Summary
The zebrafish is a widely used model animal to study the regeneration of organs, such as the fin and heart. Their average lifetime is about 3 years, and recent studies have shown that zebrafish exhibit aging-related degeneration, suggesting the possibility that aging might affect regenerative potential. In order to investigate this possibility, we compared regeneration of the fin and heart after experimental amputation in young (6–12 month old) and old (26–36 month old) fish. Comparison of recovery rate of the caudal fin, measured every two or three days from one day post amputation until 13 days post amputation, show that fins in young and old fish regenerate at a similar rate. In the heart, myocardium regeneration and cardiomyocyte proliferation occurred similarly in the two groups. Moreover, neo-vascularization, as well as activation of fibroblast growth factor signaling, which is required for neo-vascularization, occurred similarly. The epicardial tissue is a thin layer tissue that covers the heart, and starts to express several genes immediately in response to injury. The expression of epicardial genes, such as wt1b and aldh1a2, in response to heart injury was comparable in two groups. Our results demonstrate that zebrafish preserve a life-long regenerative ability of the caudal fin and heart.
Keywords: Zebrafish, Regeneration, Aging, Heart, Fin
Introduction
The zebrafish has become a popular model animal to examine a variety of biological processes (Lieschke and Currie, 2007). In particular, the high ability to regenerate a variety of organs has made the zebrafish a suitable animal model for organ regeneration research (Brittijn et al., 2009; Tal et al., 2010). Regeneration of the caudal fin after experimental amputation has been appreciated for a long period of time (Morgan, 1900; Santamaría and Becerra, 1991). Other fins, such as pectoral, pelvic, anal, caudal and dorsal fins also regenerate after amputation (Kawakami et al., 2006; Nachtrab et al., 2011). In the last decade, the zebrafish has also become a model animal for regeneration of the heart (Poss et al., 2002; Raya et al., 2003). In addition, this animal provides experimental systems to study regeneration of the liver (Sadler et al., 2007; Kan et al., 2009), mechanosensory organs (Dufourcq et al., 2006; Ma et al., 2008; LeClair and Topczewski, 2010), retina (Hitchcock and Raymond, 2004), axons in the central nervous system (Becker and Becker, 2007) and cerebellum (Liu et al., 2004).
The caudal fin is a particularly efficient model system for regeneration. Upon amputation, epidermal cells migrate and cover the wound site, and form a specialized tissue, the apical epithelial cap. Underlying mesenchymal cells communicate with the apical epithelial cap, and form the blastema, which is considered as a mass of de-differentiated cells (Akimenko et al., 2003; Poss et al., 2003). Recent analyses demonstrated that those de-differentiated cells are lineage-restricted cells, and cells in the regenerated fin are derived from the same type of cells (Knopf et al., 2011; Tu and Johnson, 2011). The blastema cells proliferate and differentiate (Lee et al., 2005), leading to addition of new segments to the distal end of the fin until the original size of the fin is restored within two weeks.
In addition to the fin, studies in the last decade have demonstrated that the zebrafish is a suitable model animal for heart regeneration studies (Raya et al., 2004; Poss, 2007; Ausoni and Sartore, 2009; Laflamme and Murry, 2011). The zebrafish heart regenerates from a variety of injuries, such as ventricular amputation (Poss et al., 2002; Raya et al., 2003), cryoprobe-induced injury (Chablais et al., 2011; González-Rosa et al., 2011; Schnabel et al., 2011) and transgenic induction of a toxin in cardiomyocytes (Wang et al., 2011). The regeneration of the heart involves rapid activation of epicardial cells, proliferation of cardiomyocytes to restore the myocardial layer, and neo-vascularization of the regenerating area (Poss et al., 2002; Raya et al., 2003; Lepilina et al., 2006; Kim et al., 2010). Prior to the finding of lineage restriction in the regenerating fins, genetic labeling studies by an inducible Cre-loxP system identified that a vast majority of cardiomyocytes in the regenerated area were derived from pre-existing cardiomyocytes that underwent de-differentiation and proliferation (Jopling et al., 2010; Kikuchi et al., 2010). Coordinated proliferation of cardiomyocytes and neo-vascularization leads to the restoration of the lost cardiac tissue and function.
Recently, zebrafish also have become a model animal for aging research (Keller and Murtha, 2004; Gerhard, 2007; Kishi et al., 2009). On average, the zebrafish life span is 3 years (Gerhard et al., 2002). Upon aging, they exhibit senescence-associated β-galactosidase activities in the skin, oxidized protein accumulation in muscle, increased accrual in the liver, and retinal atrophy (Tsai et al., 2007; Kishi et al., 2008; Kishi et al., 2009).
Compared to zebrafish, studies in mammals have shown an aging related decline in regenerative potentials, such as in skeletal muscle (Conboy et al., 2005), liver (Iakova et al., 2003), hematopoietic cells (Janzen et al., 2006), pancreas islet (Krishnamurthy et al., 2006), and neuronal cells (Kuhn et al., 1996). In the case of zebrafish, several recent studies described the relationship between aging and fin regeneration. Repeated amputation of the caudal fin, which induces continuous cell division for regeneration, did not affect regeneration (Azevedo et al., 2011). Shortening of the telomere length is associated with aging-related senescence in mammals (Harley et al., 1990). Repeated fin amputation, however, also did not affect the activity of telomerase, the enzyme that protects the telomere and allows cells to undergo continuous cell division (Azevedo et al., 2011). Similarly, zebrafish at 24 months old and 3 months old exhibited similar levels of telomere lengths in the fin as well as other regenerative organs (Lund et al., 2009). Contrary to these reports, another study showed impaired regeneration and a decrease in telomerase activities in the caudal fin of 24 month old zebrafish (Tsai et al., 2007; Anchelin et al., 2011). These reports suggest that the fin regeneration ability may involve multiple factors. The contribution of aging to the regenerative ability in zebrafish remains controversial.
In this manuscript, we re-investigated the relationship between fin regeneration and aging using the same zebrafish colony that was examined for telomere lengths (Lund et al., 2009). Analysis of regenerated length of the fin at multiple time-points shows comparable fin regeneration in young and old zebrafish, consistent with the recently reported telomere lengths (Lund et al., 2009). Furthermore, we examined whether aging affects regeneration of the heart. Our analysis shows that cardiomyocyte proliferation (Jopling et al., 2010; Kikuchi et al., 2010), neo-vascularization (Lepilina et al., 2006; Kim et al., 2010), and activation of epicardial gene expression (Lepilina et al., 2006; Kikuchi et al., 2011), known to be critical for heart regeneration, occurred similarly in young and old zebrafish. Our study shows that the regenerative ability of the fin and heart is preserved during aging in zebrafish.
