Summary
During the last phase of spermatogenesis, spermiogenesis, haploid round spermatids metamorphose towards spermatozoa. Extensive cytoplasmic reduction and chromatin remodelling together allow a dramatic decrease of cellular, notably nuclear volume. DNA packing by a nucleosome based chromatin structure is largely replaced by a protamine based one. At the cytoplasmic level among others the acrosome and perinuclear theca (PNT) are formed. In this study we describe the onset of chromatin remodelling to occur concomitantly with acrosome and PNT development. In spread human round spermatid nuclei, we show development of a DAPI-intense doughnut-like structure co-localizing with the acrosomal sac and sub acrosomal PNT. At this structure we observe the first gradual decrease of nucleosomes and several histones. Histone post-translational modifications linked to chromatin remodelling such as H4K8ac and H4K16ac also delineate the doughnut, that is furthermore marked by H3K9me2. During the capping phase of acrosome development, the size of the doughnut-like chromatin domain increases, and this area often is marked by uniform nucleosome loss and the first appearance of transition protein 2 and protamine 1. In the acrosome phase at nuclear elongation, chromatin remodelling follows the downward movement of the marginal ring of the acrosome. Our results indicate that acrosome development and chromatin remodelling are interacting processes. In the discussion we relate chromatin remodelling to the available data on the nuclear envelope and the linker of nucleoskeleton and cytoskeleton (LINC) complex of spermatids, suggesting a signalling route for triggering chromatin remodelling.
Keywords: Spermiogenesis, Chromatin remodelling, Human
Introduction
Mammalian spermatogenesis, or the production of spermatozoa, is a complex and lengthy process taking around 74 days in humans (Heller and Clermont, 1964). It is composed of the mitotic phase of spermatogonial multiplication, meiosis, and a haploid phase called spermiogenesis which comprises one third of the whole process. During spermiogenesis, haploid round spermatids gradually metamorphose towards spermatozoa. In human histological preparations and based on nuclear morphology, Clermont (Clermont, 1963) could identify six steps (Sa-Sb1-Sb2-Sc-Sd1-Sd2). In rodents, steps are defined on the basis of periodic acid Schiff staining of the developing acrosome, which is divided in the Golgi phase, capping phase, acrosome phase and maturation phase (Leblond and Clermont, 1952). In the mouse, 16 steps are distinguished (Oakberg, 1956). The steps of spermatid morphogenesis have fixed relations with the stages of the cycle of the seminiferous epithelium, which for mammals including human are denoted by Roman symbols (Clermont, 1963; Oakberg, 1956). The end product of spermiogenesis, the spermatozoon, is characterized by an extremely condensed nucleus necessary among other reasons to protect the genetic material during its journey to the oocyte (Oliva, 2006). This metamorphosis requires extensive cytoplasmic and nuclear chromatin remodelling, processes that have been studied in most detail in the mouse. However, they contain major riddles such as the induction and mechanisms of grand scale chromatin dynamics.
At the nuclear level the archetypical DNA packaging protein complexes, the nucleosomes, are largely replaced by relatively small basic arginine and cysteine rich protamines in a two step process: eviction of nucleosomes/shedding of histones is first followed by the incorporation of transition proteins that subsequently are replaced by the even more basic protamines, to finally ensure extensive nuclear condensation (for reviews, see Balhorn, 2007; Oliva, 2006; Rousseaux et al., 2008). Hence, not all nucleosomes are evicted. In mice, one percent of the DNA keeps its nucleosomes (van der Heijden et al., 2005) and for humans this estimate is higher (10–15%) (Brykczynska et al., 2010; Gatewood et al., 1987).
The actual nucleosome removal process is prepared by incorporation of testicular and haplophase specific histone variants (Boussouar et al., 2008; Rousseaux et al., 2008) in conjuncture with histone post-translational modifications (PTMs). Of these PTMs H4 acetylation and H2A ubiquitination (Baarends et al., 1999; Grimes and Henderson, 1984; Meistrich et al., 1992) are the best known. Nucleosome eviction necessitates release of torsional stress (supercoiling) which is removed by the formation of DNA strand breaks in which process TopoIIβ is involved (mouse) (Leduc et al., 2008). In the mouse, histone 4 hyperacetylation starts at step 8, around the start of nuclear orientation (the acrosomal side moves towards the cell membrane and this side orientates to the tubular basal lamina) and just before nuclear elongation (Hazzouri et al., 2000). DNA strand breaks have been observed in all elongating spermatids at step 9 and decreased during steps 10 and 11 (Marcon and Boissonneault, 2004). By immunofluorescence (IF), transition proteins in mouse have first been observed on histological sections at steps 10, 11 (Zhao et al., 2004) followed by protamine 1 (P1) at step 11 and protamine 2 (P2) at step 12 (Zhao et al., 2004) (faint signals were found one step before). In humans, H4 acetylation is already observed at the round spermatid steps Sa–Sb1 (stages I–III) (Sonnack et al., 2002). DNA strand breaks have been demonstrated at step Sb1 (stage III) (Marcon and Boissonneault, 2004). Transition protein 1 (TP1) is present at steps Sb1–Sb2 (stage III–IV) and TP2 is present from step Sa–Sc (stage I–V) (Dadoune, 2003; Steger et al., 1998). P1 and P2 are first observed in steps Sb2–Sc (Stage IV/V) of human spermiogenesis (Dadoune, 2003; Prigent et al., 1996).
P2 is transcribed as a pro form leading to pre-protamine 2 (pre-P2), that has to be proteolytically processed (for a review, see de Mateo et al., 2011a). Protamine incorporation requires a cycle of phosphorylation and dephosphorylation (reviewed by Dadoune, 2003). By in vitro radioactive labelling of protamines in mouse and rat seminiferous tubules, it has been determined that the cycle of phosphorylation and dephosphorylation occurs only after proteolytic processing of pre-P2 has been started. This suggests that unprocessed P2 is not complexed with DNA (Green et al., 1994).
During spermiogenesis, several cytoplasmic structures are formed that are in close contact with, and involved in shaping of, the nucleus (hence the sperm head) such as the acrosome (Hermo et al., 2010), the perinuclear theca (PNT) (Oko and Sutovsky, 2009) including the marginal ring and the manchette with the perinuclear ring (Kierszenbaum et al., 2007). The acrosome is a Golgi derived sac-like structure attached to the nucleus around the apical site. The PNT is an electron dense layer which almost completely surrounds the sperm nucleus except for the tail implant region (Oko and Sutovsky, 2009). Two structural and compositional different regions are present in the PNT: the sub-acrosomal layer (SAL) and post-acrosomal sheet (PAS) (Oko and Sutovsky, 2009). The SAL, also known as the acroplaxome (Kierszenbaum et al., 2003), is formed first, induced by acrosome-nuclear docking (Oko and Sutovsky, 2009) and is described as an ‘F-actin/keratin-5 containing cytoskeletal plate’ (Kierszenbaum et al., 2007). The PAS is formed in concert with a temporal cytoskeletal microtubule network structure, the manchette (Oko and Sutovsky, 2009).
Summarizing, sperm head shaping and nuclear condensation are tightly regulated processes involving proper chromatin remodelling and accurate assembly of cytoskeletal (manchette) and cytoplasmic (acrosome) structures as is demonstrated by the many mouse mutants that affect this process and hence fertility (Hermo et al., 2010; Kierszenbaum et al., 2007).
By electron microscopy it has since long been known that in mammals, nuclear condensation takes place following a gradient from cranial to caudal (Courtens et al., 1995; Courtens and Loir, 1981; Dooher and Bennett, 1973). The evidence in part has been obtained by lysine PTA staining, which is nearly absent in protamines compared to histones and TPs. In the human, the study of chromatin remodelling during spermiogenesis is not advancing quickly, which in part is due to the ethical aspects of acquiring experimental material and the absence of an in vitro system. Another aspect is that human spermatogenesis stands out as variable as to sperm production, head morphology, and motility. These aspects of variation are increased in the so called oligo-astheno-teratozoospermia syndrome (OAT) that affects 1 in 4 infertile men (Dohle et al., 2005). Male sub and infertility can be assessed at 7% of the male population trying to reproduce (Irvine, 1998), and has among others been associated with incomplete chromatin remodelling towards the protamine dominated state (de Mateo et al., 2011b; Ramos et al., 2008).
Due to developments in the treatment of male fertility (TESE-ICSI: the injection of a testicular derived spermatozoon in the oocyte), testicular biopsies are now taken on a regular basis. In many of these, a distinctive pathology in the focal spermatogenesis regions is not apparent (Steger et al., 2001). Also men with a failed vaso-vasostomy can be helped by this form of artificial reproduction technique. We have used remnants of testicular biopsies to describe chromatin remodelling in human spermatids in relation to changes in shape and structure of the nucleus. This IF based analysis has mainly been executed on spermatid nuclei spread in one focal plane. By this approach, we have been able to describe chromatin remodelling as a downstream consequence of landing of the acrosomal vesicle and granule, via the SAL-PNT/acroplaxome, on the apical surface of the nucleus. From here, a gradient as to nucleosome and histone loss could be constructed. We analysed this process using of a number of histones and histone PTMs and included pre-P2 to P2 processing.
Results
Appearance of a doughnut-like structure during spermiogenesis
The DAPI characteristics of human round spermatids after nucleus spreading have previously been established using X, Y DNA FISH for identification as only one signal is observed (de Vries et al., 2012; data not shown). A DAPI-intense doughnut-like structure was frequently observed (Fig. 1A). Several sizes were found, next to round spermatids without one, indicating a gradually developing/growing structure in which we defined four types (Fig. 1A). We also observed nuclei in which the DAPI-intense structure was located at one side of the nucleus losing the DAPI negative centre. These nuclei had a less regularly curved nuclear outline (Fig. 2A) (Type 5). We interpreted these nuclei as the transition towards the elongating spermatid stage. At this time point, a DAPI-intense cap-like structure is noticed at the apical site of the nuclei that moves towards the implantation fossa of the tail which often can be observed during nucleus elongation and condensation (the tail is usually lost in the spreading procedure) (Fig. 2A). Elongating spermatids were classified (based on nuclear size and distribution of DAPI intensity) into four types as well (Fig. 2A). From type 7 on, a sharp boundary of the cap-like structure could often not be observed anymore, as the DAPI intensity was more evenly distributed within the nucleus. In type 8 elongating spermatids the apical side is lighter, while the basal/tail side is DAPI bright. Integrating these observations, the DAPI demarcated cap-like structure is the continuation of the DAPI-intense doughnut-like structure. These morphologies were encountered in all probands (supplementary material Table S1). In this report, we present an analysis of the pooled images over probands.
Fig. 1. Round spermatid nuclei in succeeding steps of human spermiogenesis.
