Summary
Dgp71WD/Nedd1 proteins are essential for mitotic spindle formation. In human cells, Nedd1 targets γ-tubulin to both centrosomes and spindles, but in other organisms the function of Dgp71WD/Nedd1 is less clear. In Drosophila cells, Dgp71WD plays a major part in targeting γ-tubulin to spindles, but not centrosomes, while in Xenopus egg extracts, Nedd1 acts as a more general microtubule (MT) organiser that can function independently of γ-tubulin. The interpretation of these studies, however, is complicated by the fact that some residual Dgp71WD/Nedd1 is likely present in the cells/extracts analysed. Here we generate a Dgp71WD null mutant lacking all but the last 12 nucleotides of coding sequence. The complete loss of Dgp71WD has no quantifiable effect on γ-tubulin or Centrosomin recruitment to the centrosome in larval brain cells. The recruitment of γ-tubulin to spindle MTs, however, is severely impaired, and spindle MT density is reduced in a manner that is indistinguishable from cells lacking Augmin or γ-TuRC function. In contrast, the absence of Dgp71WD leads to defects in the assembly of the acentrosomal female Meiosis I spindle that are more severe than those seen in Augmin or γ-TuRC mutants, indicating that Dgp71WD has additional functions that are independent of these complexes in oocytes. Moreover, the localisation of bicoid RNA during oogenesis, which requires γ-TuRC function, is unperturbed in Dgp71WD120 mutants. Thus, Dgp71WD is not simply a general cofactor required for γ-TuRC and/or Augmin targeting, and it appears to have a crucial role independent of these complexes in the acentrosomal Meiosis I spindle.
Keywords: Dgp71WD, Centrosome, Mitosis, Meiosis
Introduction
Bipolar spindle formation depends on MT polymerisation occurring in a spatially and temporally controlled manner. In many cell types, centrosomes are the primary MT organising centres, however centrosomes are not the only location where MTs are generated. During mitosis MTs are nucleated from at least 3 main sources: (1) from centrosomes (Bettencourt-Dias and Glover, 2007); (2) from an area around the chromosomes and kinetochores (Gadde and Heald, 2004); (3) from within the spindle via the newly discovered Augmin pathway (Goshima and Kimura, 2009). Surprisingly, neither the centrosomal pathway (Basto et al., 2006; Khodjakov et al., 2000; Megraw et al., 1999) nor the Augmin pathway (Goshima et al., 2008; Meireles et al., 2009; Wainman et al., 2009) are essential for mitosis in Drosophila, although spindle assembly is severely disrupted in cells that lack both pathways (Goshima et al., 2008; Meireles et al., 2009; Wainman et al., 2009). In contrast, the chromosomal pathway appears to be essential for mitosis, as mutations in the Drosophila gene misato (which abolishes chromosome-dependant MT nucleation) result in lethality (Mottier-Pavie et al., 2011).
Complexes containing γ-tubulin are important for MT nucleation in all three pathways in mitosis (Goshima et al., 2008; Goshima et al., 2007; Hannak et al., 2002; Joshi et al., 1992; Lüders et al., 2006; Sunkel et al., 1995). These complexes broadly fall into two classes: a γ-tubulin small complex (γ-TuSC) and a larger γ-tubulin ring complex (γ-TuRC) that contains several copies of the γ-TuSC and several additional proteins such as Dgrip75, Dgrip128 and Dgrip163 in Drosophila (Lüders and Stearns, 2007; Wiese and Zheng, 2006). In Drosophila, γ-TuSC mutants exhibit severe spindle defects and are lethal (Barbosa et al., 2000; Colombie et al., 2006; Sunkel et al., 1995), whereas γ-TuRC mutants exhibit more moderate spindle defects and are viable (though male and female sterile), and have defects in male meiosis, female meiosis II, and in bicoid (bcd) mRNA localisation during oogenesis (Schnorrer et al., 2002; Verollet et al., 2006; Vogt et al., 2006). This has led to the proposal that, in flies, the γ-TuRC is only essential for the nucleation of specific subsets of MTs that are required for certain developmental processes (Schnorrer et al., 2002; Verollet et al., 2006; Vogt et al., 2006).
Unlike the formation of the mitotic spindle, the formation of the meiosis I spindle is less well understood, and occurs without centrosomes in species like Drosophila and Xenopus (McKim and Hawley, 1995; Waters and Salmon, 1997). MTs are initially nucleated around the DNA before being organised into a bipolar spindle by motor proteins such as Ncd and Subito, and bundling/stabilising proteins such as DTACC and Msps (Cullen and Ohkura, 2001; Gadde and Heald, 2004; Giunta et al., 2002; Matthies et al., 1996; McKim and Hawley, 1995). Until recently, the role of γ-tubulin 37C — the form of γ-tubulin found in the oocyte (Tavosanis et al., 1997) — in the formation of the meiosis I spindle was controversial (Tavosanis et al., 1997; Wilson and Borisy, 1998). Recent data has, however, confirmed that γ-tubulin has a crucial role in bipolar spindle formation and kinetochore MT attachment in meiosis I (Hughes et al., 2011). The Augmin complex and the γ-TuRC appear to have minor roles in meiosis I spindle assembly, as the Augmin mutant wac has robust bipolar spindles but chromosome alignment defects, whereas γ-TuRC mutants have no detectable defects in meiosis I spindle assembly (Meireles et al., 2009; Vogt et al., 2006).
Dgp71WD (Grip71)/Nedd1 is a conserved centrosomal protein that was originally identified as a component which associates with the γ-TuRC (Gunawardane et al., 2003). Dgp71WD and its human homologue Nedd1 are structurally unrelated to the γ-TuSC/γ-TuRC proteins, as they lack the conserved ‘Grip’ motifs found in these proteins (Gunawardane et al., 2003). In human cells and in Zebrafish, Nedd1/GCP-WD is essential for targeting the γ-TuRC to the centrosome (Haren et al., 2006; Lüders et al., 2006; Manning et al., 2010), and in human cells it also appears to have a role in centriole duplication (Haren et al., 2006). Nedd1 is also required to target the γ-TuRC to the spindle in human cells (Lüders et al., 2006), where it is thought to link the γ-TuRC to the Augmin complex, thus promoting MT nucleation within the spindle (Johmura et al., 2011; Uehara et al., 2009; Zhu et al., 2008b). In Drosophila cells and Xenopus extracts, however, the depletion of Dgp71WD/Nedd1 leads to only a partial depletion of γ-tubulin and other PCM components from the centrosome, and to a much stronger loss of γ-tubulin from the spindle MTs (Dobbelaere et al., 2008; Liu and Wiese, 2008; Verollet et al., 2006). In mouse oocytes, Nedd1 has recently been shown to be essential for the proper assembly of the acentrosomal Meiosis I spindle, supporting the idea that Nedd1 has roles that do not require its localisation to centrosomes (Ma et al., 2010). The interpretation of these previous studies is complicated by the fact that Dgp71WD/Nedd1 may not be completely eliminated from the cells/extracts being analysed. To overcome this problem, we have generated a Dgp71WD null mutation in Drosophila by completely deleting virtually the entire coding sequence of the gene.