Results
Comparable regeneration of the caudal fin in young and old zebrafish
A previous study has shown that telomerase activity is present in a variety of zebrafish organs after aging, and that telomere lengths are comparable in the caudal fin of young and aged fish after two successive rounds of amputation and regeneration (Lund et al., 2009). In this analysis, telomere length was analyzed at 15 days post amputation (dpa), when the caudal fin was fully regenerated. To further investigate whether aging affects fin regeneration in more detail, we used zebrafish housed in the same core facility by the same operation standards, and examined fin regeneration with time-points in young (6–12 months old, n = 12) and old (26–36 months old, n = 12) zebrafish. In addition, we compared fin regeneration at the central area and near the dorsal edge of the fin. These two areas represent the shortest and longest length of lost tissue along the fin ray (Fig. 1G,H).
Fig. 1. Comparable regeneration of the caudal fin in young and old zebrafish.
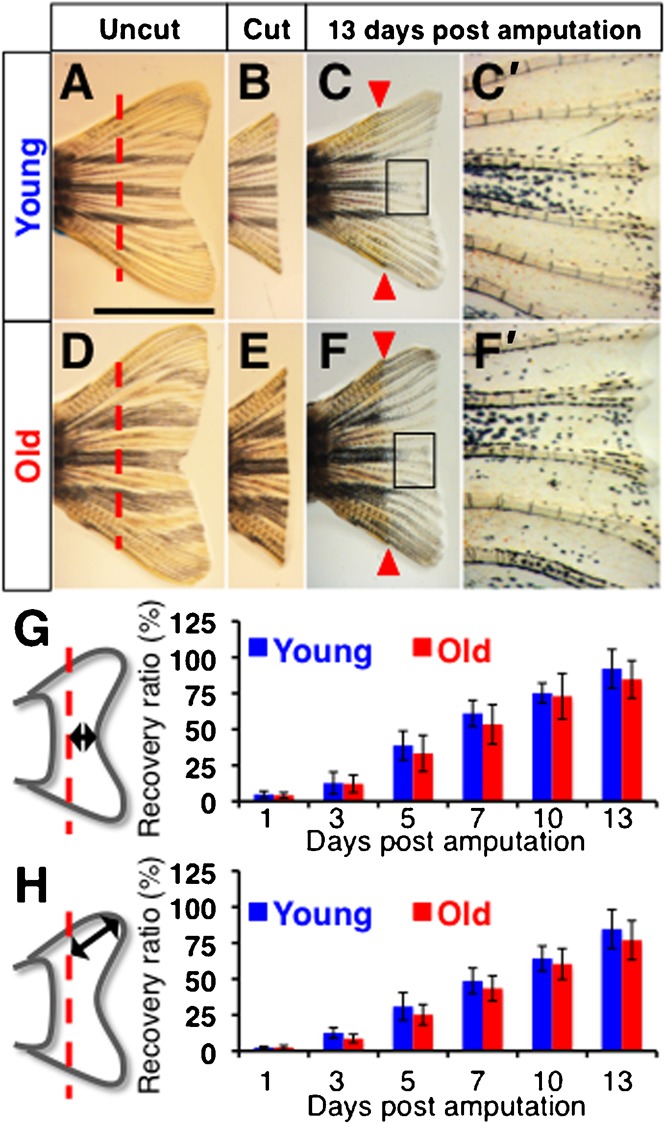
(A–F′) The caudal fin prior to amputation (A,D), immediately after amputation (B,E) and 13 days after amputation (C,C′,F,F′) in young (A–C′) and old (D–F′) fish. C′ and F′ show close ups of the boxed areas in C and F, respectively. Dashed lines in A and D indicate the amputation lines. Arrows in C and F indicate the levels of amputation in the regenerated fin, which is visible by the altered pigmentation pattern. Scale bar: 5 mm; the degree of zoom: ×5. (G,H) Schematic representations of the measured regenerative length at the center (G) and near the dorsal edge (H) of the regenerating fin. The angles between the measured lines along the dorsal edge and the center were 25.1±2.9 degrees and 27.1±4.7 degrees in young and old fish, respectively. The graphs show recovery ratio at indicated time-points from amputation in young and old fish.
To induce fin regeneration, we amputated the caudal fin with a sharp razor blade. The amputation line overlapped with the straight line connecting two bifurcation points of the longest fin rays of the dorsal and ventral side (Fig. 1A,B,D,E). From 1 dpa, we measured regenerated length every two or three days (from the amputated plane to the edge of regenerated tissue), and evaluated the degree of fin regeneration in each fish. We calculated the regeneration ratio by dividing the length of regenerated tissue by the length of the corresponding area of removed fin tissue. At all time-points examined, caudal fin at both the central area and near the dorsal edge exhibited a similar tissue regeneration ratio (Tables 1, 2; Fig. 1G,H). Although young fish showed a slightly higher recovery ratio at all time-points analyzed, the differences were not statistically significant, and the regenerated fin looked comparable in young and old fish (Fig. 1C,C′,F,F′). These results indicate that both young and old fish possess similar fin regeneration abilities, in agreement with the recent analysis of telomere length (Lund et al., 2009).
Table 1. Recovery ratios1 of the shortest fin parts were not significantly different in young and old fish.

Table 2. Recovery ratios1 of the longest fin parts were not significantly different in young and old fish.
Comparable regeneration of the myocardial layer in young and old zebrafish
The heart is one of the major organs, along with the fin, that zebrafish contribute to regeneration research (Raya et al., 2004; Poss, 2007; Ausoni and Sartore, 2009; Laflamme and Murry, 2011). Thus, we next asked whether aging affects heart regeneration in zebrafish. We injured zebrafish hearts by amputating the apex of the ventricle (Poss et al., 2002; Raya et al., 2003). The survival rate of young fish and old fish at one day after surgery was 96.7% (n = 29/30) and 95.8% (n = 23/24), respectively. All fish that survived one day after surgery did not die until analysis (3–30 dpa). Thus, old fish did not seem to be sensitive to heart surgery compared to young fish.
