function of KLHL18 beyond the early mitotic stages. It can be expected that such studies may yield similarities to what we and others have observed in time lapse studies of CUL3 knockdown cells throughout mitosis (Fig. 1C) (Sumara et al., 2007).
It is also noteworthy that we and others have identified, by mass spectrometry, CUL3 interaction with the mitotic motor protein dynein (supplementary material Table S1) (Bennett et al., 2010) which is associated with the transport of proteins along spindle microtubules towards minus ends (Kardon and Vale, 2009). It is plausible that CUL3 interaction with dynein may be responsible for CUL3 transport to and along microtubules and towards the centrosomes.
Mitotic entry regulated by the CUL3-KLHL18 ligase through Aurora-A
The importance of CUL3-KLHL18 in mitotic entry is consistently evident in our data. Furthermore the activity of Aurora-A affected by CUL3-KLHL18 corresponds to the time of mitotic entry. Our present study suggests that ubiquitylation of Aurora-A, by the CUL3-KLHL18 ligase before mitosis, might serve as a signal for its activation at mitotic entry. Although Aurora-A is abundant at the centrosome in G2 to M phase (Fukuda et al., 2005) (Fig. 6), the majority of Aurora-A only becomes active around the time of prophase. This activation process is not completely understood, but requires Aurora-A interaction with the proteins Ajuba (Hirota et al., 2003; Sabino et al., 2011), HEF1 (Pugacheva and Golemis, 2005), Arpc1b (Molli et al., 2010), Cep192 (Joukov et al., 2010), and TPX2 (Eyers et al., 2003; Tsai et al., 2003). Aurora-A ubiquitylation might promote its interactions with these activators and thus promote Aurora-A activation. Conversely Aurora-A ubiquitylation may also inhibit the interaction of Aurora-A with phosphatases, such as PP1, which inhibit Aurora-A activity (Katayama et al., 2001). Further study is required to elucidate how Aurora-A ubiquitylation promotes Aurora-A activation at the centrosome at mitotic entry.
Non-proteosomal function of the CUL3-KLHL18 ligase
It is apparent from our data that the CUL3-KLHL18 ligase regulates its substrate Aurora-A in a non-proteasomal fashion. Knockdown of CUL3 or KLHL18 did not cause any significant stabilization of Aurora-A (Fig. 6A), and over expression of CUL3 or KLHL18 did not reduce expression of Aurora-A (Fig. 8), consistent with our interpretation that CUL3-KLHL18 ubiquitylation of Aurora-A does not target Aurora-A for degradation. Rather, our data indicates that CUL3-KLHL18 has a novel, proteasome independent function of regulating the activity of Aurora-A. Non-proteasomal functions of CUL3 based E3 ligases have been previously reported in metaphase, mitotic progression and completion of mitosis, for regulating the localization of the Aurora B kinase (Sumara et al., 2007; Maerki et al., 2009). Therefore it is most intriguing that entry into mitosis is also regulated by a CUL3 based E3 ligase; furthermore the substrate in this case is yet another Aurora kinase. Our study assigns an additional role of CUL3 ligases in mitosis, and underscores the importance of CUL3 and BTB proteins in the cell cycle and mitosis.