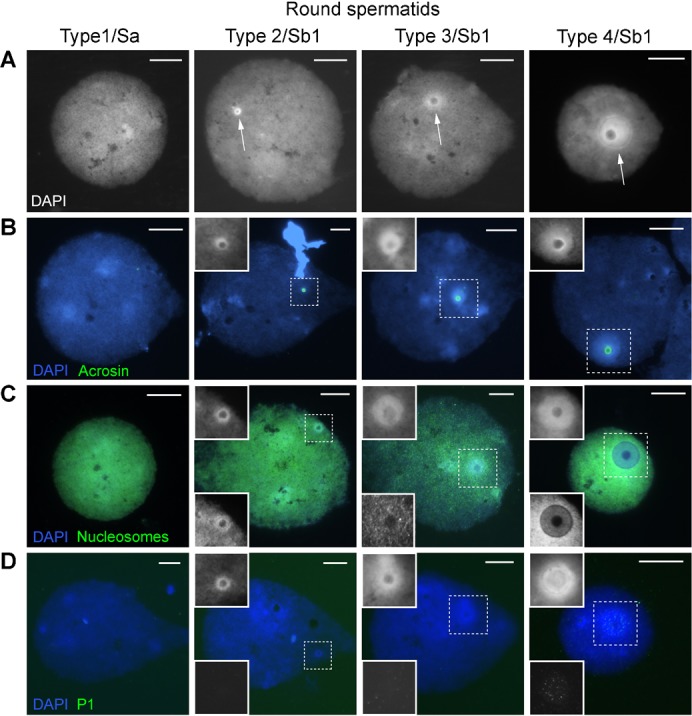
Types 1–4 have tentatively been assigned to the spermatid morphology nomenclature of Clermont (Clermont, 1963). (A) Gradual development of a DAPI-intense doughnut-like structure (arrows). (B) Localization of acrosin IF staining; the inset (dotted square) shows the DAPI image. (C) Nucleosome staining (ab #32); the upper inset shows the DAPI image and the lower inset the nucleosome staining. (D) P1 staining; the upper inset shows the DAPI image and the lower inset P1 staining. Scale bar: 10 µm.
Fig. 2. Elongating spermatid nuclei in succeeding steps of human spermiogenesis.
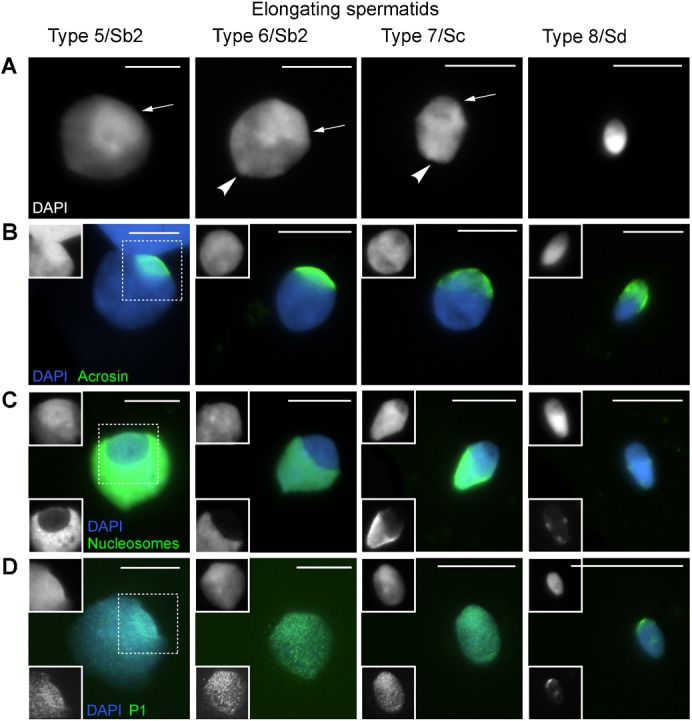
Types 5–8 have tentatively been assigned to the spermatid morphology nomenclature of Clermont (Clermont, 1963). (A) Gradual development of an initially DAPI-intense cap-like structure (arrows) with the implantation fossa indicating the nuclear posterior pole (arrow head). (B) Localization of acrosin IF staining; the inset (dotted square) shows the DAPI image. (C) Nucleosome staining (ab #32); the upper inset shows the DAPI image and the lower image the nucleosome staining. (D) P1 staining; the upper inset shows the DAPI image and the lower inset P1 staining. Scale bar: 10 µm.
Relation between the doughnut-like structure and the acrosome
The doughnut- and cap-like structure remind of the developing acrosome. We therefore stained with acrosin, a proven acrosome marker (Hermo et al., 2010) to investigate co-localisation. Fig. 1B and Fig. 2B show overlap between the acrosome and the two DAPI defined structures, demonstrating the cap-like one to evolve from the doughnut. Next to acrosin we stained for the subacrosomal part of the PNT (SAL-PNT/acroplaxome), the cytoskeletal plate attaching the acrosome to the nucleus (Fig. 3H). Supplementary material Fig. S1 clearly shows overlap between the acrosome, the SAL-PNT/acroplaxome and the doughnut-/cap-like structure. The SAL-PNT/acroplaxome covers the complete DAPI-intense doughnut-/cap-like region in contrast with the acrosomal sac stained by acrosin.
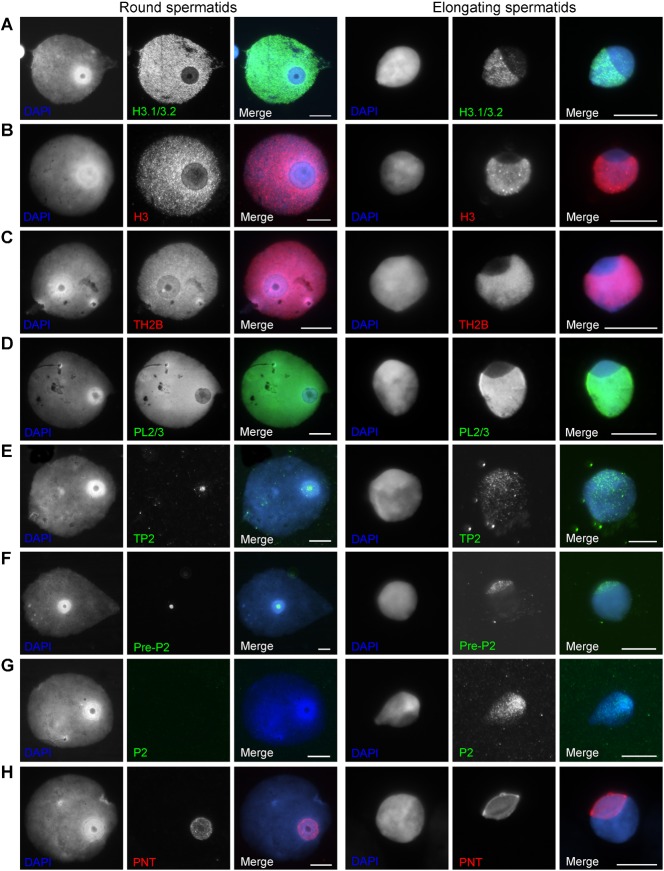
Fig. 3. Examples of round spermatid (type 4) and elongating spermatid (type 5/6) nuclei.
(A) H3.1/3.2 staining. (B) panH3 staining. (C) TH2B staining. (D) PL2/3 nucleosome staining. (E) TP2 staining. (F) Pre-P2 staining. (G) P2 staining. (H) SAL-PNT/acroplaxome staining. Scale bar: 10 µm.
Nucleosome and core histone removal at the doughnut/cap-like structure
Staining with a nucleosome specific antibody (monoclonal #32) revealed a gradual decrease of nucleosomes in the doughnut- and cap-like structure starting from type 2–3 on (Figs 1C, 2C). IF performed with another marker for nucleosomes (monoclonal PL2/3) and antibodies detecting H3.1/3.2, panH3 and TH2B (core histones) show the same decrease in signal intensity at the doughnut- and cap-like structure (Fig. 3A–D). In supplementary material Fig. S2A we have attempted to approximate the relative intensity of H3.1/3.2 staining at the doughnut in the subsequent types of round spermatids (n = 1085). In type 4 round spermatids (Fig. 1), with the largest doughnut-like structures, we observed 3 consecutive appearances of the doughnut (supplementary material Fig. S2B,C). A gradual decrease in H3.1/3.2 staining is observed over the successive doughnut types enforcing the validity of this sequence in time. These data also indicate the process of initiation of nucleosome loss to be variable in human round spermatid development.
In order to determine conservation of staining patterns between species we probed mouse round and elongating spermatids for nucleosomes and PNT. The DAPI-intense doughnut- (supplementary material Fig. S3A) and cap-like structure as identified by PNT (supplementary material Fig. S3B) were abundantly observed in mouse spermatids as well and often showed decreased intensity of nucleosomes.
Appearance of basic nuclear proteins
We observed TP2 first in doughnuts of type 4.1 round spermatids (Fig. 3E; supplementary material Fig. S2). Clear P1 signals were detected in doughnuts of type 4.2/4.3 round spermatids (Fig. 1D; supplementary material Fig. S2). In these types, P1 was sometimes also detected elsewhere in the nucleus without a signal in the doughnut-like structure. P2 was not observed in round spermatids but only in elongating spermatids from type 5 on. Staining patterns suggested localisation in the doughnut/cap area first (Fig. 3G; data not shown). Pre-P2 was observed in the doughnut-like structure from Type 2 on (Fig. 3F; data not shown). The localisations of TP2, pre-P2 and P1/2 (Figs 1-3) indicate that TP and P1 and P2 preferentially spread from the doughnut-/cap-like structure into the rest of the nucleus while pre-P2 is only present in the doughnut-/cap-like structure and never elsewhere.