Results
Dgp71WD120 is a null mutant
A Dgp71WD mutant fly line Dgp71WDGE30807 has been previously characterised (Verollet et al., 2006). This line contains a P-element insertion in the 5′UTR of the Dgp71WD gene 49bp upstream of the initiating ATG. In our hands, antibodies raised against Dgp71WD appeared to detect small amounts of residual Dgp71WD protein in both western blotting and immunofluorescence experiments (R.F.R., unpublished observations), suggesting that this mutant was not a complete null. We therefore attempted to generate a null mutant by imprecise excision of this P-element (see Materials and Methods). We screened 384 potential lines using PCR and recovered two alleles that contained deletions in the Dgp71WD CDS. PCR and sequencing showed that the entire CDS apart from the last 12bp had been deleted from the Dgp71WD120 line without affecting the UTRs or CDS of any neighbouring genes (Fig. 1A,B). Western blotting and immunofluorescence experiments confirmed the absence of detectable Dgp71WD protein in this line (Fig. 1C,D). We conclude that Dgp71WD120 is likely a null mutation.
Fig. 1. Dgp71WD120 is a null mutant.
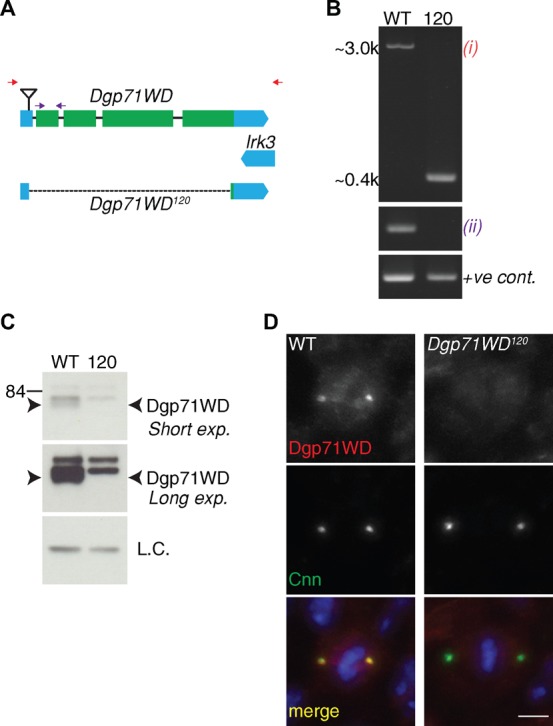
(A) Schematic view of the Dgp71WD gene. Coding sequences are indicated in green and UTRs in blue; the inverted triangle represents the insertion site of the Dgp71WDGE30807 P-element. The dotted line represents the region deleted in the Dgp71WD120 mutant. Coloured arrows refer to PCR primer pairs used for the PCRs shown in (B) - red = reaction (i), purple = reaction (ii). (B) Agarose gel of the PCR products obtained using the indicated primer pairs (the final reaction is a positive control) using either WT or Dgp71WD120 genomic DNA. (C) Western blots of WT and Dgp71WD120 3rd instar larval brain extracts at two different exposure levels. In WT extracts a double band can be seen below the 84 kDa marker. The higher band is nonspecific-background whereas the lower band represents Dgp71WD. The lower band is absent in the mutant, indicating that Dgp71WD120 is likely a protein null. (D) WT and Dgp71WD120 3rd instar larval brain cells stained with anti-Cnn (green) and anti-Dgp71WD (red) antibodies; DNA (blue). Scale bar represents 2.5 µm.
We found that, like the original Dgp71WDGE30807 mutation, Dgp71WD120 was viable but female sterile when homozygous or hemizygous over a deficiency that removed the Dgp71WD gene (Df(2L)Exel7071). It was previously reported that Dgp71WDGE30807 mutant flies have abdominal abnormalities and a dramatically shortened lifespan (Verollet et al., 2006), and this was also true in our hands. Dgp71WD120 homozygous or hemizygous adult flies, however, did not show the same reduced lifespan or any morphological abnormalities, suggesting that these phenotypes are not the result of a lesion in the Dgp71WD gene. Interestingly, Dgp71WD120 homozygous or hemizygous adult flies were male fertile, unlike known γ-TuRC mutants (Schnorrer et al., 2002; Vogt et al., 2006), suggesting that Dgp71WD cannot be required for γ-TuRC function in all circumstances.
Dgp71WD has no detectable role in centriole duplication or recruiting γ-tubulin to centrosomes in larval brain cells
In human cells, Nedd1 has a role in centriole duplication (Haren et al., 2006). In Drosophila, flies with defects in centriole duplication usually display uncoordinated behaviour due to defects in sensory neuron formation resulting from an absence of normal cilia (Basto et al., 2006). Dgp71WD120 mutant flies did not display uncoordinated behaviour, and centriole number was not detectably perturbed in larval neuroblasts (supplementary material Fig. S1).
In human cells, Nedd1 is also essential for γ-tubulin localisation to the centrosome (Haren et al., 2006; Lüders et al., 2006), and depleting Dgp71WD from Drosophila cultured cells using RNAi leads to a partial loss of both γ-tubulin and other PCM components from centrosomes (Dobbelaere et al., 2008; Verollet et al., 2006). To test whether Dgp71WD is required for γ-tubulin and/or PCM recruitment in vivo, we quantified the levels of PCM in WT and Dgp71WD120 mutant larval brain cells. We also included Dgrip75175 (a null mutation in the gene encoding the Dgrip75 component of the γ-TuRC) mutant brain cells in this analysis as this mutation strongly disrupts the formation of the γ-TuRC (Vogt et al., 2006), allowing us to test whether γ-TuRC formation is required for γ-tubulin or PCM recruitment. To our surprise, the centrosomal localisation of Cnn (Fig. 2A,D) — an upstream component in the PCM recruitment pathway; (Lucas and Raff, 2007; Megraw et al., 1999; Terada et al., 2003; Zhang and Megraw, 2007) — or γ-tubulin (Fig. 2B,D) was not detectably perturbed in either the Dgp71WD120 or Dgrip75175 mutant brain cells. We also examined the localisation of the centrosomal proteins Asterless (Asl) and Dspd2 — both upstream components in the PCM recruitment pathway (Blachon et al., 2008; Bonaccorsi et al., 2000; Conduit et al., 2010; Dobbelaere et al., 2008; Dzhindzhev et al., 2010; Gomez-Ferreria et al., 2007; Zhu et al., 2008a) — in Dgp71WD120 mutant brain cells, and found that levels of these proteins were also not perturbed (R.F.R., unpublished observations). We further confirmed the γ-tubulin result by quantifying the centrosomal levels of γ-tubulin-GFP (Hallen et al., 2008) in living WT and mutant brain cells. Again, we could detect no obvious difference in the amount of γ-tubulin recruited to centrosomes in WT, Dgp71WD120 or Dgrip75175 mutant brain cells (Fig. 2C,D). Taken together, these data suggest that Dgp71WD and Dgrip75 do not have a major role in the centrosomal recruitment of γ-tubulin or the PCM in vivo in fly larval brain cells.
Fig. 2. Centrosomal recruitment of Cnn and γ-tubulin is unperturbed in Dgp71WD120 and Dgrip75175 mutant brain cells.
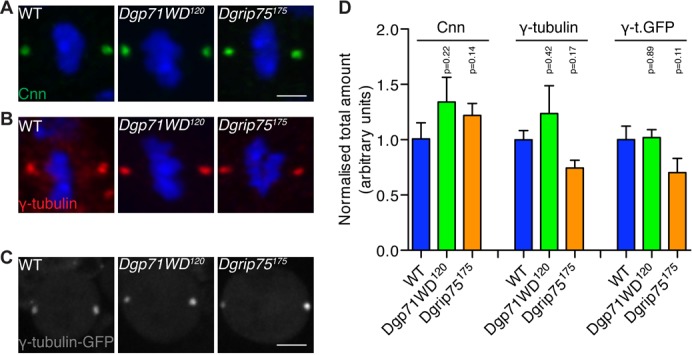
(A,B) Images show metaphase 3rd instar larval Ganglion Mother cells (GMCs) from WT, Dgp71WD120, and Dgrip75175 brains that were stained with anti-Cnn (A) (green) or anti-γ-tubulin (B) (red) antibodies; DNA (blue). (C) Images show living metaphase 3rd instar larval NBs from WT, Dgp71WD120, and Dgrip75175 brains expressing γ-tubulin GFP. (D) Quantification of the total amount of centrosomal Cnn, γ-tubulin or γ-tubulin-GFP in brain cells. Centrosomal γ-tubulin or Cnn levels in fixed cells were quantified in at least 7 cells in each of 5 brains. Centrosomal γ-tubulin-GFP levels in living cells were quantified in at least 9 NBs of each genotype. Significance testing was conducted using a Student's t-test. Error bars are S.E.M. Scale bar represents 2.5 µm (A,B) and 5 µm (C).