Materials and Methods
Plasmids, antibodies, and chemicals
Plasmids expressing CUL3 and ROC1 were described previously (Furukawa et al., 2003; Furukawa and Xiong, 2005). Full length and N-terminal 65 amino acid truncated Human KLHL18 (KLHL18 ΔBTB) expression plasmids were cloned into pcDNA3-FLAG vector by PCR amplification from the EST clone using following primers; full length forward, GGATCCAGATGGTGGAGGACGGCGCGGAGGAGCTGGAGGATCTGGTGCACTTCTCCGTGTCTGAGTTGCCTAGTCGCGGCTACGGCGTCATGGAGGAG; full length reverse, CCGCTCGAGTTAGATGGTGAGGAGAGGGATG; ΔBTB forward, CGGGATCCCTATGTTTACAAATGACATG; ΔBTB reverse, same as full length reverse. Human histone H2B-GFP expressing plasmid was obtained from Geoff Wahl (Salk Institute). The obtained clones were verified by DNA sequencing. Antibodies to CUL3 (Furukawa et al., 2003), hemagglutinin (HA) (12CA5, Boehringer-Mannheim), Myc (9E10, NeoMarker), T7 (Novagen), FLAG (M2, Sigma), KLHL18 (17229-1-AP, Protein Tech Group, Chicago, IL), GAPDH (MAB374, Chemicon), TOGp (Cassimeris and Morabito, 2004); gift from Cassimeris, L, Lehigh Univ. PA), α-tubulin (DM1A, Sigma), γ-tubulin (GTU-88 and AK-15, sigma), phospho-Chk1 Ser317 (Cell Signaling), human Aurora-A (610939, BD), Xenopus Aurora-A (Tsai et al., 2003), Aurora-A T288-P (C39D8, Cell Signaling), TPX2 (3164C6a, Santa Cruz), Xenopus TPX2 (Tsai et al., 2003), Ajuba (gift from Keith Johnson, University of Nebraska Medical Center), Arpc1b (IMG-3367, Imgenex, CA), and actin (C-11; Santa Cruz) were previously described or purchased commercially. A rabbit polyclonal antibody against Xenopus CUL3 was raised against a synthetic peptide corresponding to the conserved C-terminal region of Xenopus and human CUL3 (QGESDPERKETRQKVDDDRKHEIE). MG132 (Peptides International, Louisville, Ky.), nocodazole (Sigma), 4',6-diamidino-2-phenylindole (DAPI, Sigma), cytochalasin B (Sigma), Thymidine (Sigma) and cycloheximide (Sigma) were purchased commercially.
Cell culture, transfection, and immunoprecipitation
U2OS, HeLa and 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 unit/ml penicillin (Gibco), and 0.1 mg/ml streptomycin (Gibco) in a 37°C incubator with 5% CO2. Transfections were performed using the Lipofectamine 2000 (Invitrogen) transfection reagent according to the manufacturer's instructions (for HeLa and U2OS cells) or using calcium phosphate buffer (for 293T cells). For UV treated U2OS cell lysates, cells were washed with PBS, dishes were uncovered and exposed to UV irradiation using the germicidal UV lamp in a tissue culture hood. Except where indicated, cells were lysed with Nonidet P-40 (NP-40) lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 50 mM NaF, 0.5% NP-40, 1 mM Na3VO4, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonylfluoride [PMSF], 25 mg/leupeptin, 25 mg/aprotinin, 150 mg/benzamidine, 10 mg/ trypsin inhibitor). For examining Aurora-A T288-P, phosphatase inhibitor cocktail (Cantharidin, Bromotetramisole, and Microcystin LR) and 1 mM EDTA were added to NP-40 lysis buffer described above. All chemicals were purchased from Sigma if not specified.
BrdU assay and cell cycle analysis
For Bromodeoxyuridine (BrdU) labeling, cells were incubated for 2h with a final concentration 10 μM of BrdU, 1h and 6h following release from double thymidine block as shown in Fig. 2A. Cells were stained for BrdU and DNA using the FITC BrdU Flow kit (BD Bioscience, San Jose, CA) according to manufacturers' protocol. Cell cycle analysis was performed by flow cytometry using a BD FACScalibur (BD Bioscience, San Jose, CA) according to the manufacturers' instructions. Analysis of the list mode data was performed using Modfit software (Verity Softwarehouse, Topsham, ME) (Telford et al., 1991).
Isolation of centrosomes
Centrosomes were isolated from U2OS cells by discontinuous gradient ultracentrifugation as described previously (Mitchison and Kirschner, 1986). In brief, cells were treated with cytochalasin B (5 μg/ml) and nocodazole (10 μg/ml) for 90 minutes. After washing, cells were collected with buffer LB (1 mM Tris-Cl, 8 mM beta-mercaptoethanol). After 10 min incubation, Pipes (final 10 mM) and EDTA (final 1 mM) were added to the cells and cells were spun for 3 min at 3000 rpm at 4°C. The supernatant was overlayed onto a Ficoll cushion (20% (w/w) Ficoll (MW 400,000), 0.1% NP-40, 10 mM Pipes, 1 mM EDTA and spun 12,700 rpm for 15 min. at 4°C. The supernatant was aspirated and the interface of the Ficoll cushion was collected, then overlayed onto discontinuous (62.5% and 20% (w/w)) sucrose gradient in 10 mM PIPES, 1 mM EDTA, 8 mM beta-mercaptoethanol. The sample was spun for 90 min at 12,700 rpm at 2°C. The sample was fractionated and the fraction between 48 and 60% (w/w) sucrose was assayed.