Restoration of the myocardium is essential for heart regeneration (Poss et al., 2002; Raya et al., 2003). Thus, we first examined regeneration of the myocardium by immunostaining for myosin heavy chain (MHC) using the MF20 antibodies (Poss et al., 2002). The regenerating area of each heart can be determined by the morphological difference from the uninjured area. In young fish, cardiomyocytes were detected in the regenerating area as early as 7 dpa (Fig. 2A,A′) (n = 4). At this time-point, MHC signals in the regenerating area were weaker and sparser than those in uninjured area. At 14 dpa, the MHC signal in the regenerating area became more intense in a wider region (Fig. 2C,C′) (n = 5). At 30 dpa, the injured area was filled by cardiomyocytes with strong MHC signals (Fig. 2E,E′) (n = 5), and became nearly indistinguishable from the uninjured area. In old fish, we detected MHC-positive cardiomyocytes in the regenerating area at 7 dpa (Fig. 2B,B′) (n = 3), similar to young fish. This was followed by detection of more intense MHC signals in a wider region at 14 dpa (Fig. 2D,D′) (n = 5), and regenerated myocardium at 30 dpa (Fig. 2F,F′) (n = 4). These data indicate that regeneration of the myocardium occurred similarly in both young and old fish.
Fig. 2. Regeneration of the myocardial layer and induction of Gata4 expression in young and old fish hearts.

(A–F′) MHC staining of regenerating hearts at 7 dpa (A–B′), 14 dpa (C–D′) and 30 dpa (E–F′) in young (A,A′,C,C′,E,E′) and old (B,B′,D,D′,F,F′) fish. A′–F′ show higher magnification images of the boxed areas in A–F. Open arrowheads indicate MHC signals in the regenerating area. For simplicity, not all signals are pointed. Scale bar: 50 μm; the degree of zoom: ×5.5. (G,H) Gata4 immunoreactivity was not detected immediately after amputation. H shows a close up of the boxed areas in G. Scale bar: 50 μm; the degree of zoom: ×2. (I–J′) Gata4 staining of regenerating hearts at 7 dpa in young (I,I′) and old (J,J′) fish. Black arrowheads indicate Gata4 signals. I′ and J′ show close ups of the boxed areas in I and J. Sections were counterstained with hematoxylin. Dotted lines indicate the amputated planes of 7 and 14 dpa hearts. Brackets indicate the regenerating areas of 30 dpa hearts. Scale bar: 50 µm; the degree of zoom: ×2.
A recent report demonstrated that cardiomyocytes in the outer compact muscle layer expressed a fluorescent reporter driven by upstream sequences of the gata4 gene in response to ventricular amputation (Kikuchi et al., 2010). These cardiomyocytes were shown to contribute to the regenerated myocardium. Thus, we examined endogenous expression of Gata4 protein by immunohistochemistry after heart injury. Gata4 immunoreactivity was not detected in both the injured and non-injured areas immediately after amputation (Fig. 2G,H). At 7 dpa, we observed expression of immunoreactive Gata4 protein in the outer compact muscle layer and at the surface of the regenerating area, similarly in both young and old fish hearts (Fig. 2I,I′,J,J′) (n = 3 for both young and old fish). Consistent with the contribution of the gata4-reporter-expressing cells to the regenerated myocardium, both the Gata4 immunoreactivity-positive area (Fig. 2I,I′,J,J′) and the MHC signal-positive area (Fig. 2A,A′,B,B′) were observed at the surface of the regenerating area at 7 dpa. These data show that both young and old fish hearts respond to injury and express Gata4 protein similarly.
Similar levels of cardiomyocyte proliferation in young and old fish during heart regeneration
The proliferation of pre-existing cardiomyocytes after de-differentiation is a critical factor for successful regeneration of the myocardium (Jopling et al., 2010; Kikuchi et al., 2010). Therefore, we next analyzed proliferation events in the regenerating heart by immunofluorescent detection of proliferating cell nuclear antigen (Pcna, an S-phase marker) at 14 dpa. A previous report showed that cardiomyocytes proliferate actively at this time-point after ventricular amputation (Poss et al., 2002). Sections were also stained for Mef2, a nuclear marker of cardiomyocytes in regenerating zebrafish hearts (Wang et al., 2011), which allows proliferating cardiomyocytes to be distinguished from other proliferating cells (Fig. 3A,B). First, we counted the total number of proliferating cells (Pcna positive nuclei) in sections around the center of the regenerating area in each heart. We did not detect a significant difference in the number of proliferating cells in regenerating hearts between young and old fish (Fig. 3C) (n = 5 in both young and old fish). To analyze cardiomyocyte proliferation, we identified cells expressing both Pcna and Mef2, as proliferating cardiomyocytes (Fig. 3A,B, yellow arrowhead). The numbers of Pcna/Mef2 double positive cells in regenerating hearts of young and old fish were at similar levels (Fig. 3D) (n = 5 in both young and old fish). Thus, both young and old fish exhibited similar levels of cardiomyocyte proliferation and proliferation of other types of cells in regenerating hearts. We further compared the densities of cardiomyocytes in the regenerating area by dividing the number of Mef2 positive nuclei by the cubic volume of the regenerating area in 4 µm-thick z-stacked confocal images. Cardiomyocyte density in the regenerating area was at similar levels in young and old fish (Fig. 3E) (n = 5 in both young and old fish).
Fig. 3. Cell proliferation in regenerating young and old fish heart.

(A,B) Immunofluorescence images for Pcna (green) and Mef2 (magenta) of 14 dpa young (A) and old (B) fish hearts. Yellow arrowheads point to Pcna and Mef2 double positive, proliferating cardiomyocytes (shown as white signal). For simplicity, not all proliferating cardiomyocytes are pointed. Dotted lines indicate the amputated planes. Scale bar: 50 µm. (C,D) Number of Pcna-positive proliferating cells per section (C), and Pcna-Mef2 double positive proliferating cardiomyocytes per section (D) of young and old fish hearts at 14 dpa. (E) Densities of cardiomyocytes in the regenerating area in 14 dpa hearts. The p-values between young and old fish are shown. Same slides were examined for C, D and E.
Taken together, our analyses demonstrate that cardiomyocytes respond to heart injury and proliferate, and that the myocardial layer is regenerated similarly in both young and old fish.