The doughnut like structure in cryosections
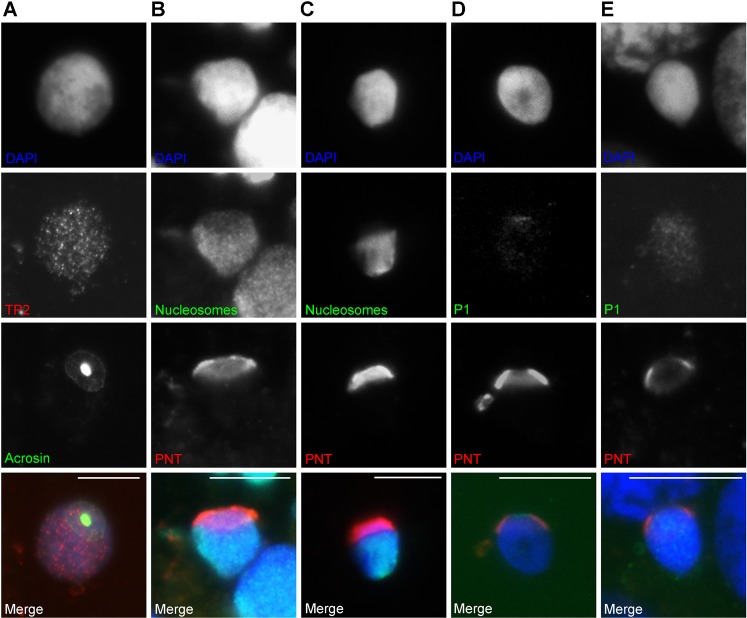
In nucleus spread preparations nuclear morphology is not original anymore. Resistant chromatin domains will be accentuated. The nuclei expand in size, but become restricted in this respect from the late round spermatid stage on. Therefore we used cryosections to confirm our results on polarity of nucleosome eviction in the capping phase of acrosome formation. Fig. 4 shows that in cryosections the doughnut- and cap-like structure could not be distinguished by DAPI. However, acrosin and SAL-PNT staining detect the apical pole of the nucleus (Fig. 4A–E). SAL-PNT/acroplaxome and acrosin gave images reminiscent to the structures found in alkaline nuclear spreads indicating that these withstand the nuclear spreading procedure. Fewer nucleosomes were found to be present at the apical side of the round/elongating spermatids similar to the nucleus spread preparations (Fig. 4B,C). P1 first appeared as a faint signal, enriched at the top of the round to elongating spermatid nucleus (Fig. 4D). This did not apply to all nuclei (of this one patient), i.e. the reverse pattern, P1 coming up over the remaining nucleus and not under the acrosome, reminiscent of EM immunolocalisations in mouse (Biggiogera et al., 1992), was also found (not shown). In slightly more condensed nuclei a P1 spread was seen throughout the nucleus (Fig. 4E). From these results we can conclude that morphologically similar representations of the SAL-PNT/acroplaxome and acrosome are present in cryosections and in alkaline spread nuclei, and that nucleosome loss follows these structures.
Fig. 4. Demonstration of spermatid nucleus polarity in cryosectioned round/elongating spermatid nuclei.
(A) The doughnut-/cap-like structure is represented by acrosin staining. TP2 is visible throughout the nucleus. (B) The doughnut-/cap-like structure is represented by SAL-PNT/acroplaxome staining; fewer nucleosomes (ab #32) are present at the SAL-PNT stained region. (C) The doughnut-/cap-like structure is represented by SAL-PNT staining; fewer nucleosomes (ab #32) are present at the SAL-PNT stained region. (D) The doughnut-/cap-like structure is represented by SAL-PNT staining; a small P1 signal is observed at the apical, SAL-PNT stained, region of the nucleus. (E) The doughnut-/cap-like structure is represented by SAL-PNT staining; P1 staining has advanced over the nucleus. Scale bar: 10 µm.
Histone acetylation marker characteristics of the doughnut- and cap-like structure
The first histone PTMs to be associated with nucleosome to protamine chromatin remodelling were visualised by using a panH4 acetyl antibody (recognising acetylation of H4 on K5,K8,K12 and K16) (Hazzouri et al., 2000). Here we have tested H4K8ac and H4K16ac with respect to staining characteristics of the doughnut-/cap-like structure, the latter recently implicated in the action of RNF8 to induce chromatin remodelling (Lu et al., 2010) and both giving a distinct signal in elongating mouse spermatids (van der Heijden et al., 2006).
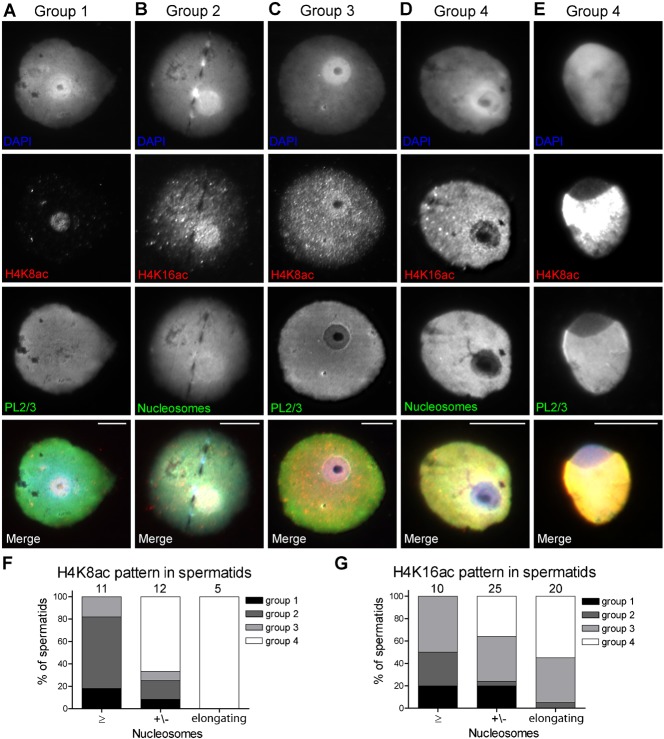
In round spermatids we could distinguish 4 staining patterns for both H4 acetylation markers: (1) staining exclusively at the doughnut; (2) staining throughout the nucleus but more intense at the doughnut; (3) even staining throughout the nucleus, and (4) staining throughout the nucleus with decreasing intensity at the doughnut (Fig. 5A–D). Although numbers of studied nuclei were low (H4K8ac n = 23, H4K16ac n = 35), we observed a trend towards pattern 4 during nucleosome removal (Fig. 5F,G). At the elongating spermatid stage, this pattern is the dominant one (Fig. 5E–G). These observations confirm that H4 acetylation is an early signal for nucleosome eviction (Sonnack et al., 2002), and also agree with the onset of nucleosome loss in the doughnut region of the nucleus.
Fig. 5. H4K8/16ac staining in round spermatids.
(A–E) Round (A–D) and elongating (E) spermatids representing the 4 groups of H4K8/16ac staining patterns as explained in the results. (F,G) Graphs displaying the relationship between H4K8/16ac staining patterns (bars) and the degree of nucleosome decrease (F: ab PL2/3, G: ab #32) on the doughnut in round spermatids and the cap-like structure of elongating spermatids (X-axis). Scale bar: 10 µm.
Chromatin characteristics of the doughnut-/cap-like structure
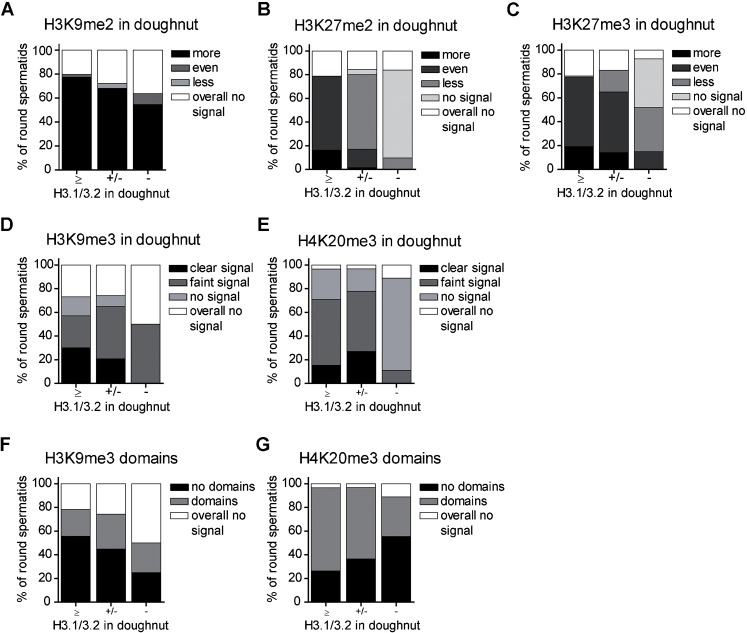
We showed loss of H3.1/3.2 (Fig. 3; supplementary material Fig. S2A) and other histone/nucleosome markers (Figs 1–3) during doughnut development/progression of spermiogenesis. Therefore, we asked ourselves whether this loss is equally represented by staining patterns for a number of histone 3 and 4 PTMs. Alternatively, if such a mark is enriched this may indicate implication in remodelling. Histone PTM markers for constitutive heterochromatin (H3K9me3 and H4K20me3) and euchromatic heterochromatin (H3K9me2 and H3K27me2/3) were visualised in spermiogenic nuclei together with H3.1/3.2 as a marker for doughnut nucleosome eviction (Figs 6–8). For all marks, a downward trend over increasing nucleosome eviction could be noticed (Fig. 6) (for significances see the legend). H3K27me2/3 followed most closely the loss of H3.1/3.2 (Fig. 6B,C, Fig. 7C,E). H3K9me2 was most conspicuously present at the doughnut, a pattern presented by up to 80% of nuclei and often this mark expanded into adjacent chromatin (Fig. 6A, Fig. 7A). H3K9me3 and H4K20me3 staining showed several domains of various size throughout the nucleus representing centric heterochromatin, as was determined by co-localisation of these domains with a centromere specific antiserum (Crest, Fig. 8E–H). H3K9me3 and H4K20me3 could show specific staining at the doughnut like structure as well, the signal decreasing together with H3.1/3.2 loss (Fig. 6D,E, Fig. 8A,C). The centric heterochromatin domains were regularly found to be associated with the doughnut like structure (Fig. 6F,G, Fig. 8A,C,E,G) which was more obvious for H4K20me3. Association of these domains with the doughnut decreased together with H3.1/3.2 loss (Fig. 6F,G).
Fig. 6. Frequency distributions of H3, H4 PTM stainings in round spermatids.
(A–C) Inactive chromatin marker characteristics at the doughnut-like structure (bars), compared to the rest of the round spermatid nucleus and in relation to the degree of H3.1/3.2 loss in the doughnut (X-axis). ‘Overall no signal’ applies to the whole nucleus. (D,E) Heterochromatin marker characteristics at the doughnut-like structure (bars) in relation to the degree of H3.1/3.2 loss at the doughnut (X-axis). (F,G) The location of heterochromatin domains near the doughnut in relation to loss of H3.1/3.2 in the doughnut (X-axis). (A–G) Chi-square analysis was performed to test for homogeneity of histone PTM scoring classes over H3.1/3.2 loss. H3.1/3.2 categories +/− and − indicating loss, were joined. (A) PTM categories even, less and overall no signal were joined χ2 = 2.6 df 1, p = 0.11 (ns) n = 147. (B) χ2 = 107 df 4, p<0.001 n = 181. (C) χ2 = 31 df 4, p<0.001 n = 176. (D) χ2 = 7.6 df 3, p = 0.056 (ns) n = 147. (E) PTM categories no signal and overall no signal were joined χ2 = 2.5 df 2, p = 0.29 n = 189. (F) χ2 = 2.7 df 2, p = 0.26 n = 197. (G) χ2 = 3.4 df 2, p = 0.18 n = 189.
Fig. 7. Staining patterns for H3 post translational methylations in round and early elongating spermatids.