RNA interference and cell synchronization
Double-stranded control non-targeting, CUL3 or KLHL18 targeting siRNAs (On-Target plus, Dharmacon) were transfected to U2OS cells by Lipofectamine RNAi Max (Invitrogen) transfection reagent according to the manufacturer's instructions. The target sequences of the siRNAs were as follows: CUL3-1, 5′-GUCGUAGACAGAGGCGCAA; CUL3-2, 5′-GAAGGAAUGUUUAGGGAUA; KLHL18-1, 5′-GGGAUGAGCUGAAUGUCAA; KLHL18-2, 5′-GAGAGAUGCCGUCCCAUGA; KLHL18-3, 5′-GUGCGUUGCUGCCACAAAU; KLHL18-4, 5′-CAGAAGAGAUGGCGAAACG. RNAi experiments in supplementary material Table S1 and Fig. 1, used individual CUL3 siRNAs. Other experiments used a pool of two CUL3 siRNAs for CUL3 depletion or a pool of four KLHL18 siRNAs for KLHL18 depletion.
Xenopus egg extract preparation, immunopurification and mass spectrometric analysis
CSF-arrested Xenopus egg extracts were prepared as described previously (Tsai et al., 2003). CSF-arrested Xenopus egg extracts (300 μl/IP) was pre-cleaned with protein A beads for 30 min at 4°C and cleared by centrifugation. The cleared extract was diluted with 0.1% NP-40 lysis buffer (1:2), then immunoprecipitated by anti-Xenopus CUL3 antibody (20 μg) or pre-immune serum overnight. The reaction was cleared by centrifugation then incubated with protein A beads (20 μl/IP) for 1 hr. CUL3 immunocomplexes were resolved by SDS–PAGE, stained with Coomassie blue, and the protein bands in the CUL3 IP and corresponding gel bands in pre-immune IP were excised and subjected to LC/MS at the University of Nebraska Proteomics Core Facility (Lincoln, NE) as described previously (Kayser et al., 2004). Briefly, gel pieces were digested by trypsin (no.V5111, Promega, Madison, WI) and digested peptides were extracted in 5% formic acid / 50% acetonitrile and separated using C18 reversed phase LC column (75 micron×15 cm, BEH 130, 1.7 micron Waters, Milford, MA). A Q-TOF Ultima tandem mass spectrometer couple with a Nanoaquity HPLC system (Waters) with electrospray ionization was used to analyze the eluting peptides. The system was user controlled employing MassLynx software (v 4.1, Waters) in data-dependant acquisition mode with the following parameters: 0.9-sec survey scan (380–1900 Da) followed by up to three 1.4-sec MS/MS acquisitions (60–1900 Da). The instrument was operated at a mass resolution of 8000. The instrument was calibrated using the fragment ion masses of doubly protonated Glu-fibrinopeptide. The peak lists of MS/MS data were generated using Distiller (Matrix Science, London, UK) using charge state recognition and de-isotoping with the other default parameters for Q-TOF data. Data base searches of the acquired MS/MS spectra were performed using Mascot (Matrix Science, v2.2.0, London, UK). The NCBI database (20100701) was used in the searches. Search parameters used were: no restrictions on protein molecular weight or pI, enzymatic specificity was set to trypsin with up to 3 missed cleavage sites, carbamidomethylation of C was selected as a fixed modification, and methionine oxidaztion was selected as a variable modification. Mass accuracy settings were 0.15 daltons for peptide mass and 0.12 daltons for fragment ion masses.