Neo-vascularization occurs similarly in regenerating hearts in young and old fish
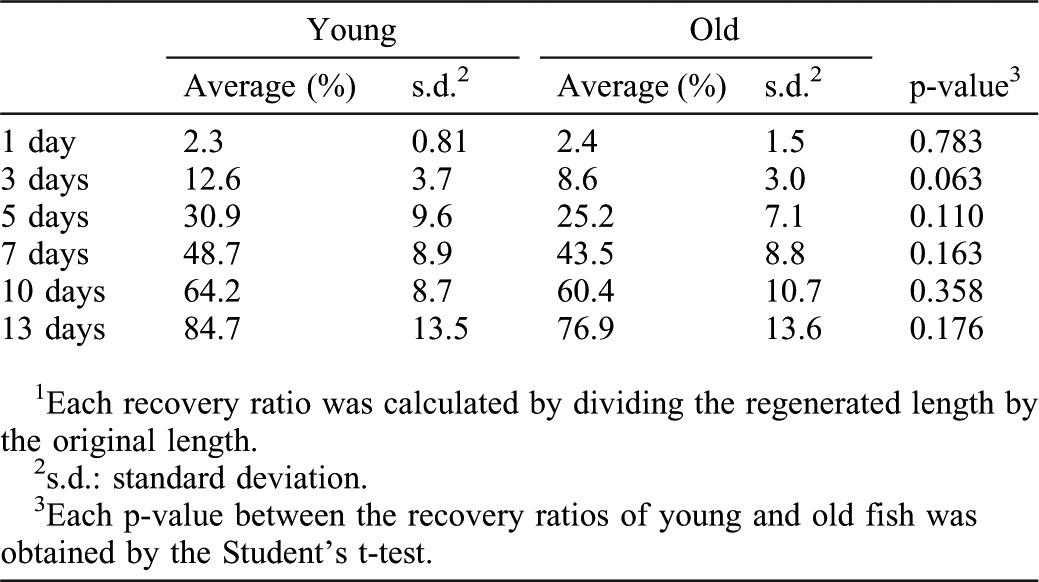
It has been shown that neo-vascularization occurs in the regenerating area concomitantly with myocardial regeneration (Lepilina et al., 2006), and that vascularizing the regenerating myocardial layer is necessary for heart regeneration (Lepilina et al., 2006; Kim et al., 2010). In order to examine whether neo-vascularization is affected by aging, we used a transgenic reporter fish line, fli1:EGFP, which reports vascular endothelial cells to monitor vascularization (Lawson and Weinstein, 2002).
We detected patchy EGFP signal in the regenerating area at 7 dpa in both young and old fish (Fig. 4A,B) (n = 4 in young fish, n = 3 in old fish). This contrasts to the signals of MHC and Gata4 at the surface of the regenerating area (Fig. 2A–B′,I–J′). At 14 dpa, the signal spread in the regenerating area similarly in both young and old fish (Fig. 4C,D) (n = 3 in both young and old fish). In both young and old fish, the EGFP signals were clustered in the regenerating area, compared to the uninjured area at 14 dpa (Fig. 4E,E′,F,F′). To further compare neo-vascularization, we measured EGFP positive areas in the regenerating area using ImageJ software. The ratios of the neo-vascularized area in the regenerating area were 14.6±2.7% and 14.3±1.2% in young and old fish hearts, respectively (Fig. 4G) (n = 3 in both young and old fish). These ratios are similar to the ratios of the vascularized area in the uninjured area in the same images (14.6±3.6% and 16.0±2.2% for young and old fish hearts, respectively). These results indicate that neo-vascularization progressed similarly in both young and old fish heart, and that the regenerating area vascularized to a similar level as the uninjured area by 14 dpa.
Fig. 4. Comparable vascularization and activation of FGF signaling in regenerating hearts of young and old fish.
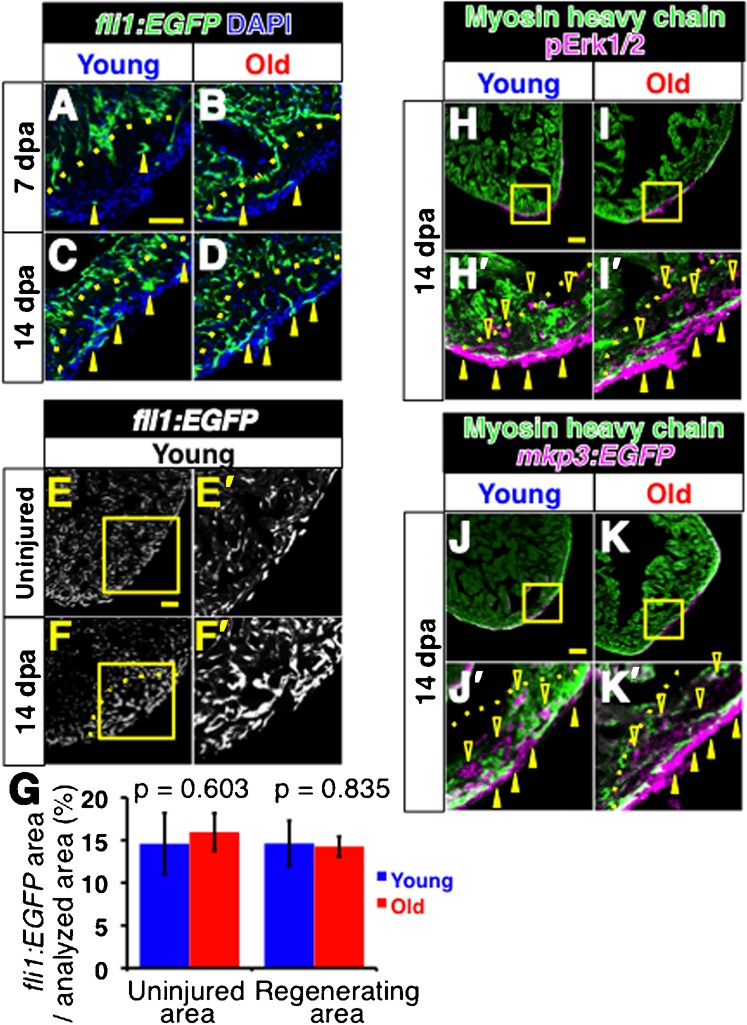
(A–D) Fluorescent images of fli1:EGFP signals of regenerating young (A,C) and old (B,D) fish hearts at 7dpa (A,B) and 14 dpa (C,D). Arrowheads point to the fli1:EGFP signals in the regenerating area. DAPI was used for counterstaining. In C and D, not all signals were labeled for simplicity. (E–F′) fli1: EGFP signals in the uninjured heart (E,E′) and in regenerating heart (F,F′) at 14 dpa in young fish. The signals were detected as small clusters throughout the heart. In the regenerating area, the fli1: EGFP signals formed larger clusters than those in uninjured area. E′ and F′ are close up images of the boxed areas in E and F, respectively. Scale bar: 50 μm; the degree of zoom: ×2. (G) Degree of vascularized areas in the regenerating and uninjured areas. The degree was quantified by the ratio of fli1:EGFP signal-positive area in the regenerating area and the uninjured area from single confocal plane. The p-values between young and old fish are shown. (H–I′) Immunofluorescence images of pErk1/2 (magenta) and MHC (green). Closed and open arrowheads point to the pErk1/2 positive signals at the surface of the heart and inside the regenerating area, respectively. H′ and I′ show close ups of the boxed areas in H and I. Scale bar: 50 μm; the degree of zoom: ×4. (J–K′) Immunofluorescence images of mkp3: EGFP (magenta) and MHC (green). Arrowheads point to the mkp3: EGFP signal positive signals at the surface of the heart and inside the regenerating area, respectively. J′ and K′ show close ups of the boxed areas in J and K. Dotted lines indicate the amputated planes. Scale bar: 50 µm; the degree of zoom: ×4.