Dgp71WD is required for robust spindle assembly, but it is not essential for cell division in vivo
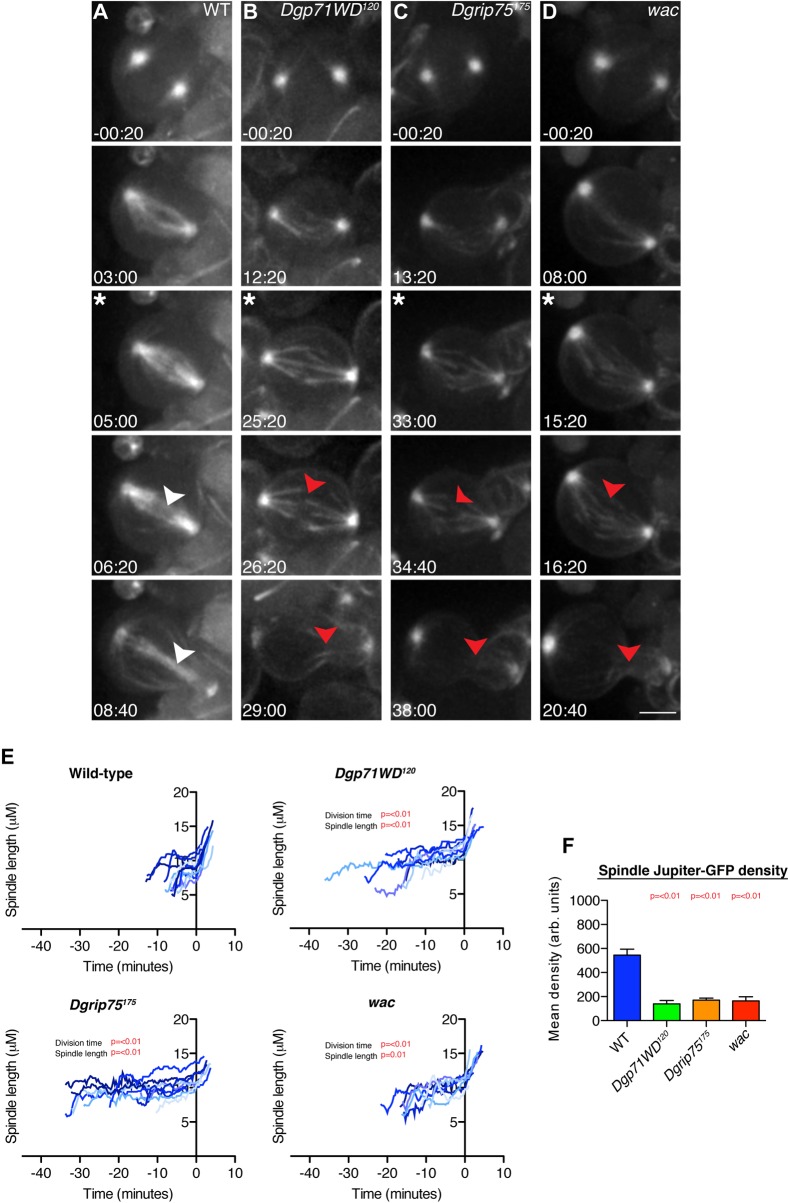
In human cells, Nedd1 also targets the γ-TuRC to the spindle MTs (Lüders et al., 2006) where it seems to cooperate with the Augmin complex to nucleate MTs within the spindle (Johmura et al., 2011; Uehara et al., 2009; Zhu et al., 2008b). In fly cells, Dgp71WD is also required to recruit γ-tubulin to the mitotic spindle (Verollet et al., 2006) (supplementary material Fig. S2), and the localisation of Dgp71WD to the spindle also appears to require the Augmin complex (Wainman et al., 2009). To confirm the potential relationship between Dgp71WD, the γ-TuRC and the Augmin complex in vivo we examined MT behaviour in living WT, Dgp71WD120, Dgrip75175 and wac — a gene encoding a component of the Augmin complex (Meireles et al., 2009) — mutant 3rd instar larval brain neuroblasts that expressed Jupiter-GFP — a MT-associated protein whose behaviour has been shown to accurately reflect MT behaviour (Karpova et al., 2006). Importantly, all of these alleles are reported to be null alleles (Meireles et al., 2009; Schnorrer et al., 2002).
We filmed neuroblasts from before the onset of Nuclear Envelope Breakdown (NEB) until late telophase. Upon NEB, WT neuroblasts rapidly established a robust metaphase spindle with well-defined kinetochore (k)-fibres surrounded by a more diffuse array of MTs (Fig. 3A; supplementary material Movie 1). These cells then underwent a characteristic asymmetric division, with prominent central spindle microtubules clearly visible between the dividing halves of the spindle (white arrowheads, Fig. 3A). In contrast, Dgp71WD120, Dgrip75175 and wac mutant neuroblasts took much longer to establish a metaphase plate, and the MTs appeared to have difficulty in capturing and aligning the chromosomes (Fig. 3B–D; supplementary material Movies 2–4). Even when formed, the metaphase spindle was clearly abnormal; the k-fibres did not appear to be reinforced by the formation of additional MTs within the spindle, and MT density within the spindle remained low (Fig. 3F). The whole spindle structure was unstable, often flexing and bending, and in all cases the metaphase spindles were longer than normal (Fig. 3E; WT = 9.0+/−0.34 mm; Dgp71WD120 = 11.4+/−0.33 mm (p<0.001); Dgrip75175 = 10.7+/−0.43 mm (p = 0.006); wac = 10.6+/−0.40 mm (p = 0.01)). Unsurprisingly, the mutant cells were delayed in exiting mitosis (Fig. 3E); time from NEB - Anaphase B onset in WT = 8.0+/−0.7 mins; Dgp71WD120 = 21.3+/−2.3 mins (p<0.001); Dgrip75175 = 22.4+/−3.14 mins (p<0.001); wac = 15.1+/−1.6 mins (p<0.001). Although wac mutant spindles appeared to divide slightly faster than Dgp71WD120 or Dgrip75175 mutants, this difference was on the border of statistical significance (p = 0.058 and p = 0.078, respectively). When the mutant spindles eventually exited mitosis they underwent an apparently normal asymmetric division, although the formation of a robust central spindle in late anaphase was severely impaired in the mutant neuroblasts (red arrows, Fig. 3B–D). This spindle phenotype is similar to that reported in S2 cells depleted of Dgp71WD, Augmin components or γ-TuRC components (Goshima et al., 2008; Verollet et al., 2006).
Fig. 3. Dgp71WD120, Dgrip75175 and wac NBs have severe spindle defects, yet successfully complete bipolar asymmetric divisions.