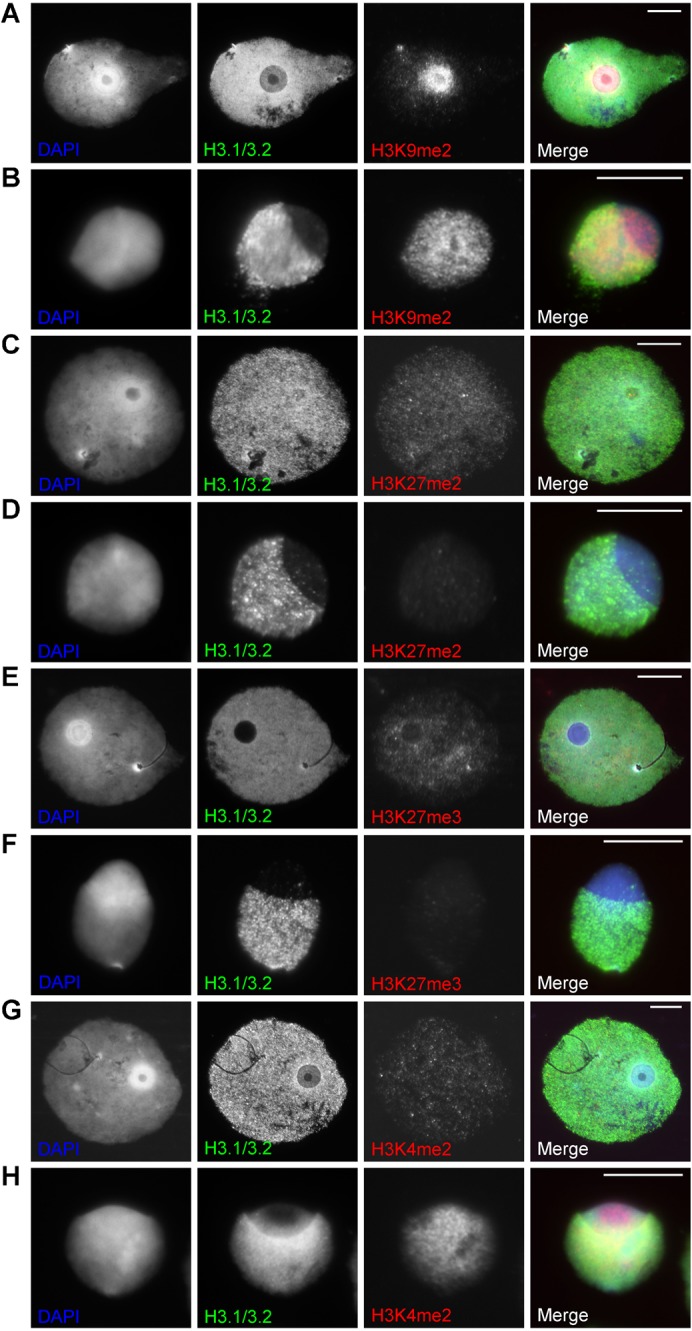
(A,B) H3K9me2 and H3.1/3.2 staining in a round (A) and elongating (B) spermatid. (C,D) H3K27me2 and H3.1/3.2 staining in a round (C) and elongating (D) spermatid. (E,F) H3K27me3 and H3.1/3.2 staining in a round (E) and elongating (F) spermatid. (G,H) H3K4me2 and H3.1/3.2 staining in a round (G) and elongating (H) spermatid. Scale bar: 10 µm.
Fig. 8. Staining patterns for constitutive heterochromatin histone PTMs in round and early elongating spermatids.
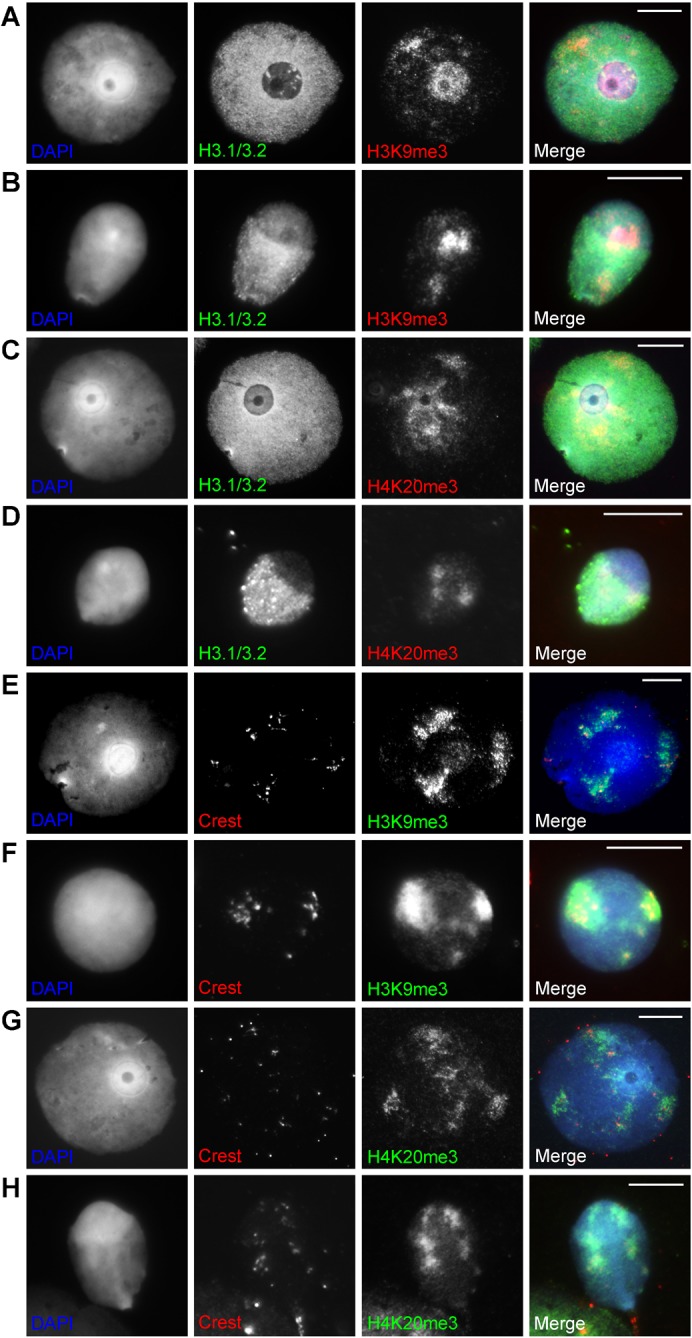
(A,B) H3K9me3 and H3.1/3.2 staining in a round (A) and elongating (B) spermatid. (C,D) H4K20me3 and H3.1/3.2 staining in a round (C) and elongating (D) spermatid. (E,F) H3K9me3 and Crest staining in a round (E) and elongating (F) spermatid. (G,H) H4K20me3 and Crest staining in a round (G) and elongating (H) spermatid. Scale bar: 10 µm.
Elongating spermatids stained for H3K27me2/3 showed in most cases no or a very faint signal throughout the nucleus (Fig. 7D,F). When there is staining, the cap-like structure shows less staining following the decrease of H3.1/3.2 (not shown). In a few cases H3.1/3.2 was strongly decreased while H3K27me2/3 was still present at the cap (not shown). H3K9me2 often showed signal, sometimes strong, in the cap while H3.1/3.2 is decreased like in round spermatids (Fig. 7B). H3K9me3 and H4K20me3 show centric heterochromatin domains similar to the round spermatids. Domains were often found at the border of the cap annex other nuclear parts and do not form one distinct chromocentre (Fig. 8B,D,F,H).
Next to ‘heterochromatin’ modifications, we stained with H3K4me2, a marker for active chromatin. This marker showed in the majority of nuclei no difference between the doughnut- and cap-like structure and other nuclear parts, even when the H3.1/3.2 signal was decreasing (Fig. 7G,H).
Discussion
In this study we describe the gradual development of a DAPI-intense doughnut-like structure in human spread round spermatids metamorphosing towards a cap-like structure in elongating spermatids, a pattern also found in mouse. Our results show the decrease of nucleosomes and the appearance of protamines at this region. Staining with acrosome and SAL-PNT markers showed co-localisation with the doughnut/cap like structure. In cryosections we could not identify DAPI-intense domains. In the alkaline nuclear spreads, spreading of chromatin can be restricted by structural components of the nucleus such as the nuclear matrix, leading to identification of the doughnut like structure in this type of preparation. To localise this area in cryosections, we applied acrosin and SAL-PNT staining to follow docking and capping of the acrosomal vesicle on/over the spermatid nucleus. The behaviour of the acrosome and associated structures in spread nuclei and cryosections were the same as to concomitant nucleosome loss and initial protamine localisation.
Kinetics of remodelling
Nuclear spreading provides higher spatial resolution compared to testicular sections. Therefore, we were able to show that histone/nucleosome removal starts early in type 3/4 round spermatids, which probably corresponds to part of Sa (stage II) and Sb1 (stage III) spermatids. TP2 and P2 were in most nuclei first noted in the doughnut, quickly spreading over the nucleus. P1 is more variable in this respect. In round and elongating spermatids, pre-P2 was exclusively found in this region of the nucleus. Hence, we assume the doughnut to be the initiation site of remodelling. However, we did not observed a decrease in histone/nucleosome staining outside the doughnut-/cap-like region (in elongating spermatids) (Sc, stages V,VI). This may be caused by the relative abundance of these proteins. Although there is a decline in nucleosomes when the nuclear volume decreases, the epitopes become compacted as well, leading to an intense signal.
We showed incorporation of P1 before P2. P1 is already present in round spermatids (Step Sb1) while P2 is only observed from the early elongating steps on (Step Sb2). Compared to timing of P1/2 incorporation reported in the literature (see introduction) we show these proteins to be present earlier, likely resulting from the higher spatial resolution our preparation technique offers. Also the order appearance, P1 before P2, is the same as in mouse (Zhao et al., 2004).
Sub-acrosomal nuclear organisation at the Golgi and capping phases
Transmission electron microscopy studies in both man and mouse show that landing of the acrosome vesicle induces a change in nuclear envelope and underlying chromatin: the inner and outer membrane are visible as one layer and the underlying chromatin changes into an electron dense configuration (De Kretser, 1969; Horstmann, 1961). Also nuclear pore complexes are removed from this area (Fawcett and Chemes, 1979; Loir and Courtens, 1979). In the human, landing of the acrosomal vesicle induces a depression in the nucleus that at the capping phase restores to a gentle round curve (Holstein and Roosen-Runge, 1981). The ‘little hole’ we observed in the doughnut types 2,3, and 4.1 could well represent this depression following from the acrosomal granule pressing away the chromatin. When the acrosomal granule at the capping phase spreads over the nucleus, the hole disappears (within type 4).
In the somatic nucleus, chromatin is attached to the nuclear envelope (NE) via lamins B and A/C, being the main constituents of the nuclear lamina (Kind and van Steensel, 2010; Wilson and Foisner, 2010). This chromatin is also labelled by H3K9me2, classifying it as ‘inactive’ euchromatin (Wen et al., 2009) and is AT rich (65% compared to 55% of non lamina associated domains as determined by sequencing; B. van Steensel, personal communication). Our images show a higher intensity of AT preferring DAPI and H3K9me2 at the doughnut where a more detailed chromatin structure is found (supplementary material Fig. S2B,C). Could this DAPI intensity due to resistance to chromatin relaxation procedures be more pronounced by the slight AT enrichment? To gain more insight into this matter we searched the literature for the composition of the nuclear lamina in round to elongating spermatids, knowing that lamin AC is absent during spermatogenesis (mouse (Moss et al., 1993); human, data not shown).