Immunofluorescence microscopy
For CUL3 immunofluorescence staining, cells were fixed in 3% paraformaldehyde for 10 min followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min and blocking with 10% donkey serum in PBS for 1 hour. For Aurora-A or T288 phosphorylated Aurora-A immunofluorescence staining, cells were fixed with methanol for 10 min at −20°C followed by permeabilization with 1% triton X-100 in TBS for 5 min. After permeabilization, cells were incubated with blocking solution containing 10% donkey serum and 0.1% Triton X-100 in TBS for 1 hour. After blocking, cells were stained with primary antibodies followed by incubation with secondary antibody conjugated with Alexa 488 or Alexa 594 for 1 hour. Cells were counterstained with DAPI for DNA. Images were captured using a 40X oil objective with the Olympus IX81 inverted fluorescence microscope equipped spinning disc confocal system. For time lapse- experiments Histone H2B-GFP stably expressing HeLa cells were transfected with either control or CUL3 siRNAs. 48h after transfection, cells were analyzed by time-lapse microscopy. An Olympus IX81 inverted fluorescence live microscope equipped with a spinning disc confocal system and a humidified CO2 incubator, was used to take snapshots every 5 minutes using an ORCA-ER camera (Hamamatsu Photonics).
GST fusion protein pull-down assay
Full-length human KLHL18 and Aurora-A cDNA were cloned into pGEX and pET-His vectors, respectively. The six-histidine (His6)-tagged fusion proteins were purified with Ni-nitrilotriacetic acid(NTA) agarose (QIAGEN) after induction with 0.4 mM isopropyl-1-ß-D-galactopyranoside (IPTG, Sigma) for 4 h in exponentially growing Escherichia coli BL21(DE3) cells cultured at 30°C. GST or GST fusion proteins were purified with glutathione agarose (Sigma) after induction with 0.4 mM IPTG for 4 h in exponentially growing E. coli XL1-Blue cells cultured at 30°C. One microgram of purified His6-tagged fusion proteins were incubated with 1 μg of purified GST or GST-KLHL18 fusion proteins immobilized on the glutathione agarose in buffer A, which contains 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 120 mM NaCl, 10% glycerol, 0.5% NP-40, 1 mM DTT, 0.5 mM PMSF, and 1 mg of bovine serum albumin. After 1 h incubation at room temperature, the glutathione agarose beads were washed with buffer A three times and resolved by SDS-PAGE, followed by Ponceau S staining or WB with anti-Aurora-A antibody.
Ubiquitin ligase activity assay
The procedures for the in vivo and in vitro ubiquitin ligase activity assays were performed as previously described (Furukawa et al., 2003; Furukawa et al., 2005; Furukawa and Xiong, 2005). Briefly, for in vivo ubiquitylation assays, Myc-tagged Aurora-A ΔD, FLAG-tagged CUL3, FLAG-tagged KLHL18, and HA-tagged Ubiquitin expressing plasmids were transfected to 293T cells as indicated. Twenty four hours after transfection, cells were lysed with a 1% SDS lysis buffer (50 mMTris-HCl, pH 7.5, 0.5 mM EDTA, 1% SDS, 1 mM DTT) and boiled for 15 min to dissociate any protein-protein interaction except ubiquitylation. Lysates were then diluted ten times with 0.5% NP-40 lysis buffer and immunoprecipitated with anti-Myc antibody. The washed immunoprecipitates were resolved by SDS-PAGE, followed by WB with anti-HA antibody to examine ubiquitylated Aurora-A. For in vitro ubiquitin ligase assays, 293T cells were cotransfected with plasmids expressing Myc-tagged CUL3, FLAG-tagged KLHL18, and Myc-tagged ROC1. Twenty-four hours after transfection, cells were lysed and immunoprecipitated with an anti-FLAG antibody. FLAG-KLHL18 immunocomplexes immobilized on anti-FLAG antibody conjugated-agarose beads were incubated with 1 μg of purified His-tagged Aurora-A in a ubiquitin ligation reaction mixture (final volume, 30 μl) containing 50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 1 μg of HA-tagged ubiquitin (Furukawa et al., 2002), 60 ng of E1 (Boston Biochem), and 300 ng of E2-UbcH5c (Furukawa et al., 2002) at 37°C for 1 h. The reaction was terminated by boiling for 15 min in a 1% SDS lysis buffer then diluted ten times with 0.5% NP-40 lysis buffer and IP with anti-Myc antibody. The washed immunoprecipitates were resolved by SDS-PAGE, followed by WB with anti-HA antibody to examine ubiquitylated Aurora-A.