(A–D) WT and mutant 3rd instar larval neuroblasts (NBs) expressing Jupiter-GFP were filmed from before NEB to the onset of cytokinesis. Representative WT (A), Dgp71WD120 (B), Dgrip75175 (C) and wac (D) NBs are shown. Time is shown as min:sec. Time point −00:20 represents one time point before NEB; the asterisk (*) marks the time of the metaphase to anaphase transition. Mutant spindles at the metaphase-anaphase transition consist of only a few k-fibre bundles unlike WT cells, where spindle density is higher (third row). Central spindle formation is strongly impaired in mutant spindles (red arrowheads) compared to WT spindles (white arrowheads). (E) Spindle length was measured over time from NEB to the onset of cytokinesis in WT (n = 11), Dgp71WD120 (n = 9), Dgrip75175 (n = 10), and wac (n = 7) NBs. Data was aligned to Anaphase B onset (time = 0 on the graphs). Mutant cells take significantly longer to undergo division, and have significantly longer spindles than WT NBs at Anaphase-B onset. (F) The density of Jupiter-GFP (as a proxy for MT density) was calculated at spindles in WT (n = 11), Dgp71WD120 (n = 9), Dgrip75175 (n = 8) and wac (n = 5) NBs. Spindle Jupiter-GFP intensity is significantly lower in mutant neuroblasts. Significance testing was conducted using a Student's t-test. Scale bar represents 5 µm.
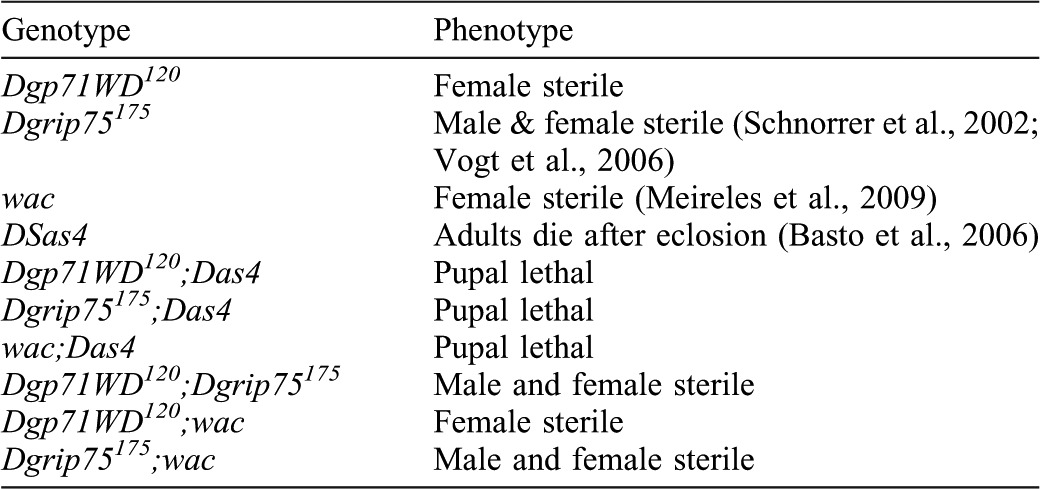
The remarkable similarity in these phenotypes strongly suggests that Dgp71WD, Dgrip75 and Wac all function in the same pathway to drive robust spindle and central spindle formation in larval brain cells. This pathway is clearly not essential for viability in flies, as Dgp71WD mutants (this study and (Verollet et al., 2006), γ-TuRC mutants (Schnorrer et al., 2002; Verollet et al., 2006; Vogt et al., 2006) and Augmin mutants (Meireles et al., 2009; Wainman et al., 2009) are all viable. It has previously been shown, however, that the Augmin pathway is essential for spindle formation in the absence of properly functioning centrosomes, indicating that the chromatin pathway cannot support robust spindle formation in the absence of both the centrosome and Augmin pathways (Goshima et al., 2008; Wainman et al., 2009). We confirmed that Dgp71WD and Dgrip75 are also essential for viability in the absence of centrosomes by combining the Dgp71WD120 and Dgrip75175 alleles with a mutation in the DSas4 gene, which is essential for centriole duplication (Basto et al., 2006). Dgp71WD120;Dsas4S2214, Dgrip75175;DSas4S2214 and wac;DSas4S2214 double mutants were all not viable and died during pupal stages of development (Table 1). In contrast, Dgp71WD120;Dgrip75175 and Dgp71WD120;wac double mutant combinations were all viable but female sterile (Table 1; see Materials and Methods), supporting the hypothesis that these proteins act together in the same pathway.
Table 1. Phenotypic analysis of Dgp71WD120, Dgrip75175 and wac mutant combinations.
Dgp71WD is not an essential cofactor for the γ-TuRC
Dgp71WD was initially identified as a component which associates with the γ-TuRC (Gunawardane et al., 2003), and our results in brain cells are consistent with the previous reports that Dgp71WD normally cooperates with the γ-TuRC and Augmin complex to drive robust spindle assembly (Uehara et al., 2009; Verollet et al., 2006; Wainman et al., 2009). We wondered, therefore, whether Dgp71WD might act as an essential co-factor for the γ-TuRC. Two previous studies have shown that the γ-TuRC components Dgrip75 and Dgrip128 (as well as γ-tubulin 37C) are required to localise bicoid (bcd) mRNA to the anterior of the oocytes from stage 10b/11 onwards (Schnorrer et al., 2002; Vogt et al., 2006). The correct localisation of bcd mRNA is required to help pattern the anterior-posterior axis of the Drosophila embryo (Riechmann and Ephrussi, 2001), and it has been suggested that the γ-TuRC nucleates a specific subset of MTs (that cannot be nucleated by the γ-TuSC) that are required for proper bcd localisation. We therefore examined whether Dgp71WD was also required for the proper localisation of bcd. In WT oocytes, bcd was localised to the anterior end of stage 11/12 oocytes (Fig. 4A,D), but this localisation was disrupted in Dgrip75175 mutant oocytes (Fig. 4C,D), as expected (Schnorrer et al., 2002; Vogt et al., 2006). In contrast, bcd localised normally in Dgp71WD120 mutant oocytes at stage 11/12 (Fig. 4B,D). Thus, Dgp71WD cannot be an essential co-factor for all aspects of γ-TuRC function.
Fig. 4. bicoid mRNA localises normally in Dgp71WD120 oocytes.
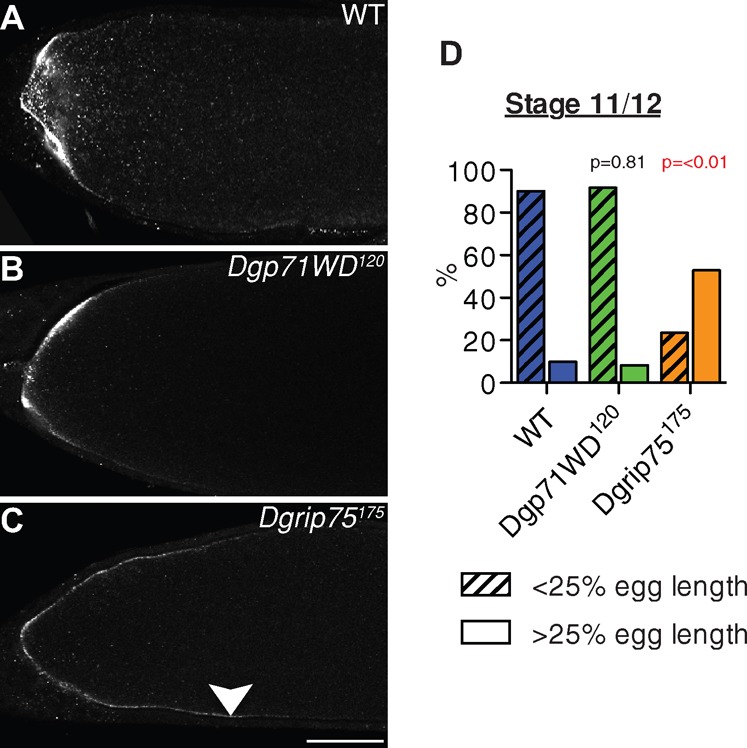
(A–C) bcd mRNA fluorescent in situ hybridisation in representative stage 11/12 WT, Dgp71WD120 and Dgrip75175 oocytes. bcd mRNA localises normally to the anterior end of the oocyte in WT and Dgp71WD120 mutants but is mislocalised towards the posterior in Dgrip75175 mutants (white arrowhead). (D) Quantification of bcd mRNA localisation defects in WT, Dgp71WD120 and Dgrip75175 oocytes. Oocytes were scored as having bcd localised either at <25% or >25% egg length. WT, n = 30; Dgp71WD120, n = 36, Dgrip75175, n = 17. Significance testing was conducted using a Chi-squared test. Scale Bar = 50 µm.