Neo-vascularization in the regenerating heart requires activation of fibroblast growth factor (FGF) signaling, and the expression of FGF receptor 2 gene (fgfr2) is highest at 14 dpa in amputated hearts (Lepilina et al., 2006). Thus, we examined activation of FGF signaling during heart regeneration by means of phosphorylation of Erk1/2 (pErk1/2) and expression of map kinase phosphatase 3 (mkp3, also known as dual specificity phosphatase 6, dusp6).
It has been demonstrated that pErk1/2 reports active sites of FGF signaling in vivo (Sawada et al., 2001; Shinya et al., 2001; Corson et al., 2003). We detected pErk1/2 signals at the surface and inside of the regenerating area at 14 dpa in both young and old fish hearts (Fig. 4H,H′,I,I′) (n = 4 in young fish, n = 3 in old fish), consistent with the expression pattern of fgfr2 (Lepilina et al., 2006). mkp3 is a transcriptional target of FGF signaling (Kawakami et al., 2003; Tsang et al., 2004). We monitored expression of mkp3 by means of mkp3-EGFP reporter fish, in which an EGFP cassette is inserted near the mkp3 gene by the sleeping beauty transposon. The EGFP signal in this line has been shown to report mkp3 expression, and hence, active FGF signaling (Balciunas et al., 2004). We detected mkp3-EGFP signal at the surface and inside of the regenerating area of 14 dpa hearts, similarly in both young and old fish (Fig. 4J,J′,K,K′) (n = 2 in both young and old fish).
Taken together, these results indicate that activation of FGF signaling and neo-vascularization occurs similarly during heart regeneration in young and old fish.
Activation of epicardial gene expression occurs similarly in regenerating hearts of young and old fish
The epicardium is a thin tissue that envelops the entire heart. It has been shown that the expression of several developmental genes is activated in epicardial cells during the early phase of heart regeneration, prior to myocardial regeneration and neo-vascularization (Lepilina et al., 2006). In order to further investigate the effects of aging on heart regeneration, we examined activation of expression of wilms tumor 1b (wt1b) and aldehyde dehydrogenase 1a2 (aldh1a2, previously known as retinaldehyde dehydrogenase 2, raldh2) in epicardial cells in response to injury.
It has been shown that expression of wt1b, a gene encoding a zinc finger transcription factor, is induced in the epicardial cells upon heart injury (Schnabel et al., 2011). No wt1b expression was observed immediately after amputation in both young and old fish (Fig. 5A,B) (n = 2 in both young and old fish). At 3 dpa and 7 dpa, we detected strong wt1b expression similarly in the regenerating area in both young and old fish hearts (Fig. 4C–F) (n = 4 for young fish and n = 3 for old fish). Expression of aldh1a2, which encodes a rate-limiting enzyme for retinoic acid synthesis, has been shown to be activated in epicardial cells upon heart injury (Lepilina et al., 2006; Kikuchi et al., 2011). Similar to wt1b expression, in both young and old fish, aldh1a2 was up-regulated at 3 dpa in a wide region of the heart, such as the epicardial cells of the regenerating area and uninjured area, and endocardial cells close to the amputated plane (Fig. 5G,G′,H,H′) (n = 4 in both young and old fish).
Fig. 5. Comparable expression of epicardial genes in regenerating hearts of young and old fish.
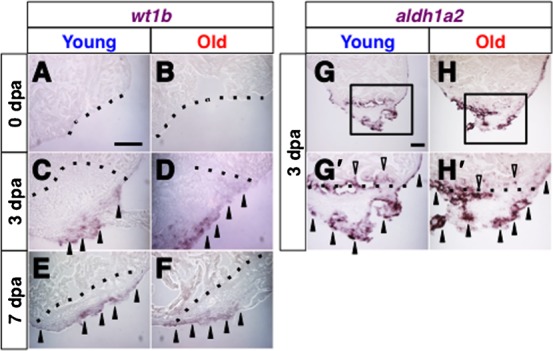
(A–F) In situ hybridization of wt1b immediately after amputation at 0 dpa (A,B), at 3 dpa (C,D) and at 7 dpa (E,F) in regenerating hearts of young (A,C,E) and old (B,D,F) fish. Arrowheads point to the wt1b signals. (G–H′) in situ hybridization of aldh1a2 at 3 dpa in the regenerating hearts of young (G,G′) and old (H,H′) fish. Black arrowheads and open arrowheads point to the aldh1a2 signals in the epicardial tissue and endocardial tissue, respectively. G′ and H′ shows close up images of the boxed areas in G and H. Dotted lines indicate the amputated planes. Scale bar: 50 µm; the degree of zoom: ×2.
These results indicate that gene expression characteristic to the epicardial cells in response to heart injury occurs similarly in young and old fish.