Of the known lamin B splicing products, B1 and the spermatogenesis specific B3 are present in the spermatid nucleus (Schütz et al., 2005). By a specific antibody, lamin B can be found over the whole nuclear periphery in purified round spermatids (steps 1–8) and in mature sperm after DTT and CTAB treatment (Moss et al., 1993). Lamin B3 is not detected in the acrosomal region using standard cryo- and paraffin-sections (Göb et al., 2010; Schütz et al., 2005).
Lamin B interacts with many proteins (Wilson and Foisner, 2010) among which the lamin B receptor (LBR) (Olins et al., 2010). LBR candidates to play a role in the phosphorylation of P1 by SR kinase (Mylonis et al., 2004; Papoutsopoulou et al., 1999) which is required to avoid precocious complexing with DNA (Balhorn, 2007) (as also applies to P2 in which case CaMKIV is involved (Wu et al., 2000) and to TP2 (Pradeepa and Rao, 2007)). In their active chromatin bound states, P1 is unphosphorylated and P2 predominantly so (Pruslin et al., 1987). CaMKIV is targeted to the nuclear matrix in elongating spermatids only (Wu et al., 2000). The fact that we exclusively localised pre-P2 in the doughnut- and cap-like structure is additional evidence for the fact that indeed CaMKIV is localised to an insoluble nuclear component and that (pre-)P2 phosphorylation and proteolytic cleavage are interrelated processes (Green et al., 1994).
Human LBR has a Tudor domain (recognizing specific histone methylation PTMs) that by analogy to the Tudor domain of the double strand DNA break repair protein 53BP1 is predicted to bind H4K20me2 (Olins et al., 2010). Additional to H3K9me2, we found H4K20me2 fibrillar staining at the doughnut- and cap-like structure in round and elongating spermatids (pilot experiment, data not shown). The marking of doughnut chromatin by both H3K9me2 and H4K20me2 adds evidence to a role of the nuclear lamina in chromatin remodelling under the acrosomal sac as evidenced by nucleosome and histone loss.
LINC complex proteins (among others the SUN domain transmembrane proteins (for a review, see Simon and Wilson, 2011)) provide the link between the nucleoskeleton (of which the nuclear lamina is an important part) and the cytoskeleton. In mouse spermatids, a polarized distribution of these proteins was found from the round spermatid stage on (Göb et al., 2010). Sun 1 localises both caudally at the spermatid tail region as well as apically under the acrosome while Sun 3 is present only laterally, associating with the manchette area. Recently, another SUN domain protein, Spag4l-2, was identified in mouse spermatids, specifically localizing at the apical, acrosomal region (Frohnert et al., 2011). Conclusively, the LINC complex is indicated to function as a communicator between spermatid chromatin and the acroplaxome by which signals nucleosome remodelling could be initiated.
Another indication for a link between the spermatid nuclear lamina and chromatin remodelling during spermiogenesis is provided by Mgcl-1 mutant mice (Kimura et al., 2003). Morphological abnormalities in the majority of sperm and an extended presence of TP2, lower P1,P2 levels and accumulation of pre-P2 were observed. In wild-type mice, MGCL-1 locates to the nuclear lamina, where it associates with LAP2β (lamin associated polypeptide) (Nili et al., 2001), which binds lamin B. In knockout mice, NE abnormalities are found from the spermatocyte stage on. In histological preparations of elongating spermatids Lamin B and LAP2β are both found at the posterior side of the nucleus (LAP2β (Alsheimer et al., 1998; Göb et al., 2010) and Lamin B3 (Göb et al., 2010; Schütz et al., 2005)). This could mean that when chromatin remodelling continues, the post-acrosomal NE is also involved with the initiation of chromatin remodelling (Biggiogera et al., 1992).
In humans with acrosomeless spermatozoa, a syndrome called globozoospermia, sperm heads are round-shaped. Several clinical reports indicate chromatin condensation to be abnormal and higher percentages of DNA fragmentation are found (Dam et al., 2007). Lower protamine levels were observed as well, but not in all men (Carrell et al., 1999). In line with this observation and the insight we present here, is the observation that in some globozoospermic men the subacrosomal change in the NE and underlaying chromatin specialisation had not developed (Escalier, 1990).
Chromatin aspects of polar initiation of remodelling
Chromatin remodelling in elongating spermatids involves massive breakdown of nucleosomes, induced by histone ubiquitination (Baarends et al., 1999; Chen et al., 1998; Lu et al., 2010). H4 acetylation is a key step to open up chromatin (Lu et al., 2010) and previously we localised H4K5,8,12 and 16ac in mouse elongating spermatids (van der Heijden et al., 2006). We found H4K16ac and H4K8ac to be enriched in the doughnut prior to staining in the remaining nucleus, confirming chromatin remodelling to start in the doughnut area. We were not able to demonstrate the presence of DNA DSBs in Sb1 round spermatids by using the DNA DSB marker γH2AX (not shown). In a pilot study, we identified the chromatin remodelling and DNA repair implicated protein PARP1 (Maymon et al., 2006; Meyer-Ficca et al., 2011; Meyer-Ficca et al., 2005) in the doughnut- and cap-like structure (not shown).
In the mouse, two chromatin proteins are known to demonstrate polarity of the round spermatid nucleus: H1T2 and HMG4 are present at opposite poles of the round spermatid nucleus (H1T2 apical from step 5 on (capping phase)) (Catena et al., 2006; Martianov et al., 2005) and HMG4 distal from step 9 on (at elongation) (Catena et al., 2009). In the absence of H1T2, chromatin in late elongated spermatids and sperm is less compacted and sperm heads are morphologically abnormal. Electronmicrographs show an abnormal chromatin structure under the acrosomal cap, linking H1T2 to the onset of remodeling.
Although the images obtained with the available nucleosome and histone type and histone PTMs were largely congruent with each other and mostly complementary to those obtained with P1 and P2, some noteworthy observations were made. We found H3K9me2 and especially H3K4me2 to be present in the apical region of the nucleus in the absence of a signal for nucleosomes. Both nucleosome antibodies were mouse monoclonals from LUPUS mice. The exact epitope is unknown and therefore detection of all nucleosomes may be unlikely. However, loss of signals for H3.1/3.2, panH3, and TH2B supported nucleosome eviction. It could be possible that both PTMs are so abundant on remaining nucleosomes/histones that detection sensitivity is at stake. Alternatively, there could be a conformational change, induced by the eviction process, which impedes recognition by the antibodies. Knowledge about nucleosome eviction in round and elongating spermatids is scant. Sperm specific histone subtypes such as H1T2 and HILS1 are known to play a role (for reviews, see Gaucher et al., 2010; de Boer et al., 2011). Alternative nucleosome structures have been encountered in compacting elongated mouse spermatids (steps 12–16) containing only H2A/H2B like histones (H2AL1/L2 plus TH2B) (Govin et al., 2007). Possibly, the ‘normal’ nucleosome structure is disrupted and replaced by an alternative nucleosome structure in which process H3K9me2 and H3K4me2 may play a role. Another explanation might be found in the presence of several H3 variants (at the protein level four are known). The exact epitopes of the polyclonal pan H3 antibody are not known to us. The immunogen peptide used (amino acid residues 100–135, see Materials and Methods, Antibodies) differs at only 1 amino acid from H3t (H3.4) and H3.5 (for H3.5, only RNA in human testis has been shown) (Schenk et al., 2011). However, it differs at 6 and 7 amino acids from H3.X and H3.Y, respectively (in human testis only RNA has been shown) (Wiedemann et al., 2010). The epitope of the H3.1/3.2 antibody is known (residues 28–32) and this region is not different in H3t, but several amino acid differences are present in H3.X, H3.Y and H3.5 suggesting these not to be recognized. Therefore, the H3K4me2 and H3K9me2 PTMs could be present on these variants specifically.
Pre-P2 processing
We found pre-P2 to accumulate at the doughnut-/cap-like structure (and never outside this region) suggesting this to be the site of proteolytic processing. This indicates that only processed P2 is incorporated into chromatin, as was suggested before (Green et al., 1994). However, other reports showed the presence of pre-P2 in a subset of mature spermatozoa (de Mateo et al., 2011a), the more so when the P1/P2 ratio is higher, which is related to infertility (reviewed by Carrell et al., 2008; Steger et al., 2011). This suggests the possibility of incorporation of pre-P2 at a later elongated spermatid stage (as a consequence of defective/incomplete remodelling) as pre-P2 can complex with DNA (Balhorn, 2007).
Concluding remarks
Summarizing, in this study we could connect the onset of chromatin remodeling in human round spermatids with the descent of the acrosomal vesicle on the nucleus. We have interpreted our findings using the available literature on protamine processing, the communication between the nucleus and cytoplasm in round and elongating spermatids, and current knowledge on spermatid chromatin remodeling. Our data fit in the concept by which chromatin remodelling is initiated by environmental signals likely derived from the Sertoli cell as the Sertoli cell Jam-C cell adhesion protein has been shown to be implicated in acrosome biogenesis (Gliki et al., 2004).
Materials and Methods
Ethics Statement
On October 18th, 2006, the CCMO (Central Committee on Research involving Human Subjects, the Hague) approved of the research protocol entitled ‘Intracytoplasmatic Sperm Injection using testicular spermatozoa in men with azoospermia: an observational study’. CCMO - NL12408.000.06
Human testis material
Testis material was obtained from 15 men willing to conceive who underwent a testicular biopsy for sperm retrieval (TESE: testicular sperm extraction). For proband details see supplementary material Table S1. Some men were of proven fertility, but now diagnosed with obstructive azoospermia (OA) because of a failed vaso-vasostomy. The others were classified as non obstructive azoospermia (NOA). All probands showed a normal karyotype and AZF deletions were not present. Biopsies were taken following the procedure of Silber (Silber, 2000). After biopsy processing, retrieved testicular spermatozoa were used for oocyte fertilization via ICSI (intra cytoplasmic sperm injection). From most biopsies a drop of spermatogenic cell suspension was smeared on a microscope slide. Cells were Giemsa stained and pachytene spermatocytes, cells with a sperm morphology (collectively called sperm) and Sertoli cells were counted. In supplementary material Table S1, ratios between pachytene cells and sperm, and between sperm and Sertoli cells are given. The latter ratio indicates the spermatogenic activity of the tissue sampled, the former ratio the efficiency of the production of mature spermatids per meiotic cell. Although not strictly comparable with histological studies, our data are in line with those published (McVicar et al., 2005; Rowley and Heller, 1971; Zhengwei et al., 1998). For probands 11 and 13 (failed vaso-vasostomy), spermatogenesis was evaluated by histology using the Johnsen score, (Johnsen, 1970) and found to be normal (scores of 9.1 and 9.5 respectively with tubule scores of 9 and 10 only). Remnants of the testicular samples were available for research after successful sperm retrieval. All men signed an informed consent for participation in this project. Mouse testis material was used from a wild-type CBA/B6 hybrid mouse (2 months old). The procedure involving the use of overcomplete animals is approved by the animal ethics committee of our university in conformance with the Dutch law on the use of experimental animals.