Dgp71WD has a crucial role in the formation of the Meiosis I spindle
Embryos laid by mothers lacking γ-TuRC or Augmin components fail to develop, and this appears to be due to earlier defects in the assembly of the acentrosomal female meiotic spindles – although in neither case is spindle assembly completely suppressed (Meireles et al., 2009; Vogt et al., 2006; Wainman et al., 2009). We examined embryos laid by Dgp71WD120 homozygous mothers and found that they also failed to show any signs of development (R.F.R., unpublished observations), although the embryos appeared to have been fertilised. We therefore examined Meiosis I spindles in mature WT and Dgp71WD120/Df(2L)ED119 oocytes stained to reveal the distribution of D-TACC — a marker for the acentrosomal poles of the meiosis I spindle (Cullen and Ohkura, 2001) — α-tubulin and DNA. Visual examination of WT (n = 38) and mutant (n = 40) Meiosis I spindles showed that Dgp71WD120 spindles had much weaker α-tubulin spindle labelling compared to WT spindles (Fig. 5A) and quantification revealed that mutant spindles had a significantly reduced spindle area and spindle width (Fig. 5B; p≤0.001 in both cases). Surprisingly, whereas the majority of WT spindles exhibited strong D-TACC localisation, ∼85% of mutant spindles had weak or no D-TACC staining (Fig. 5D; p≤0.001). Despite these defects, spindle length was unaffected relative to WT, and there were no obvious defects in chromosome alignment (Fig. 5C).
Fig. 5. Dgp71WD120 mutant oocytes have defective Meiosis I spindles.
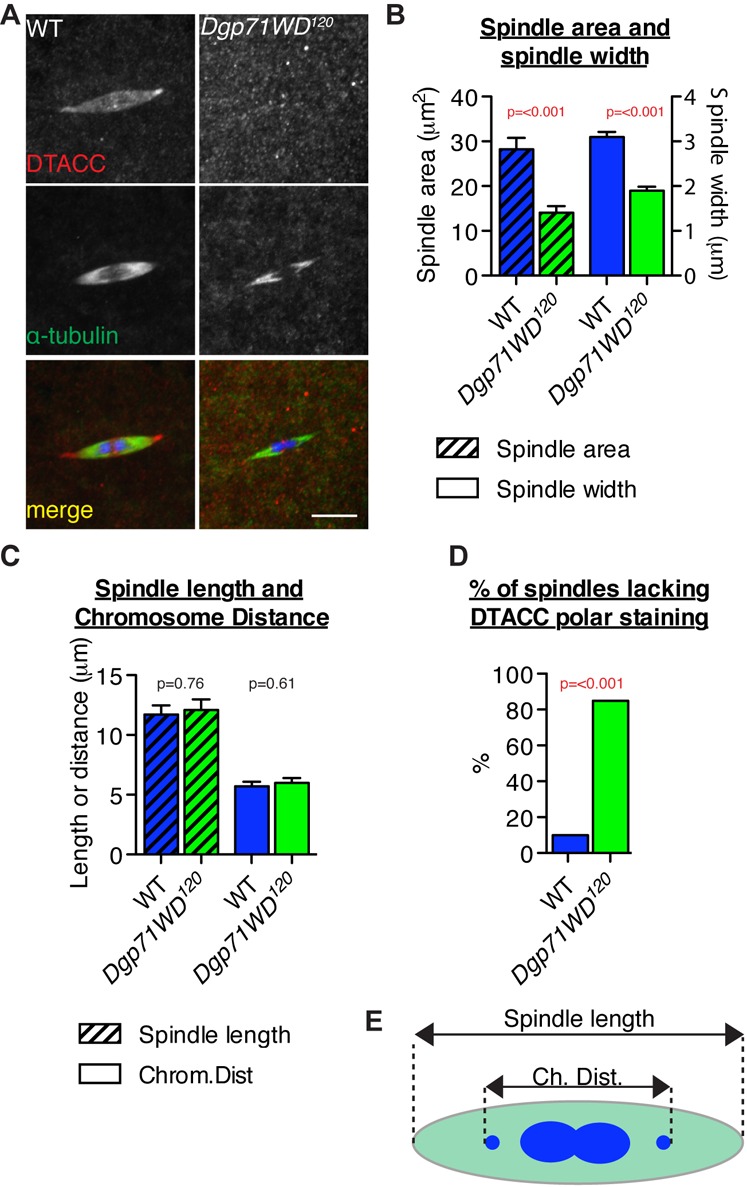
(A) WT (n = 38) and hemizygous Dgp71WD120/Df(2L)ED119 Meiosis I spindles (n = 40) from non-activated oocytes were fixed and stained with anti-DTACC (red) and anti-α-tubulin (green) antibodies; DNA (blue). (B) Dgp71WD120 meiosis I spindles have a significantly reduced spindle area and width compared to WT spindles. (C) Dgp71WD120 meiosis I spindles do not have any significant alteration in spindle length or chromosome positioning. (D) Quantification of the percentage of WT or Dgp71WD120 spindles lacking DTACC polar staining or showing only very weak staining. Significance testing was conducted using Student's t-tests (B,C) and a Chi-squared test (D). Error bars are SEM. Scale bar represents 5 µm. (E) Schematic showing how spindle length and chromosome distance were measured in (C).
Interestingly, these phenotypes are quite distinct from those observed in the Augmin mutant wac, which does not appear to have a reduced spindle area, where no defects in D-TACC localisation are apparent, but where chromosome alignment is disrupted (Meireles et al., 2009). They are also more severe than those observed in the γ-TuRC mutants Dgrip75 and Dgrip128, where there are no detectable defects in Meiosis I spindle assembly but there are defects in Meiosis II spindle assembly (Vogt et al., 2006). Indeed the Meiosis I spindle phenotype in the Dgp71WD120 mutant is almost as severe as that seen in γ-tub37C mutants (Hughes et al., 2011; Tavosanis et al., 1997). Taken together, these observations suggest that Dgp71WD may have functions in the Meiosis I spindle that are independent of Augmin and γ-TuRC function, while Augmin may have a role in aligning meiotic chromosomes that is independent of Dgp71WD and the γ-TuRC.
Discussion
In this study we have generated a near complete deletion of the Dgp71WD gene, allowing us to analyse how cells in vivo cope with the absence of this protein. We find that Dgp71WD has no discernable role at the centrosome in larval brain cells. Instead, Dgp71WD is required for normal spindle and central spindle formation in these cells, where it seems to cooperate with the γ-TuRC and the Augmin complex to nucleate MTs within the forming mitotic spindle. Dgp71WD is not, however, an essential cofactor for the γ-TuRC, as, in contrast to other γ-TuRC components (Schnorrer et al., 2002; Vogt et al., 2006), it is not required for the proper localisation of bcd mRNA during oogenesis, and a Dgp71WD null mutant is male fertile, unlike γ-TuRC mutants. Although Dgp71WD is not essential for cell division in larval brain cells, it is essential for the proper formation of the acentrosomal female meiosis I spindle, where it seems to have functions that are independent of the γ-TuRC and Augmin complexes.