Discussion
Fin regeneration and aging in zebrafish
Several studies showed different results in relation to aging and fin regeneration in zebrafish. For instance, one study shows that telomere length, which should be maintained for continued cell division (Harley et al., 1990), did not change in young (3, 6, 12 month old) and old (24 month old) zebrafish fins (Lund et al., 2009). This study also shows that telomere length is maintained after consecutive fin regeneration. Similarly, another study shows that fin regeneration is not affected by animal aging, and expression of msxb and fgf20a, which are characteristic to regenerating fins (Akimenko et al., 1995; Whitehead et al., 2005), was also not altered in old fish (Shao et al., 2011). In agreement with these reports, our study shows that young and old fish possess the similar ability to regenerate amputated fins. The outgrowth process during fin regeneration was comparable between young (6–12 month old) and old (26–36 month old) fish in our study. Thus, our data support the idea that zebrafish maintains regenerative ability throughout the lifespan.
Contrary to these observations, conflicting results were also reported. A study shows that fin regeneration was impaired and telomerase activity in the regenerating fin was reduced in old fish (24 month old) (Anchelin et al., 2011). Moreover, there is a report of an intermediate observation, showing that some old fish exhibited fin regeneration defects, while other old fish exhibited normal fin regeneration, compared to young fish (Tsai et al., 2007). Such discrepancy suggests that aging itself would not be a primary factor that affects fin regeneration. From this point of view, it is interesting to note that genotoxic stress, such as ionizing radiation, has been suggested to enhance symptoms of senescence (Tsai et al., 2007). Animals are continuously exposed to genotoxic stress from endogenous and environmental sources, which would cause accumulation of DNA damage (Pollycove and Feinendegen, 2003). Moreover, chronic stress that could be caused by specific housing conditions and feeding conditions could cause DNA damage (Antoni et al., 2006; Flint et al., 2007; Hara et al., 2011). Thus, such environmental factors might play a role in affecting the preservation of the regenerative ability of the fin, and aging might modulate stress caused by those factors.
Heart regeneration is comparable in young and old zebrafish
Our analysis of heart regeneration in young and old zebrafish shows that critical processes for heart regeneration occur in both groups in a similar manner, and injured hearts regenerate similarly. Major processes for successful heart regeneration include rapid activation of wound or regeneration-response in the epicardial layer (Lepilina et al., 2006), regeneration of the myocardial layer (Poss et al., 2002; Raya et al., 2003), and neo-vascularization (Lepilina et al., 2006; Kim et al., 2010). These processes occurred similarly in both young and old fish after amputation-induced injury. Thus, similar to the case of fin regeneration, our data of heart regeneration support the idea that zebrafish maintains regenerative ability throughout the lifespan.
One difference between our study and a previous report is the expression pattern of Gata4 in regenerating hearts. The gata4-EGFP reporter signal in the transgenic line was detected only in the outer compact muscle layer, but not in the regenerating area, at 7 dpa in the previous study (Kikuchi et al., 2010). In contrast, we detected Gata4 immunoreactivity in the outer compact muscle layer and in the regenerating area (Fig. 4). This difference in the distribution of signals is unlikely to be due to non-specific signals in our study, since Gata4 signal was not detected in hearts immediately after amputation (Fig. 2G,H). The gata4-EGFP reporter line was generated with upstream sequences of the gata4 gene, which were characterized in comparison to embryonic gata4 expression pattern (Heicklen-Klein and Evans, 2004). These upstream sequences might lack other regulatory sequence(s). Thus, it is likely that the difference was caused by detecting endogenous Gata4 protein (this study) and detecting the reporter EGFP.
Similar to the fin (Tal et al., 2010), heart regeneration is a highly orchestrated system (Raya et al., 2004; Poss, 2007; Ausoni and Sartore, 2009; Laflamme and Murry, 2011), thus, comparable heart regeneration in young and old fish could be achieved by the progression of key processes in both groups. Unlike the case of fin regeneration, our report is the first case of examining the effects of aging on heart regeneration in zebrafish. Further studies, such as examining how genotoxic treatment affects regenerative ability along with aging, would provide a comparative analysis between heart regeneration and fin regeneration.
Aging and regeneration in zebrafish
Our study shows that zebrafish possess the ability to regenerate the fin and heart after aging. Interestingly, recent reports have shown that regeneration of these two organs is achieved mainly by de-differentiation and proliferation of lineage restricted cells rather than activation of stem cells or progenitor cells (Jopling et al., 2010; Kikuchi et al., 2010; Knopf et al., 2011; Tu and Johnson, 2011). The molecular changes during de-differentiation that cells undergo are still to be elucidated. However, the preserved ability to regenerate after animal aging might be related to de-differentiation-based regeneration. In this regard, it would be interesting to examine in the future whether other organs known to regenerate in zebrafish utilize stem/progenitor cells or de-differentiation of lineage restricted cells or both, and whether aging affects their regeneration.
Materials and Methods
Zebrafish maintenance and surgery
Zebrafish were raised under standard conditions that were not changed during the course of the study. Zebrafish were maintained in a standard environment in an Aquaneering recirculating system in a core facility. Air and water temperatures were maintained at 27–28°C with a 14 hours light and 10 hours dark cycle. The housing density was at 15–20 fish in a nine liter tank. Zebrafish were fed 1–2 ml brine shrimp twice a day, morning and afternoon. Fish were derived from zebrafish originally obtained from Segrest Farms (Gibsonton, FL) as previously described (Lund et al., 2009). Six to 12 month-old and 26 to 36 month-old fish were used as young and old fish, respectively. The body lengths of not all fish were recorded. However, of the fish measured, body lengths of fish used in this study and of fish maintained in the same groups were 2.92±0.16 cm (n = 15) and 3.40±0.35 cm (n = 15) for young and old fish, respectively. Male fish were used for fin regeneration studies. Ventricular amputation and caudal fin amputation were performed as previously published (Raya et al., 2003; Kawakami et al., 2006). Care and experimentation were done in accordance with the Institutional Animal Care and Use Committee of the University of Minnesota.