Surface spread preparations
Nucleus spreads were made as described by Peters (Peters et al., 1997), with minor modifications. Briefly, a suspension of spermatogenic cells was made by crushing the remnant seminiferous tubuli with two ribbed forceps in a drop of SIM (spermatocyte isolation medium) (Heyting and Dietrich, 1991). Remnants were separated from the cell suspension by a quick spin (25 g). The supernatant was transferred to a clean tube, centrifuged for 7 minutes (159 g), and the pellet was resuspended in 1 ml SIM. An equal volume of a hypotonic solution (17 mM sodium citrate, 50 mM sucrose, 30 mM Tris-HCl pH 8.2) was added for 7 minutes. Cells were centrifuged again (7 minutes, 159 g) and resuspended in 100 mM sucrose (pH 8.2) at a concentration of 10–15×106/ml. Two 5 µl drops were pipetted onto a paraformaldehyde (1% PFA, 0.15% Triton-X-100 pH 9.2) fluid coated microscope slide, placed in a levelled humid box for about 75 minutes, rinsed twice in 0.08% photoflow (Kodak), and air dried. Slides were stored at −80°C until use.
Cryosections
Testis tissue was prepared for cryosections according to Soper et al. (Soper et al., 2008). Tissue was embedded in Tissue Tek and 8-µm sections were cut with a Cryostat and transferred onto polysine coated slides (Menzel-gläser, Germany). Slides were stored at −80°C. Prior to immunofluorescence cells were permeabilised in PBS with 0.15% Triton-X-100 for 10 minutes.
Immunofluorescence
Surface spread preparations were washed twice in PBS containing 0.05% Triton-X-100 and blocked for 1 hour at 37°C (blocking buffer: 1% bovine serum albumin, 10% normal goat serum or normal donkey serum in PBS containing 0.05% Triton-X-100). Primary antibodies were diluted in blocking buffer and slides were incubated for 20 minutes at 37°C, followed by overnight incubation at 4°C extended by 20 minutes at 37°C. Then slides were rinsed and washed once in PBS containing 0.05% Triton-X-100 and afterwards rinsed and washed once in PBS. A second 30 minute blocking step was applied in blocking buffer without Triton-X-100, followed by a 2 hour incubation with the secondary antibodies diluted in blocking buffer without Triton-X-100. After rinsing and washing once in PBS, nuclei were stained with DAPI (10 min, 0.3 µg/ml in PBS) and mounted with Vectashield (Vector).
Antibodies
All somatic H3 isoforms were detected with a polyclonal pan H3 antibody (Abcam, 1791) at a dilution of 1∶500. Testis specific H3t and H3.5 are most likely detected as well because of the only 1 amino acid difference in the immunogen sequence (110 A vs V) (Albig et al., 1996), (104 L vs F), (Schenk et al., 2011). For this antibody, epitopes were first uncovered by a quick dip in 4 M HCl, followed by extensive washing in PBS. A monoclonal anti-nucleosome antibody (#32) (Kramers et al., 1996), provided by J. van der Vlag, was used at a 1∶2000 dilution. To localize H3.1/3.2 (also recognizing H3t), monoclonal antibody #34 (van der Heijden et al., 2005), provided by J. van der Vlag, was used at a dilution of 1∶1500. Nucleosomes were also detected at 1∶2000 dilution by the monoclonal antibody PL2/3 (Dieker et al., 2005; Losman et al., 1992), provided by J. van der Vlag. TH2B was stained with a polyclonal antibody (Abcam, ab23913) at a 1∶5000 dilution. Protamine 1 and 2 were detected with the monoclonal antibodies Hub1N and Hub2B respectively, used at a 1∶500 dilution and provided by R. Balhorn. A polyclonal antibody recognizing pre-P2 was provided by M. Meistrich and was used at 1∶200. TP2 was stained at 1∶200 dilution with a polyclonal antibody (TP2 303), provided by W.S. Kistler. A monoclonal anti-acrosin antibody (AMC-ACRO-C5F10-AS, Biosonda, Chile) was used to detect the acrosome at 1∶40 dilution. The subacrosomal layer of the perinuclear theca was demonstrated using the rabbit polyclonal antibody PNT-1449, provided by R. Oko, at a 1∶200 dilution. A rabbit polyclonal antibody against H3K9me2, provided by T. Jenuwein was used at 1∶150. To detect H3K9me3, two rabbit polyclonal antibodies were used, Abcam (ab8898) at 1∶1000, and one generated by A.H.F.M. Peters at 1∶750. Polyclonal rabbit antibodies against H3K27me2 and H3K27me3, provided by T. Jenuwein, were used at 1∶350. Two rabbit polyclonal antibodies against H3K4me2 were used, Abcam (ab7766) at 1∶100 and Upstate (07-030) at 1∶500. H4K20me2 was detected at 1∶50 dilution by a polyclonal antibody provided by T. Jenuwein. H4K20me3 was detected by a polyclonal antibody (Upstate 07-463) and used at 1∶500 dilution. H4K8ac and H4K16ac were detected by polyclonal antibodies (Upstate 06-760 (1∶1000) and Upstate 06-762 (1∶500), respectively). A monoclonal γH2AX antibody (Upstate 05-636) was used at 1∶1000. PARP1 was detected using a polyclonal antibody (Abcam ab2168) at 1∶300. Centromeres were stained by a human autoantibody (Crest) at a dilution of 1∶500 (ImmunoVision, Springdale AR). Primary antibodies were detected by using goat anti-human, goat or donkey anti-mouse and goat or donkey anti-rabbit secondaries with respectively a red or a green fluorochrome at 1∶500 dilution (Invitrogen Alexa 488; A11001, A21202, Alexa 594; A11014, A11012, A21207).
Image capture and analysis
The number of probands used for each antibody combination varied, but was always larger than 2 (except for H4K8ac (1), H4K16ac (2) and TH2B (2)). Series of representative photographs were taken, based on the inspection of a much larger sample as especially round spermatids are abundantly present in these preparations. For histone PTMs (4*), pan H3 (7), #32 (7), #34 (4), pre-P2 (6) and P1 (6) at least 50 round spermatids per proband were photographed (*number of probands). We aimed at collecting as many elongating spermatid images as possible. Their numbers often being low, we sampled on average 15 per staining per proband. Nuclei were captured at an exposure time reflecting the microscopic image by a Zeiss AxioCam MR camera on a Zeiss Axioplan fluorescence microscope using Axiovision 3.1 software (Carl Zeiss).
The relative intensity (compared to remaining nucleus) of H3.1/3.2 in the doughnut was subjectively categorized in three groups (Fig. 6; supplementary material Fig. S2): more intense or equally intense H3.1/3.2 signal in the doughnut (≥), less intense H3.1/3.2 signal in the doughnut (+/−), no H3.1/3.2 signal in the doughnut (−). Doughnut histone PTM staining in Fig. 6A,B,C was subjectively determined relative to PTM staining in the remaining nucleus. The category ‘overall no staining’ means no PTM staining in the entire nucleus (Fig. 6A–G). Doughnut PTM staining intensity in Fig. 6D,E was subjectively determined and categorized in four groups. The relative intensity of PL2/3 (Fig. 5F) and #32 (Fig. 5G) nucleosome markers in the doughnut was categorized in two groups: more intense or equally intense nucleosome signal in the doughnut (≥) and less intense or no H3.1/3.2 signal in the doughnut (+/−). H4K8/K16ac PTM marker staining patterns (Fig. 5F,G) were categorized into 4 groups as described in the legend of Fig. 5A–E.
Statistics
Deviations from random class distributions have been tested by Chi square analysis.
Supplementary Material
Acknowledgments
We would like to thank K. D'Hauwers department of Urology, Radboud University Nijmegen Medical Centre, responsible for the testis biopsies. We thank the following colleagues for providing antibodies: J. van der Vlag, Nephrology Research Laboratory and J. Martens, Department of Molecular Biology, NCMLS, Nijmegen, The Netherlands. D.R. de Bruin, Department of Human Genetics, Radboud University Nijmegen Medical Centre, The Netherlands. E. Ahmed, formerly at Department of Endocrinology, Utrecht University, Utrecht, The Netherlands. A.H.F.M. Peters, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland. T. Jenuwein, Max-Planck Institute of Immunobiology, Freiburg, Germany. G.W. van der Heijden, formerly at Carnegie Institute, Baltimore, Maryland, USA. R. Balhorn, Lawrence Livermore National Laboratory, Livermore, California, USA. M. Meistrich, M.D. Anderson Cancer Center, University of Texas, Houston, Texas, USA. W.S. Kistler, University of South Carolina, Columbia, USA. R. Oko, Queen's University, Kingston, Canada and K. Steger, Justus-Liebig University of Giessen, Giessen, Germany.
Footnotes
Competing interests: The authors declare that there are no competing interests.