Our finding that Dgp71WD has no detectable function at centrosomes in fly larval brain cells is perhaps surprising. In humans, Nedd1 is essential for recruiting γ-tubulin to centrosomes (Haren et al., 2006; Lüders et al., 2006), and this is also the case in Zebrafish (Manning et al., 2010). Previous studies in Drosophila have indicated that γ-tubulin recruitment to centrosomes is partially disrupted when Dgp71WD is depleted from S2 cells or in brain cells from the original Dgp71WDGE30807 mutant (Dobbelaere et al., 2008; Verollet et al., 2006). While our assays cannot rule out that Dgp71WD has a minor role in γ-tubulin or PCM recruitment in these cells, it seems clear that Dgp71WD cannot act as an essential factor for targeting γ-tubulin to centrosomes in flies. In this light, it is interesting to note that we could also detect no decrease in the amount of γ-tubulin recruited to centrosomes in Dgrip75175 mutant cells, where the formation of the γ-TuRC is disrupted (Vogt et al., 2006). Interpreting this result is not straightforward, as although Dgrip75 depletion abolishes γ-TuRC formation in S2 cells (Verollet et al., 2006), γ-TuRC formation appears to be only partially disrupted in the Dgrip75175 mutant in vivo (Vogt et al., 2006); thus, some residual γ-TuRC complexes may still be present in Dgrip75175 mutant cells. Nevertheless, it is an intriguing possibility that a significant fraction of the centrosomal γ-tubulin in flies may be in the form of the γ-TuSC, rather than the γ-TuRC.
Although it plays at most only a minor part at the centrosome in fly brain cells, Dgp71WD is required to recruit γ-tubulin to the spindle MTs in these cells and it clearly has an important role in forming the mitotic spindle. During mitosis in living WT cells, k-fibres appear to be rapidly reinforced by the formation of additional spindle MTs, a process that appears to be lacking in Dgp71WD, Augmin and γ-TuRC mutant cells. Similar observations have been made in Drosophila S2 cells (Goshima et al., 2008). These observations support the hypothesis that the role of Dgp71WD at the mitotic spindle in flies is to target the γ-TuRC to the spindle Augmin complex, thus reinforcing spindle MT density (Wainman et al., 2009), similar to the role of Nedd1 in human cells (Uehara et al., 2009; Zhu et al., 2008b). In addition to their role in bipolar spindle formation, our data suggests that Dgp71WD, Wac and Dgrip75 are necessary for the formation of a normal central spindle, as is also the case in human cells in culture (Uehara and Goshima, 2010) and Drosophila S2 cells (J.G. Wakefield, personal communication).
Our data shows that Dgp71WD has an important role in the assembly of the acentrosomal female meiosis I spindle. Interestingly, the absence of Dgp71WD results in a more severe spindle phenotype than that caused by the absence of either the Augmin component Wac, or the γ-TuRC components Dgrip75 or Dgrip128 (Meireles et al., 2009; Vogt et al., 2006). Whilst Meiosis I spindles in the Augmin mutant wac have a relatively normal MT density but show defects in chromosome alignment, (Meireles et al., 2009), the Dgp71WD120 mutant spindles have a low MT density and normal chromosome alignment. It is important to note, however, that while the wac and Dgrip75175 mutations are nulls, the lack of either protein may not completely abolish Augmin or γ-TuRC function, respectively. It is possible, therefore, that meiosis I spindles that completely lacked Augmin or γ-TuRC function would have a similar phenotype to that seen in the Dgp71WD or γ-tub37C null mutants.
Intriguingly, a recent study has shown that γ-tub37C mutant spindles have both a low MT density and chromosome alignment defects (Hughes et al., 2011). It is thus tempting to speculate that γ-tubulin 37C may act primarily with Dgp71WD to increase spindle MT density, and with Augmin to drive chromosome alignment. It is also striking that an absence of Dgp71WD (this study) or γ-tubulin 37C (Hughes et al., 2011) but not Wac (Meireles et al., 2009) leads to a severe loss of D-TACC from the meiosis I spindle. TACC proteins are known to stabilise MTs in several situations through their association with the Msps/ch-TOG family of MAPs (Cullen et al., 1999; Gergely et al., 2003; Lee et al., 2001). This could suggest that Dgp71WD or γ-tubulin are necessary to recruit D-TACC to the spindle, although we could find no evidence of any interaction between Dgp71WD and DTACC via co-immunoprecipitation (R.F.R., unpublished observations). Alternatively, the very low density of spindle MTs in Dgp71WD120 mutants may mean that only a limited amount of D-TACC is recruited to the poles, which is not detectable in our analysis. Interestingly, whilst DTACC and Msps mutants often have tripolar spindles, we did not note the presence of tripolar spindles in Dgp71WD120 mutant spindles. This may support the hypothesis that limited amounts of these proteins are still recruited to the poles in Dgp71WD120 mutant meiosis I spindles, and that this is enough to maintain spindle bipolarity. Clearly further work will be required to resolve this important issue.
Materials and Methods
Fly stocks
Standard fly techniques were used (Ashburner et al., 2005). w67 flies were used as controls in all experiments. Dgp71WD mutants were generated by remobilisation of a P-element inserted upstream of the Dgp71WD gene in the Dgp71WDGE30807 stock (Verollet et al., 2006). γ-tubulin-GFP (Hallen et al., 2008) and Jupiter-GFP (Karpova et al., 2006), were introduced into the Dgp71WD120 (this study), Dgrip75175 (Schnorrer et al., 2002) and wac (Meireles et al., 2009) genetic backgrounds using standard genetic crosses. Double mutants with Dsas4S2214 (Basto et al., 2006), Dgrip75175 (Schnorrer et al., 2002; Vogt et al., 2006) and wac were created using standard crosses or recombinations. Most double mutant combinations were examined as homozygotes, however Dgp71WD120;wac was also examined as hemizygotes over the appropriate deficiencies (Df(2L)Exel7071 or Df(2L)ED119 which uncover Dgp71WD, and Df(3L)BSC125 which uncovers wac; Bloomington stock centre, Bloomington, Indiana, USA).
Western blotting and immunofluorescence analysis
Samples were subjected to standard SDS-PAGE using 3–8% pre-cast NuPAGE polyacrylamide gels (Invitrogen, Paisley, UK) and a Mini Trans-Blot cell (BioRad, Hemel Hempstead, Herts, UK). Western blotting was conducted using PVDF membrane (GE Healthcare, Amersham, UK). Membranes were probed with appropriate primary antibodies, washed, probed with appropriate HRP-conjugated secondary antibodies (GE Healthcare), incubated with Supersignal chemiluminescent reagent (Thermo Fisher Scientific/Pierce, Rockford, IL, USA) and then exposed to X-ray film (GE Healthcare).
Preparation, fixation, and staining of squashed third instar larval brains was carried out as described previously (Martinez-Campos et al., 2004). Appropriate Alexa Fluor secondary antibodies were obtained from Invitrogen, and DNA was labelled with Hoechst 33342 (Invitrogen). The following primary antibodies were used for western blotting and immunofluorescence: 1:400 anti-Dgp71WD (Verollet et al., 2006), 1:500 anti-γ-tubulin (GTU88; Sigma, St Louis, MO, USA), 1:500 anti-Cnn (Lucas and Raff, 2007), 1:500 anti-DSpd2 (Dix and Raff, 2007), 1:500 anti-Asl (Conduit et al., 2010), 1:1000 anti-actin (Sigma), and 1:500 Dm1α anti-α-tubulin (Sigma). Imaging of fixed brains was performed on an Olympus Fluoview FV1000 IX81 confocal microscope system using a 60× oil immersion lens and Olympus FV1000 software (Olympus, Southend-on-Sea, Essex, UK).