Immunostaining and in situ hybridization
Immunostaining on 14 µm thickness sections was performed according to a standard procedure to detect specific protein expression (Itou et al., 2011; Kawakami et al., 2011). Briefly, the section corresponding to around the center of regenerating area was washed with PBS with 0.1% Triton X-100, and heat-induced antigen retrieval was performed with 10 mM sodium citrate buffer (pH6.0) by boiling for 40 min. The primary antibodies used were anti-myosin heavy chain antibody (Developmental Studied Hybridoma Bank, Iowa City, IA, USA; MF20, 5.14 µg/ml), anti-GATA4 antibody (Abcam, Cambridge, MA, USA; ab61170, 1∶500), anti-PCNA antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA; sc-56, 1∶100), anti-MEF2 antibody (Santa Cruz Biotechnology; sc-313, 1∶50), anti-pErk1/2 antibody (Cell Signaling Technology, Beverly, MA, USA; 9101, 1∶1000) and anti-GFP antibody (Molecular Probes, Eugene, CA, USA; A11122, 1∶2000). Secondary antibodies used were Alexa 488 anti-mouse IgG (Invitrogen, Carlsbad, CA, USA; A11001, 1∶1000), Alexa 594 goat anti-rabbit IgG (Invitrogen; A11012, 1∶1,000), biotinylated anti-mouse IgG (Vector laboratories, Burlingame, CA, USA; BA-9200, 1∶500), and biotinylated rabbit IgG (Vector laboratories; BA-1000, 1∶500). To visualize the signals in immunohistochemistry, we used a horseradish peroxidase streptavidin system (Vector laboratories). Counterstaining was done with DAPI for immunofluorescence or with hematoxylin for immunohistochemistry. To analyze gene transcriptions, in situ hybridization on 14 µm thickness sections was performed as previously described (Raya et al., 2003). Sections located at around the center of the regenerating area from each heart were analyzed.
Imaging and quantification
Bright field images of heart sections were taken by using Nikon ACT1 software (Nikon, Melville, NY, USA) with Zeiss Axioscope 2 microscope (Carl Zeiss Microscopy, Thornwood, NY, USA) equipped with Nikon DXM1200 digital camera (Nikon). Fluorescent confocal images were obtained by using Zeiss LSM 710 laser scanning microscope system (Carl Zeiss Microscopy), and analyzed by ZEN2009 software (Carl Zeiss Microscopy). The number of immunofluorescent signals of each section was counted manually. Regenerating areas and EGFP positive areas were measured by ImageJ software. Zeiss SteREO Discovery V8 stereoscope (Carl Zeiss Microscopy) and iSolution Lite software version 8.3 (IMT iSolution, Vancouver, BC, Canada) were used to obtain bright field images of regenerating fins, and to measure regenerated lengths.
Statistical analysis
All error bars on graphs are standard deviation. The Student's t-test was used to analyze statistical significance between young and old fish.
Competing Interests
The authors declare that there are no competing interests.
Acknowledgments
We thank the zebrafish core facility at the University of Minnesota for general help with breeding and maintenance of zebrafish lines. We thank Drs Stephen C. Ekker, Michael O'Connor, Thomas Neufeld, Yasushi Nakagawa, Angel Raya and Isao Oishi for sharing equipment or materials. We thank members of the Kawakami laboratory and the Gammill laboratory for discussion, Austin Johnson for editing assistance, and the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. J.I. and Y.K. conceived and designed experiments. J.I., H.K., T.B. and Y.K. performed experiments. J.I. and Y.K. analyzed and interpreted results. J.I. and Y.K. wrote the manuscript.
Footnotes
Competing interests: The authors declare that there are no competing interests.
References
- Akimenko M.-A., Johnson S. L., Westerfield M., Ekker M. (1995). Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development 121, 347–357. [DOI] [PubMed] [Google Scholar]
- Akimenko M.-A., Marí-Beffa M., Becerra J., Géraudie J. (2003). Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn. 226, 190–201 10.1002/dvdy.10248 [DOI] [PubMed] [Google Scholar]
- Anchelin M., Murcia L., Alcaraz-Pérez F., García-Navarro E. M., Cayuela M. L. (2011). Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS ONE 6, e16955 10.1371/journal.pone.0016955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni M. H., Lutgendorf S. K., Cole S. W., Dhabhar F. S., Sephton S. E., McDonald P. G., Stefanek M., Sood A. K. (2006). The influence of bio-behavioural factors on tumour biology: pathways and mechanisms. Nat. Rev. Cancer 6, 240–248 10.1038/nrc1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausoni S., Sartore S. (2009). From fish to amphibians to mammals: in search of novel strategies to optimize cardiac regeneration. J. Cell Biol. 184, 357–364 10.1083/jcb.200810094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo A. S., Grotek B., Jacinto A., Weidinger G., Saúde L. (2011). The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS ONE 6, e22820 10.1371/journal.pone.0022820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Davidson A. E., Sivasubbu S., Hermanson S. B., Welle Z., Ekker S. C. (2004). Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genomics 5, 62 10.1186/1471-2164-5-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C. G., Becker T. (2007). Growth and pathfinding of regenerating axons in the optic projection of adult fish. J. Neurosci. Res. 85, 2793–2799 10.1002/jnr.21121 [DOI] [PubMed] [Google Scholar]
- Brittijn S. A., Duivesteijn S. J., Belmamoune M., Bertens L. F., Bitter W., de Bruijn J. D., Champagne D. L., Cuppen E., Flik G., Vandenbroucke-Grauls C. M. et al. (2009). Zebrafish development and regeneration: new tools for biomedical research. Int. J. Dev. Biol. 53, 835–850 10.1387/ijdb.082615sb [DOI] [PubMed] [Google Scholar]
- Chablais F., Veit J., Rainer G., Jaźwińska A. (2011). The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev. Biol. 11, 21 10.1186/1471-213X-11-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy I. M., Conboy M. J., Wagers A. J., Girma E. R., Weissman I. L., Rando T. A. (2005). Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764 10.1038/nature03260 [DOI] [PubMed] [Google Scholar]
- Corson L. B., Yamanaka Y., Lai K. M., Rossant J. (2003). Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development 130, 4527–4537 10.1242/dev.00669 [DOI] [PubMed] [Google Scholar]
- Dufourcq P., Roussigné M., Blader P., Rosa F., Peyrieras N., Vriz S. (2006). Mechano-sensory organ regeneration in adults: the zebrafish lateral line as a model. Mol. Cell. Neurosci. 33, 180–187 10.1016/j.mcn.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Flint M. S., Baum A., Chambers W. H., Jenkins F. J. (2007). Induction of DNA damage, alteration of DNA repair and transcriptional activation by stress hormones. Psychoneuroendocrinology 32, 470–479 10.1016/j.psyneuen.2007.02.013 [DOI] [PubMed] [Google Scholar]