References
- Albig W., Ebentheuer J., Klobeck G., Kunz J., Doenecke D. (1996). A solitary human H3 histone gene on chromosome 1. Hum. Genet. 97, 486–491 10.1007/BF02267072 [DOI] [PubMed] [Google Scholar]
- Alsheimer M., Fecher E., Benavente R. (1998). Nuclear envelope remodelling during rat spermiogenesis: distribution and expression pattern of LAP2/thymopoietins. J. Cell Sci. 111, 2227–2234. [DOI] [PubMed] [Google Scholar]
- Baarends W. M., Hoogerbrugge J. W., Roest H. P., Ooms M., Vreeburg J., Hoeijmakers J. H., Grootegoed J. A. (1999). Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207, 322–333 10.1006/dbio.1998.9155 [DOI] [PubMed] [Google Scholar]
- Balhorn R. (2007). The protamine family of sperm nuclear proteins. Genome Biol. 8, 227 10.1186/gb-2007-8-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggiogera M., Muller S., Courtens J. L., Fakan S., Romanini M. G. (1992). Immunoelectron microscopical distribution of histones H2B and H3 and protamines in the course of mouse spermiogenesis. Microsc. Res. Tech. 20, 259–267 10.1002/jemt.1070200305 [DOI] [PubMed] [Google Scholar]
- Boussouar F., Rousseaux S., Khochbin S. (2008). A new insight into male genome reprogramming by histone variants and histone code. Cell Cycle 7, 3499–3502 10.4161/cc.7.22.6975 [DOI] [PubMed] [Google Scholar]
- Brykczynska U., Hisano M., Erkek S., Ramos L., Oakeley E. J., Roloff T. C., Beisel C., Schübeler D., Stadler M. B., Peters A. H. (2010). Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- Carrell D. T., Emery B. R., Liu L. (1999). Characterization of aneuploidy rates, protamine levels, ultrastructure, and functional ability of round-headed sperm from two siblings and implications for intracytoplasmic sperm injection. Fertil. Steril. 71, 511–516 10.1016/S0015-0282(98)00498-1 [DOI] [PubMed] [Google Scholar]
- Carrell D. T., Emery B. R., Hammoud S. (2008). The aetiology of sperm protamine abnormalities and their potential impact on the sperm epigenome. Int. J. Androl. 31, 537–545 10.1111/j.1365-2605.2008.00872.x [DOI] [PubMed] [Google Scholar]
- Catena R., Ronfani L., Sassone-Corsi P., Davidson I. (2006). Changes in intranuclear chromatin architecture induce bipolar nuclear localization of histone variant H1T2 in male haploid spermatids. Dev. Biol. 296, 231–238 10.1016/j.ydbio.2006.04.458 [DOI] [PubMed] [Google Scholar]
- Catena R., Escoffier E., Caron C., Khochbin S., Martianov I., Davidson I. (2009). HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol. Reprod. 80, 358–366 10.1095/biolreprod.108.070243 [DOI] [PubMed] [Google Scholar]
- Chen H. Y., Sun J. M., Zhang Y., Davie J. R., Meistrich M. L. (1998). Ubiquitination of histone H3 in elongating spermatids of rat testes. J. Biol. Chem. 273, 13165–13169 10.1074/jbc.273.21.13165 [DOI] [PubMed] [Google Scholar]
- Clermont Y. (1963). The cycle of the seminiferous epithelium in man. Am. J. Anat. 112, 35–51 10.1002/aja.1001120103 [DOI] [PubMed] [Google Scholar]
- Courtens J. L., Loir M. (1981). Ultrastructural detection of basic nucleoproteins: alcoholic phosphotungstic acid does not bind to arginine residues. J. Ultrastruct. Res. 74, 322–326 10.1016/S0022-5320(81)80123-2 [DOI] [PubMed] [Google Scholar]
- Courtens J. L., Kistler W. S., Plöen L. (1995). Ultrastructural immunolocalisation of histones (H2B, H3, H4), transition protein (TP1) and protamine in rabbit spermatids and spermatozoa nuclei. Relation to condensation of the chromatin. Reprod. Nutr. Dev. 35, 569–582 10.1051/rnd:19950509 [DOI] [PubMed] [Google Scholar]
- de Boer P., de Vries M., Gochhait S. (2011). Histone variants during gametogenesis and early development. Epigenetics And Human Reproduction (ed. Rousseaux S, Khochbin S.), pp. 187–212 Heidelberg; Dordrecht; London; New York: Springer Verlag. [Google Scholar]
- De Kretser D. M. (1969). Ultrastructural features of human spermiogenesis. Z. Zellforsch. Mikrosk. Anat. 98, 477–505 10.1007/BF00347027 [DOI] [PubMed] [Google Scholar]
- de Mateo S., Ramos L., de Boer P., Meistrich M., Oliva R. (2011a). Protamine 2 precursors and processing. Protein Pept. Lett. 18, 778–785. [DOI] [PubMed] [Google Scholar]
- de Mateo S., Ramos L., van der Vlag J., de Boer P., Oliva R. (2011b). Improvement in chromatin maturity of human spermatozoa selected through density gradient centrifugation. Int. J. Androl. 34, 256–267 10.1111/j.1365-2605.2010.01080.x [DOI] [PubMed] [Google Scholar]
- de Vries M., Vosters S., Merkx G., D’Hauwers K., Wansink D. G., Ramos L., de Boer P. (2012). Human male meiotic sex chromosome inactivation. PLoS ONE 7, e31485 10.1371/journal.pone.0031485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadoune J. P. (2003). Expression of mammalian spermatozoal nucleoproteins. Microsc. Res. Tech. 61, 56–75 10.1002/jemt.10317 [DOI] [PubMed] [Google Scholar]
- Dam A. H., Feenstra I., Westphal J. R., Ramos L., van Golde R. J., Kremer J. A. (2007). Globozoospermia revisited. Hum. Reprod. Update 13, 63–75 10.1093/humupd/dml047 [DOI] [PubMed] [Google Scholar]
- Dieker J. W., Sun Y. J., Jacobs C. W., Putterman C., Monestier M., Muller S., van der Vlag J., Berden J. H. (2005). Mimotopes for lupus-derived anti-DNA and nucleosome-specific autoantibodies selected from random peptide phage display libraries: facts and follies. J. Immunol. Methods 296, 83–93 10.1016/j.jim.2004.10.010 [DOI] [PubMed] [Google Scholar]
- Dohle G. R., Colpi G. M., Hargreave T. B., Papp G. K., Jungwirth A., Weidner W. EAU Working Group on Male Infertility(2005). EAU guidelines on male infertility. Eur. Urol. 48, 703–711 10.1016/j.eururo.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Dooher G. B., Bennett D. (1973). Fine structural observations on the development of the sperm head in the mouse. Am. J. Anat. 136, 339–361 10.1002/aja.1001360307 [DOI] [PubMed] [Google Scholar]
- Escalier D. (1990). Failure of differentiation of the nuclear-perinuclear skeletal complex in the round-headed human spermatozoa. Int. J. Dev. Biol. 34, 287–297. [PubMed] [Google Scholar]
- Fawcett D. W., Chemes H. E. (1979). Changes in distribution of nuclear pores during differentiation of the male germ cells. Tissue Cell 11, 147–162 10.1016/0040-8166(79)90015-6 [DOI] [PubMed] [Google Scholar]
- Frohnert C., Schweizer S., Hoyer-Fender S. (2011). SPAG4L/SPAG4L-2 are testis-specific SUN domain proteins restricted to the apical nuclear envelope of round spermatids facing the acrosome. Mol. Hum. Reprod. 17, 207–218 10.1093/molehr/gaq099 [DOI] [PubMed] [Google Scholar]
- Gatewood J. M., Cook G. R., Balhorn R., Bradbury E. M., Schmid C. W. (1987). Sequence-specific packaging of DNA in human sperm chromatin. Science 236, 962–964 10.1126/science.3576213 [DOI] [PubMed] [Google Scholar]
- Gaucher J., Reynoird N., Montellier E., Boussouar F., Rousseaux S., Khochbin S. (2010). From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 277, 599–604 10.1111/j.1742-4658.2009.07504.x [DOI] [PubMed] [Google Scholar]
- Gliki G., Ebnet K., Aurrand-Lions M., Imhof B. A., Adams R. H. (2004). Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature 431, 320–324 10.1038/nature02877 [DOI] [PubMed] [Google Scholar]
- Göb E., Schmitt J., Benavente R., Alsheimer M. (2010). Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS ONE 5, e12072 10.1371/journal.pone.0012072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J., Escoffier E., Rousseaux S., Kuhn L., Ferro M., Thévenon J., Catena R., Davidson I., Garin J., Khochbin S. et al. (2007). Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 10.1083/jcb.200604141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green G. R., Balhorn R., Poccia D. L., Hecht N. B. (1994). Synthesis and processing of mammalian protamines and transition proteins. Mol. Reprod. Dev. 37, 255–263 10.1002/mrd.1080370303 [DOI] [PubMed] [Google Scholar]
- Grimes S. R., Jr, Henderson N. (1984). Hyperacetylation of histone H4 in rat testis spermatids. Exp. Cell Res. 152, 91–97 10.1016/0014-4827(84)90232-5 [DOI] [PubMed] [Google Scholar]
- Hazzouri M., Pivot-Pajot C., Faure A. K., Usson Y., Pelletier R., Sèle B., Khochbin S., Rousseaux S. (2000). Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur. J. Cell Biol. 79, 950–960 10.1078/0171-9335-00123 [DOI] [PubMed] [Google Scholar]
- Heller C. H., Clermont Y. (1964). Kinetics of the germinal epithelium in man. Recent Prog. Horm. Res. 20, 545–575. [PubMed] [Google Scholar]
- Hermo L., Pelletier R. M., Cyr D. G., Smith C. E. (2010). Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 2: changes in spermatid organelles associated with development of spermatozoa. Microsc. Res. Tech. 73, 279–319 10.1002/jemt.20783 [DOI] [PubMed] [Google Scholar]
- Heyting C., Dietrich A. J. (1991). Meiotic chromosome preparation and protein labeling. Methods Cell Biol. 35, 177–202 10.1016/S0091-679X(08)60573-7 [DOI] [PubMed] [Google Scholar]
- Holstein A. F., Roosen-Runge E. C. (1981). Atlas Of Human Spermatogenesis Berlin, Germany: Grosse Verlag. [Google Scholar]
- Horstmann E. (1961). [Electron microscopic study of spermio-histogenesis in man]. Z. Zellforsch. Mikrosk. Anat. 54, 68–89[Article in German] 10.1007/BF00384199 [DOI] [PubMed] [Google Scholar]
- Irvine D. S. (1998). Epidemiology and aetiology of male infertility. Hum. Reprod. 13, Suppl 133–44 10.1093/humrep/13.1.33 [DOI] [PubMed] [Google Scholar]
- Johnsen S. G. (1970). Testicular biopsy score count-a method for registration of spermatogenesis in human testes: normal values and results in 335 hypogonadal males. Hormones 1, 2–25 10.1159/000178170 [DOI] [PubMed] [Google Scholar]
- Kierszenbaum A. L., Rivkin E., Tres L. L. (2003). Acroplaxome, an F-actin-keratin-containing plate, anchors the acrosome to the nucleus during shaping of the spermatid head. Mol. Biol. Cell 14, 4628–4640 10.1091/mbc.E03-04-0226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum A. L., Rivkin E., Tres L. L. (2007). Molecular biology of sperm head shaping. Soc. Reprod. Fertil. Suppl. 65, 33–43. [PubMed] [Google Scholar]
- Kimura T., Ito C., Watanabe S., Takahashi T., Ikawa M., Yomogida K., Fujita Y., Ikeuchi M., Asada N., Matsumiya K. et al. (2003). Mouse germ cell-less as an essential component for nuclear integrity. Mol. Cell. Biol. 23, 1304–1315 10.1128/MCB.23.4.1304-1315.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J., van Steensel B. (2010). Genome-nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol. 22, 320–325 10.1016/j.ceb.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Kramers K., Stemmer C., Monestier M., van Bruggen M. C., Rijke-Schilder T. P., Hylkema M. N., Smeenk R. J., Muller S., Berden J. H. (1996). Specificity of monoclonal anti-nucleosome auto-antibodies derived from lupus mice. J. Autoimmun. 9, 723–729 10.1006/jaut.1996.0094 [DOI] [PubMed] [Google Scholar]