To quantify the amount of centrosomal PCM components, optical sections were reconstructed in 3D using Volocity software (Perkin Elmer/Improvision, Waltham, MA, USA). Centrosomes were identified semi-automatically using Volocity by finding objects above a minimum volume with an intensity >3 standard deviations from the mean intensity of the image. The intensity of every centrosomal pixel was then summed to produce an intensity score. To quantify spindle fluorescence, maximum intensity Z projections were first generated using Volocity. A triangle was then drawn around each half spindle, and pixel intensities for the relevant channel in this area summed. These two values were then averaged to produce an intensity score for that spindle. A Student's t-test was then used to test for significant differences between genotypes. Centrosomes or spindles in at least 7 cells per a minimum of 5 brains were quantified per genotype.
Live cell imaging
Live imaging of live 3rd instar neuroblasts was performed as described previously (Basto et al., 2006), but with a Perkin Elmer ERS Spinning Disk Confocal system (Perkin Elmer, Waltham, MA, USA) mounted on a Zeiss Axiovert 200M microscope using a 63×, 1.4NA oil objective (Zeiss, Welwyn Garden City, Herts, UK). The entire depth of the cell was imaged by taking Z-stacks spaced 0.5 mm apart at 20 second intervals from before Nuclear Envelope Breakdown (NEB) until telophase. To quantify the amount of γ-tubulin-GFP at the centrosome, sections were reconstructed in 3D using Volocity software. Centrosomes were identified semi-automatically based on their difference from the background value, and all centrosome voxels were then summed to produce an intensity score. To measure the density of Jupiter-GFP at the spindles (as a proxy for MT density), we used a method adapted from Goshima et al. (Goshima et al., 2008). Fiji software (EMBL, Heidelberg, Germany) was used to draw a line intensity profile between the poles. The values at 1/4 and 3/4 along the line were taken to be the spindle values. These values were then averaged to create a value for that individual spindle, and these values averaged across spindles. To measure the length of spindles, we used Fiji to draw a line between the two poles. Student's t-tests were used to test for significances between genotypes.
Analysis of bicoid localisation in oocytes
RNA probes were generated from in vitro transcription of linearised full length bcd cDNA plasmid, obtained from (Driever et al., 1990). T7 was then used with a Digoxigenin RNA labelling mix (Roche, Basel, Switzerland) to in vitro transcribe labelled RNA. Ovaries were dissected in PBT (PBS + 0.1% Triton), then fixed in 8% formaldehyde for 10 minutes, before being washed with PBT and methanol. For the hybridisation, oocytes were rehydrated in 1:1 methanol:PBT for 5 minutes followed by several washes in PBT. They were then washed for 15 minutes in 1:1 PH:PBT, followed by a 20 minute wash in PH at 70°C (PH = Prehybridisation solution: 50% deionised Formamide, 5× SSC in ddH2O, adjusted to pH 6.8 with HCl). Oocytes were then prehybridised in 100 µl hybridisation solution (10 ml PH plus 20 µl tRNA (20 mg/ml), 10 µl ssDNA (10 mg/ml), 5 µl Heparin (50 mg/ml) at 70°C for one hour before the addition of 1 µl of RNA probe. Hybridisation was allowed to take place at 70°C overnight, followed by 30 minute washes at 70°C with PH, 1:1 PH:PBT and then several 20 minute washes of PBT at room temperature. Oocytes were then incubated with 1:200 mouse anti-Digoxigenin (Roche) conjugated with Cy5 in PBT for 1 hour at room temperature, or overnight at 4°C. Antibody solution was then washed off with PBT before addition of Vectashield (Vectorlabs, Peterborough, UK) mounting media and storage at −20°C prior to imaging. Imaging was conducted on a Zeiss 510 Meta Multiphoton Confocal using LSM 510 image capture software (Zeiss, Welwyn Garden City, Herts, UK). bcd localisation along the length of the oocyte axis was measured using the ruler function in ImageJ (NIH, Bethesda, MD, USA).
Analysis of Meiosis I spindles
Preparation, fixation and immunostaining of Meiosis I spindles in non-activated oocytes was described previously (Cullen and Ohkura, 2001). Images were taken as z-stacks and maximum intensity projections were generated using a LSM 510 Zeiss confocal microscope and software (Zeiss). Volocity was used for quantification. The line measurement tool was used to measure spindle length and width and chromosome distance (chromosome mass length, including the 4th chromosome). The freehand ROI tool was used to measure spindle area.
Supplementary Material
Acknowledgments
We thank Brigitte Raynaud-Messina for generously providing antibodies, and Alan Wainman for advice on microscopy and critical comments on the manuscript. Supported by grants from the MRC to R.F.R., from CR-UK to J.W.R. and from the Wellcome Trust to H.O. and N.C. (081849, 092076). J.D. was supported by a postdoctoral fellowship from the Human Frontier Science Program and the Austrian Science Fund (FWF) M1293. L.W. and D.St.J were supported by the Wellcome Trust.
Footnotes
Competing interests: No competing interests declared.