Supplementary Material
Acknowledgments
We thank Ken Kono, Jun Tian and other members of the Furukawa lab and the Tsai lab for discussion throughout this work. We thank Geoff Wahl, Keith Johnson, and Lynne Cassimeris for reagents. This study was supported in part by an NIH grant P20RR018759 to M.F. S.M was supported by fellowships from UNMC and the Nebraska Center for Cellular Signaling.
References
- Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010). Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143, 951–965 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. (2004). TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization and bipolar spindle assembly. Mol. Biol. Cell 15, 1580–1590 10.1091/mbc.E03-07-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane R., Kloepfer A., Ruderman J. V. (2004). Requirements for the destruction of human Aurora-A. J. Cell Sci. 117, 5975–5983 10.1242/jcs.01418 [DOI] [PubMed] [Google Scholar]
- Cummings C. M., Bentley C. A., Perdue S. A., Baas P. W., Singer J. D. (2009). The Cul3/Klhdc5 E3 ligase regulates p60/katanin and is required for normal mitosis in mammalian cells. J. Biol. Chem. 284, 11663–11675 10.1074/jbc.M809374200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Lavia P., Guarguaglini G. (2006). A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle 5, 296–303 10.4161/cc.5.3.2392 [DOI] [PubMed] [Google Scholar]
- Eyers P. A., Erikson E., Chen L. G., Maller J. L. (2003). A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13, 691–697 10.1016/S0960-9822(03)00166-0 [DOI] [PubMed] [Google Scholar]
- Fujiyama-Nakamura S., Ito S., Sawatsubashi S., Yamauchi Y., Suzuki E., Tanabe M., Kimura S., Murata T., Isobe T., Takeyama K. et al. (2009). BTB protein, dKLHL18/CG3571, serves as an adaptor subunit for a dCul3 ubiquitin ligase complex. Genes Cells 14, 965–973 10.1111/j.1365-2443.2009.01323.x [DOI] [PubMed] [Google Scholar]
- Fukuda T., Mishina Y., Walker M. P., DiAugustine R. P. (2005). Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein. Mol. Cell. Biol. 25, 5270–5281 10.1128/MCB.25.12.5270-5281.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., Xiong Y. (2005). BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25, 162–171 10.1128/MCB.25.1.162-171.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., Ohta T., Xiong Y. (2002). Activation of UBC5 ubiquitin-conjugating enzyme by the RING finger of ROC1 and assembly of active ubiquitin ligases by all cullins. J. Biol. Chem. 277, 15758–15765 10.1074/jbc.M108565200 [DOI] [PubMed] [Google Scholar]
- Furukawa M., He Y. J., Borchers C., Xiong Y. (2003). Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001–1007 10.1038/ncb1056 [DOI] [PubMed] [Google Scholar]
- Furukawa M., Andrews P. S., Xiong Y. (2005). Assays for RING family ubiquitin ligases. Methods Mol. Biol. 301, 37–46 10.1385/1-59259-895-1:037 [DOI] [PubMed] [Google Scholar]
- Geyer R., Wee S., Anderson S., Yates J., Wolf D. A. (2003). BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783–790 10.1016/S1097-2765(03)00341-1 [DOI] [PubMed] [Google Scholar]
- Hansen D. V., Loktev A. V., Ban K. H., Jackson P. K. (2004). Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol. Biol. Cell 15, 5623–5634 10.1091/mbc.E04-07-0598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. (2003). Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Joukov V., De Nicolo A., Rodriguez A., Walter J. C., Livingston D. M. (2010). Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl. Acad. Sci. USA 107, 21022–21027 10.1073/pnas.1014664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon J. R., Vale R. D. (2009). Regulators of the cytoplasmic dynein motor. Nature reviews. Mol. Cell. Biol. 10, 854–865 10.1038/nrm2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Zhou H., Li Q., Tatsuka M., Sen S. (2001). Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J. Biol. Chem. 276, 46219–46224 10.1074/jbc.M107540200 [DOI] [PubMed] [Google Scholar]