- Leblond C. P., Clermont Y. (1952). Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the periodic acid-fuchsin sulfurous acid technique. Am. J. Anat. 90, 167–215 10.1002/aja.1000900202 [DOI] [PubMed] [Google Scholar]
- Leduc F., Maquennehan V., Nkoma G. B., Boissonneault G. (2008). DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol. Reprod. 78, 324–332 10.1095/biolreprod.107.064162 [DOI] [PubMed] [Google Scholar]
- Loir M., Courtens J. L. (1979). Nuclear reorganization in ram spermatids. J. Ultrastruct. Res. 67, 309–324 10.1016/S0022-5320(79)80031-3 [DOI] [PubMed] [Google Scholar]
- Losman M. J., Fasy T. M., Novick K. E., Monestier M. (1992). Monoclonal autoantibodies to subnucleosomes from a MRL/Mp(-)+/+ mouse. Oligoclonality of the antibody response and recognition of a determinant composed of histones H2A, H2B, and DNA. J. Immunol. 148, 1561–1569. [PubMed] [Google Scholar]
- Lu L. Y., Wu J., Ye L., Gavrilina G. B., Saunders T. L., Yu X. (2010). RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell 18, 371–384 10.1016/j.devcel.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon L., Boissonneault G. (2004). Transient DNA strand breaks during mouse and human spermiogenesis new insights in stage specificity and link to chromatin remodeling. Biol. Reprod. 70, 910–918 10.1095/biolreprod.103.022541 [DOI] [PubMed] [Google Scholar]
- Martianov I., Brancorsini S., Catena R., Gansmuller A., Kotaja N., Parvinen M., Sassone-Corsi P., Davidson I. (2005). Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc. Natl. Acad. Sci. USA 102, 2808–2813 10.1073/pnas.0406060102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maymon B. B., Cohen-Armon M., Yavetz H., Yogev L., Lifschitz-Mercer B., Kleiman S. E., Botchan A., Hauser R., Paz G. (2006). Role of poly(ADP-ribosyl)ation during human spermatogenesis. Fertil. Steril. 86, 1402–1407 10.1016/j.fertnstert.2006.03.063 [DOI] [PubMed] [Google Scholar]
- McVicar C. M., O’Neill D. A., McClure N., Clements B., McCullough S., Lewis S. E. (2005). Effects of vasectomy on spermatogenesis and fertility outcome after testicular sperm extraction combined with ICSI. Hum. Reprod. 20, 2795–2800 10.1093/humrep/dei138 [DOI] [PubMed] [Google Scholar]
- Meistrich M. L., Trostle-Weige P. K., Lin R., Allis C. D., Bhatnagar Y. M. (1992). Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol. Reprod. Dev. 31, 170–181 10.1002/mrd.1080310303 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca M. L., Scherthan H., Bürkle A., Meyer R. G. (2005). Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma 114, 67–74 10.1007/s00412-005-0344-6 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca M. L., Lonchar J. D., Ihara M., Meistrich M. L., Austin C. A., Meyer R. G. (2011). Poly(ADP-ribose) polymerases PARP1 and PARP2 modulate topoisomerase II beta (TOP2B) function during chromatin condensation in mouse spermiogenesis. Biol. Reprod. 84, 900–909 10.1095/biolreprod.110.090035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss S. B., Burnham B. L., Bellvé A. R. (1993). The differential expression of lamin epitopes during mouse spermatogenesis. Mol. Reprod. Dev. 34, 164–174 10.1002/mrd.1080340208 [DOI] [PubMed] [Google Scholar]
- Mylonis I., Drosou V., Brancorsini S., Nikolakaki E., Sassone-Corsi P., Giannakouros T. (2004). Temporal association of protamine 1 with the inner nuclear membrane protein lamin B receptor during spermiogenesis. J. Biol. Chem. 279, 11626–11631 10.1074/jbc.M311949200 [DOI] [PubMed] [Google Scholar]
- Nili E., Cojocaru G. S., Kalma Y., Ginsberg D., Copeland N. G., Gilbert D. J., Jenkins N. A., Berger R., Shaklai S., Amariglio N. et al. (2001). Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J. Cell Sci. 114, 3297–3307. [DOI] [PubMed] [Google Scholar]
- Oakberg E. F. (1956). A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am. J. Anat. 99, 391–413 10.1002/aja.1000990303 [DOI] [PubMed] [Google Scholar]
- Oko R., Sutovsky P. (2009). Biogenesis of sperm perinuclear theca and its role in sperm functional competence and fertilization. J. Reprod. Immunol. 83, 2–7 10.1016/j.jri.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Olins A. L., Rhodes G., Welch D. B., Zwerger M., Olins D. E. (2010). Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus 1, 53–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R. (2006). Protamines and male infertility. Hum. Reprod. Update 12, 417–435 10.1093/humupd/dml009 [DOI] [PubMed] [Google Scholar]
- Papoutsopoulou S., Nikolakaki E., Chalepakis G., Kruft V., Chevaillier P., Giannakouros T. (1999). SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res. 27, 2972–2980 10.1093/nar/27.14.2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H. F. M., Plug A. W., van Vugt M. J., de Boer P. (1997). A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 10.1023/A:1018445520117 [DOI] [PubMed] [Google Scholar]
- Pradeepa M. M., Rao M. R. (2007). Chromatin remodeling during mammalian spermatogenesis: role of testis specific histone variants and transition proteins. Soc. Reprod. Fertil. Suppl. 63, 1–10. [PubMed] [Google Scholar]
- Prigent Y., Muller S., Dadoune J. P. (1996). Immunoelectron microscopical distribution of histones H2B and H3 and protamines during human spermiogenesis. Mol. Hum. Reprod. 2, 929–935 10.1093/molehr/2.12.929 [DOI] [PubMed] [Google Scholar]
- Pruslin F. H., Imesch E., Winston R., Rodman T. C. (1987). Phosphorylation state of protamines 1 and 2 in human spermatids and spermatozoa. Gamete Res. 18, 179–190 10.1002/mrd.1120180208 [DOI] [PubMed] [Google Scholar]
- Ramos L., van der Heijden G. W., Derijck A., Berden J. H., Kremer J. A., van der Vlag J., de Boer P. (2008). Incomplete nuclear transformation of human spermatozoa in oligo-astheno-teratospermia: characterization by indirect immunofluorescence of chromatin and thiol status. Hum. Reprod. 23, 259–270 10.1093/humrep/dem365 [DOI] [PubMed] [Google Scholar]
- Rousseaux S., Reynoird N., Escoffier E., Thevenon J., Caron C., Khochbin S. (2008). Epigenetic reprogramming of the male genome during gametogenesis and in the zygote. Reprod. Biomed. Online 16, 492–503 10.1016/S1472-6483(10)60456-7 [DOI] [PubMed] [Google Scholar]
- Rowley M. J., Heller C. G. (1971). Quantitation of the cells of the seminiferous epithelium of the human testis employing the sertoli cell as a constant. Z. Zellforsch. Mikrosk. Anat. 115, 461–472 10.1007/BF00335713 [DOI] [PubMed] [Google Scholar]
- Schenk R., Jenke A., Zilbauer M., Wirth S., Postberg J. (2011). H3.5 is a novel hominid-specific histone H3 variant that is specifically expressed in the seminiferous tubules of human testes. Chromosoma 120, 275–285 10.1007/s00412-011-0310-4 [DOI] [PubMed] [Google Scholar]
- Schütz W., Alsheimer M., Ollinger R., Benavente R. (2005). Nuclear envelope remodeling during mouse spermiogenesis: postmeiotic expression and redistribution of germline lamin B3. Exp. Cell Res. 307, 285–291 10.1016/j.yexcr.2005.03.023 [DOI] [PubMed] [Google Scholar]
- Silber S. J. (2000). Microsurgical TESE and the distribution of spermatogenesis in non-obstructive azoospermia. Hum. Reprod. 15, 2278–2284 10.1093/humrep/15.11.2278 [DOI] [PubMed] [Google Scholar]
- Simon D. N., Wilson K. L. (2011). The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat. Rev. Mol. Cell Biol. 12, 695–708 10.1038/nrm3207 [DOI] [PubMed] [Google Scholar]
- Sonnack V., Failing K., Bergmann M., Steger K. (2002). Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia 34, 384–390 10.1046/j.1439-0272.2002.00524.x [DOI] [PubMed] [Google Scholar]
- Soper S. F., van der Heijden G. W., Hardiman T. C., Goodheart M., Martin S. L., de Boer P., Bortvin A. (2008). Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 15, 285–297 10.1016/j.devcel.2008.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger K., Failing K., Klonisch T., Behre H. M., Manning M., Weidner W., Hertle L., Bergmann M., Kliesch S. (2001). Round spermatids from infertile men exhibit decreased protamine-1 and -2 mRNA. Hum. Reprod. 16, 709–716 10.1093/humrep/16.4.709 [DOI] [PubMed] [Google Scholar]
- Steger K., Cavalcanti M. C., Schuppe H. C. (2011). Prognostic markers for competent human spermatozoa: fertilizing capacity and contribution to the embryo. Int. J. Androl. 34, 513–527 10.1111/j.1365-2605.2010.01129.x [DOI] [PubMed] [Google Scholar]
- Steger K., Klonisch T., Gavenis K., Drabent B., Doenecke D., Bergmann M. (1998). Expression of mRNA and protein of nucleoproteins during human spermiogenesis. Mol. Hum. Reprod. 4, 939–945 10.1093/molehr/4.10.939 [DOI] [PubMed] [Google Scholar]
- van der Heijden G. W., Derijck A. A., Ramos L., Giele M., van der Vlag J., de Boer P. (2006). Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev. Biol. 298, 458–469 10.1016/j.ydbio.2006.06.051 [DOI] [PubMed] [Google Scholar]
- van der Heijden G. W., Dieker J. W., Derijck A. A., Muller S., Berden J. H., Braat D. D., van der Vlag J., de Boer P. (2005). Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 122, 1008–1022 10.1016/j.mod.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Wen B., Wu H., Shinkai Y., Irizarry R. A., Feinberg A. P. (2009). Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250 10.1038/ng.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann S. M., Mildner S. N., Bönisch C., Israel L., Maiser A., Matheisl S., Straub T., Merkl R., Leonhardt H., Kremmer E. et al. (2010). Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol. 190, 777–791 10.1083/jcb.201002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. L., Foisner R. (2010). Lamin-binding Proteins. Cold Spring Harb. Perspect. Biol. 2, a000554 10.1101/cshperspect.a000554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Y., Ribar T. J., Cummings D. E., Burton K. A., McKnight G. S., Means A. R. (2000). Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat. Genet. 25, 448–452 10.1038/78153 [DOI] [PubMed] [Google Scholar]
- Zhao M., Shirley C. R., Mounsey S., Meistrich M. L. (2004). Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol. Reprod. 71, 1016–1025 10.1095/biolreprod.104.028191 [DOI] [PubMed] [Google Scholar]
- Zhengwei Y., Wreford N. G., Royce P., de Kretser D. M., McLachlan R. I. (1998). Stereological evaluation of human spermatogenesis after suppression by testosterone treatment: heterogeneous pattern of spermatogenic impairment. J. Clin. Endocrinol. Metab. 83, 1284–1291 10.1210/jc.83.4.1284 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.