References
- Ashburner M., Golic K. G., Hawley R. S. (2005). Drosophila: A Laboratory Handbook Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press. [Google Scholar]
- Barbosa V., Yamamoto R. R., Henderson D. S., Glover D. M. (2000). Mutation of a Drosophila gamma tubulin ring complex subunit encoded by discs degenerate-4 differentially disrupts centrosomal protein localization. Genes Dev. 14, 3126–3139 10.1101/gad.182800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., Khodjakov A., Raff J. W. (2006). Flies without centrioles. Cell 125, 1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Glover D. M. (2007). Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463 10.1038/nrm2180 [DOI] [PubMed] [Google Scholar]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., Avidor-Reiss T. (2008). Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics 180, 2081–2094 10.1534/genetics.108.095141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M. G., Gatti M. (2000). Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat. Cell Biol. 2, 54–56 10.1038/71378 [DOI] [PubMed] [Google Scholar]
- Colombie N., Verollet C., Sampaio P., Moisand A., Sunkel C., Bourbon H. M., Wright M., Raynaud-Messina B. (2006). The Drosophila gamma-tubulin small complex subunit Dgrip84 is required for structural and functional integrity of the spindle apparatus. Mol. Biol. Cell. 17, 272–282 10.1091/mbc.E05-08-0722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P. T., Brunk K., Dobbelaere J., Dix C. I., Lucas E. P., Raff J. W. (2010). Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20, 2178–2186 10.1016/j.cub.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Cullen C. F., Ohkura H. (2001). Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3, 637–642 10.1038/35083025 [DOI] [PubMed] [Google Scholar]
- Cullen C. F., Deak P., Glover D. M., Ohkura H. (1999). mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol. 146, 1005–1018 10.1083/jcb.146.5.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix C. I., Raff J. W. (2007). Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759–1764 10.1016/j.cub.2007.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Josue F., Suijkerbuijk S., Baum B., Tapon N., Raff J. (2008). A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Siegel V., Nusslein-Volhard C. (1990). Autonomous determination of anterior structures in the early Drosophila embryo by the bicoid morphogen. Development 109, 811–820. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N. S., Yu Q. D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D. M. (2010). Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Gadde S., Heald R. (2004). Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14, R797–R805 10.1016/j.cub.2004.09.021 [DOI] [PubMed] [Google Scholar]
- Gergely F., Draviam V. M., Raff J. W. (2003). The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17, 336–341 10.1101/gad.245603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta K. L., Jang J. K., Manheim E. A., Subramanian G., McKim K. S. (2002). subito encodes a kinesin-like protein required for meiotic spindle pole formation in Drosophila melanogaster. Genetics 160, 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria M. A., Rath U., Buster D. W., Chanda S. K., Caldwell J. S., Rines D. R., Sharp D. J. (2007). Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17, 1960–1966 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Goshima G., Kimura A. (2009). New look inside the spindle: microtubule-dependent microtubule generation within the spindle. Curr. Opin. Cell Biol. 22, 44–49 10.1016/j.ceb.2009.11.012 [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. (2007). Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316, 417–421 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., Vale R. D. (2008). Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane R. N., Martin O. C., Zheng Y. X. (2003). Characterization of a new gamma TuRC subunit with WD repeats. Mol. Biol. Cell 14, 1017–1026 10.1091/mbc.E02-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallen M. A., Ho J., Yankel C. D., Endow S. A. (2008). Fluorescence recovery kinetic analysis of gamma-tubulin binding to the mitotic spindle. Biophys. J. 95, 3048–3058 10.1529/biophysj.108.134593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E., Oegema K., Kirkham M., Gonczy P., Habermann B., Hyman A. A. (2002). The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J. Cell Biol. 157, 591–602 10.1083/jcb.200202047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Remy M. H., Bazin I., Callebaut I., Wright M., Merdes A. (2006). NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172, 505–515 10.1083/jcb.200510028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. E., Beeler J. S., Seat A., Slaughter B. D., Unruh J. R., Bauerly E., Matthies H. J., Hawley R. S. (2011). Gamma-tubulin is required for bipolar spindle assembly and for proper kinetochore microtubule attachments during prometaphase I in Drosophila oocytes. PLoS Genet. 7, e1002209 10.1371/journal.pgen.1002209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y., Soung N. K., Park J. E., Yu L. R., Zhou M., Bang J. K., Kim B. Y., Veenstra T. D., Erikson R. L., Lee K. S. (2011). Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl. Acad. Sci. USA 108, 11446–11451 10.1073/pnas.1106223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi H. C., Palacios M. J., McNamara L., Cleveland D. W. (1992). Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature 356, 80–83 10.1038/356080a0 [DOI] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., Debec A. (2006). Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton 63, 301–312 10.1002/cm.20124 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Cole R. W., Oakley B. R., Rieder C. L. (2000). Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10, 59–67 10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Lee M. J., Gergely F., Jeffers K., Peak-Chew S. Y., Raff J. W. (2001). Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3, 643–649 10.1038/35083033 [DOI] [PubMed] [Google Scholar]
- Liu L., Wiese C. (2008). Xenopus NEDD1 is required for microtubule organization in Xenopus egg extracts. J. Cell Sci. 121, 578–589 10.1242/jcs.018937 [DOI] [PubMed] [Google Scholar]
- Lucas E. P., Raff J. W. (2007). Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila Centrosomin. J. Cell Biol. 178, 725–732 10.1083/jcb.200704081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Stearns T. (2007). Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8, 161–167 10.1038/nrm2100 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U. K., Stearns T. (2006). GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Ma W., Baumann C., Viveiros M. M. (2010). NEDD1 is crucial for meiotic spindle stability and accurate chromosome segregation in mammalian oocytes. Dev. Biol. 339, 439–450 10.1016/j.ydbio.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Manning J. A., Lewis M., Koblar S. A., Kumar S. (2010). An essential function for the centrosomal protein NEDD1 in zebrafish development. Cell Death Differ. 17, 1302–1314 10.1038/cdd.2010.12 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J. W. (2004). The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673–683 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H. J., McDonald H. B., Goldstein L. S., Theurkauf W. E. (1996). Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134, 455–464 10.1083/jcb.134.2.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Hawley R. S. (1995). Chromosomal control of meiotic cell division. Science 270, 1595–1601 10.1126/science.270.5242.1595 [DOI] [PubMed] [Google Scholar]
- Megraw T. L., Li K., Kao L. R., Kaufman T. C. (1999). The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829–2839. [DOI] [PubMed] [Google Scholar]
- Meireles A. M., Fisher K. H., Colombie N., Wakefield J. G., Ohkura H. (2009). Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J. Cell Biol. 184, 777–784 10.1083/jcb.200811102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier-Pavie V., Cenci G., Verni F., Gatti M., Bonaccorsi S. (2011). Phenotypic analysis of misato function reveals roles of noncentrosomal microtubules in Drosophila spindle formation. J. Cell Sci. 124, 706–717 10.1242/jcs.072348 [DOI] [PubMed] [Google Scholar]
- Riechmann V., Ephrussi A. (2001). Axis formation during Drosophila oogenesis. Curr. Opin. Genet. Dev. 11, 374–383 10.1016/S0959-437X(00)00207-0 [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Luschnig S., Koch I., Nusslein-Volhard C. (2002). Gamma-tubulin37C and gamma-tubulin ring complex protein 75 are essential for bicoid RNA localization during drosophila oogenesis. Dev. Cell 3, 685–696 10.1016/S1534-5807(02)00301-5 [DOI] [PubMed] [Google Scholar]
- Sunkel C. E., Gomes R., Sampaio P., Perdigao J., Gonzalez C. (1995). Gamma-tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO J. 14, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavosanis G., Llamazares S., Goulielmos G., Gonzalez C. (1997). Essential role for gamma-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J. 16, 1809–1819 10.1093/emboj/16.8.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y., Uetake Y., Kuriyama R. (2003). Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 162, 757–763 10.1083/jcb.200305048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Goshima G. (2010). Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol. 191, 259–267 10.1083/jcb.201004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R., Nozawa R. S., Tomioka A., Petry S., Vale R. D., Obuse C., Goshima G. (2009). The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 106, 6998–7003 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verollet C., Colombie N., Daubon T., Bourbon H. M., Wright M., Raynaud-Messina B. (2006). Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172, 517–528 10.1083/jcb.200511071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N., Koch I., Schwarz H., Schnorrer F., Nusslein-Volhard C. (2006). The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963–3972 10.1242/dev.02570 [DOI] [PubMed] [Google Scholar]
- Wainman A., Buster D. W., Duncan T., Metz J., Ma A., Sharp D., Wakefield J. G. (2009). A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 23, 1876–1881 10.1101/gad.532209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Salmon E. (1997). Pathways of spindle assembly. Curr. Opin. Cell Biol. 9, 37–43 10.1016/S0955-0674(97)80149-4 [DOI] [PubMed] [Google Scholar]
- Wiese C., Zheng Y. (2006). Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 119, 4143–4153 10.1242/jcs.03226 [DOI] [PubMed] [Google Scholar]
- Wilson P. G., Borisy G. G. (1998). Maternally expressed gamma Tub37CD in Drosophila is differentially required for female meiosis and embryonic mitosis. Dev. Biol. 199, 273–290 10.1006/dbio.1998.8900 [DOI] [PubMed] [Google Scholar]
- Zhang J., Megraw T. L. (2007). Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell 18, 4037–4049 10.1091/mbc.E07-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Lawo S., Bird A., Pinchev D., Ralph A., Richter C., Muller-Reichert T., Kittler R., Hyman A. A., Pelletier L. (2008a). The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18, 136–141 10.1016/j.cub.2007.12.055 [DOI] [PubMed] [Google Scholar]
- Zhu H., Coppinger J. A., Jang C. Y., Yates J. R., 3rd, Fang G. (2008b). FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 183, 835–848 10.1083/jcb.200807046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.