- Kayser J. P., Vallet J. L., Cerny R. L. (2004). Defining parameters for homology-tolerant database searching. J. Biomol. Tech. 15, 285–295 [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004). Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24, 7130–7139 10.1128/MCB.24.16.7130-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J. J., Hoffmann M., Rettig W. J., Kraut N., Peters J. M. (2007). The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17, 304–315 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Littlepage L. E., Ruderman J. V. (2002). Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16, 2274–2285 10.1101/gad.1007302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L., Lindqvist A., Lim D., Lampson M. A., Klompmaker R., Freire R., Clouin C., Taylor S. S., Yaffe M. B., Medema R. H. (2008). Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 10.1038/nature07185 [DOI] [PubMed] [Google Scholar]
- Maerki S., Olma M. H., Staubli T., Steigemann P., Gerlich D. W., Quadroni M., Sumara I., Peter M. (2009). The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J. Cell Biol. 187, 791–800 10.1083/jcb.200906117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marumoto T., Honda S., Hara T., Nitta M., Hirota T., Kohmura E., Saya H. (2003). Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J. Biol. Chem. 278, 51786–51795 10.1074/jbc.M306275200 [DOI] [PubMed] [Google Scholar]
- Marumoto T., Zhang D., Saya H. (2005). Aurora-A-a guardian of poles. Nat. Rev. Cancer 5, 42–50 10.1038/nrc1526 [DOI] [PubMed] [Google Scholar]
- McEvoy J. D., Kossatz U., Malek N., Singer J. D. (2007). Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol. Cell. Biol. 27, 3651–3666 10.1128/MCB.00720-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Kirschner M. W. (1986). Isolation of mammalian centrosomes. Meth. Enzymol. 134, 261–268 10.1016/0076-6879(86)34094-1 [DOI] [PubMed] [Google Scholar]
- Molli P. R., Li D. Q., Bagheri-Yarmand R., Pakala S. B., Katayama H., Sen S., Iyer J., Chernoff J., Tsai M. Y., Nair S. S. et al. (2010). Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora A. J. Cell Biol. 190, 101–114 10.1083/jcb.200908050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W. (2004). Recycling the cell cycle: cyclins revisited. Cell 116, 221–234 10.1016/S0092-8674(03)01080-8 [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Pintard L., Willis J. H., Willems A., Johnson J. L., Srayko M., Kurz T., Glaser S., Mains P. E., Tyers M., Bowerman B. et al. (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316 10.1038/nature01959 [DOI] [PubMed] [Google Scholar]
- Pugacheva E. N., Golemis E. A. (2005). The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat. Cell Biol. 7, 937–946 10.1038/ncb1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino D., Brown N. H., Basto R. (2011). Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J. Cell Sci. 124, 1156–1166 10.1242/jcs.076711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A., Coppinger J. A., Jang C. Y., Yates J. R., Fang G. (2008). Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science 320, 1655–1658 10.1126/science.1157425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J. D., Gurian-West M., Clurman B., Roberts J. M. (1999). Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13, 2375–2387 10.1101/gad.13.18.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I., Quadroni M., Frei C., Olma M. H., Sumara G., Ricci R., Peter M. (2007). A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev. Cell 12, 887–900 10.1016/j.devcel.2007.03.019 [DOI] [PubMed] [Google Scholar]
- Telford W. G., King L. E., Fraker P. J. (1991). Evaluation of glucocorticoid-induced DNA fragmentation in mouse thymocytes by flow cytometry. Cell Prolif. 24, 447–459 10.1111/j.1365-2184.1991.tb01173.x [DOI] [PubMed] [Google Scholar]
- Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. (2003). A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5, 242–248 10.1038/ncb936 [DOI] [PubMed] [Google Scholar]
- Walter A. O., Seghezzi W., Korver W., Sheung J., Lees E. (2000). The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 19, 4906–4916 10.1038/sj.onc.1203847 [DOI] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T. H., Vidal M., Elledge S. J., Harper J. W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321 10.1038/nature01985 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.