- Gerhard G. S. (2007). Small laboratory fish as models for aging research. Ageing Res. Rev. 6, 64–72 10.1016/j.arr.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Gerhard G. S., Kauffman E. J., Wang X., Stewart R., Moore J. L., Kasales C. J., Demidenko E., Cheng K. C. (2002). Life spans and senescent phenotypes in two strains of Zebrafish (Danio rerio). Exp. Gerontol. 37, 1055–1068 10.1016/S0531-5565(02)00088-8 [DOI] [PubMed] [Google Scholar]
- González-Rosa J. M., Martín V., Peralta M., Torres M., Mercader N. (2011). Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 138, 1663–1674 10.1242/dev.060897 [DOI] [PubMed] [Google Scholar]
- Hara M. R., Kovacs J. J., Whalen E. J., Rajagopal S., Strachan R. T., Grant W., Towers A. J., Williams B., Lam C. M., Xiao K. et al. (2011). A stress response pathway regulates DNA damage through β2-adrenoreceptors and β-arrestin-1. Nature 477, 349–353 10.1038/nature10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Futcher A. B., Greider C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- Heicklen-Klein A., Evans T. (2004). T-box binding sites are required for activity of a cardiac GATA-4 enhancer. Dev. Biol. 267, 490–504 10.1016/j.ydbio.2003.09.042 [DOI] [PubMed] [Google Scholar]
- Hitchcock P. F., Raymond P. A. (2004). The teleost retina as a model for developmental and regeneration biology. Zebrafish 1, 257–271 10.1089/zeb.2004.1.257 [DOI] [PubMed] [Google Scholar]
- Iakova P., Awad S. S., Timchenko N. A. (2003). Aging reduces proliferative capacities of liver by switching pathways of C/EBPalpha growth arrest. Cell 113, 495–506 10.1016/S0092-8674(03)00318-0 [DOI] [PubMed] [Google Scholar]
- Itou J., Taniguchi N., Oishi I., Kawakami H., Lotz M., Kawakami Y. (2011). HMGB factors are required for posterior digit development through integrating signaling pathway activities. Dev. Dyn. 240, 1151–1162 10.1002/dvdy.22598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen V., Forkert R., Fleming H. E., Saito Y., Waring M. T., Dombkowski D. M., Cheng T., DePinho R. A., Sharpless N. E., Scadden D. T. (2006). Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443, 421–426 10.1038/nature05159 [DOI] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Martí M., Raya A., Izpisúa Belmonte J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609 10.1038/nature08899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan N. G., Junghans D., Izpisua Belmonte J. C. (2009). Compensatory growth mechanisms regulated by BMP and FGF signaling mediate liver regeneration in zebrafish after partial hepatectomy. FASEB J. 23, 3516–3525 10.1096/fj.09-131730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Rodríguez-León J., Koth C. M., Büscher D., Itoh T., Raya A., Ng J. K., Esteban C. R., Takahashi S., Henrique D. et al. (2003). MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat. Cell Biol. 5, 513–519 10.1038/ncb989 [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez Esteban C., Raya M., Kawakami H., Martí M., Dubova I., Izpisúa Belmonte J. C. (2006). Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 20, 3232–3237 10.1101/gad.1475106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Marti M., Kawakami H., Itou J., Quach T., Johnson A., Sahara S., O'Leary D. D., Nakagawa Y., Lewandoski M. et al. (2011). Islet1-mediated activation of the β-catenin pathway is necessary for hindlimb initiation in mice. Development 138, 4465–4473 10.1242/dev.065359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. T., Murtha J. M. (2004). The use of mature zebrafish (Danio rerio) as a model for human aging and disease. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 138, 335–341 10.1016/j.cca.2004.04.001 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Werdich A. A., Anderson R. M., Fang Y., Egnaczyk G. F., Evans T., Macrae C. A., Stainier D. Y., Poss K. D. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464, 601–605 10.1038/nature08804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G., Poss K. D. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404 10.1016/j.devcel.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Wu Q., Zhang Y., Wiens K. M., Huang Y., Rubin N., Shimada H., Handin R. I., Chao M. Y., Tuan T. L. et al. (2010). PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl. Acad. Sci. USA 107, 17206–17210 10.1073/pnas.0915016107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S., Bayliss P. E., Uchiyama J., Koshimizu E., Qi J., Nanjappa P., Imamura S., Islam A., Neuberg D., Amsterdam A. et al. (2008). The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet. 4, e1000152 10.1371/journal.pgen.1000152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi S., Slack B. E., Uchiyama J., Zhdanova I. V. (2009). Zebrafish as a genetic model in biological and behavioral gerontology: where development meets aging in vertebrates–a mini-review. Gerontology 55, 430–441 10.1159/000228892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S. et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713–724 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J., Ramsey M. R., Ligon K. L., Torrice C., Koh A., Bonner-Weir S., Sharpless N. E. (2006). p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443, 453–457 10.1038/nature05092 [DOI] [PubMed] [Google Scholar]
- Kuhn H. G., Dickinson-Anson H., Gage F. H. (1996). Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J. Neurosci. 16, 2027–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M. A., Murry C. E. (2011). Heart regeneration. Nature 473, 326–335 10.1038/nature10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D., Weinstein B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- LeClair E. E., Topczewski J. (2010). Development and regeneration of the zebrafish maxillary barbel: a novel study system for vertebrate tissue growth and repair. PLoS ONE 5, e8737 10.1371/journal.pone.0008737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Grill S., Sanchez A., Murphy-Ryan M., Poss K. D. (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173–5183 10.1242/dev.02101 [DOI] [PubMed] [Google Scholar]
- Lepilina A., Coon A. N., Kikuchi K., Holdway J. E., Roberts R. W., Burns C. G., Poss K. D. (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619 10.1016/j.cell.2006.08.052 [DOI] [PubMed] [Google Scholar]
- Lieschke G. J., Currie P. D. (2007). Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353–367 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- Liu Q., Azodi E., Kerstetter A. E., Wilson A. L. (2004). Cadherin-2 and cadherin-4 in developing, adult and regenerating zebrafish cerebellum. Brain Res. Dev. Brain Res. 150, 63–71 10.1016/j.devbrainres.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Lund T. C., Glass T. J., Tolar J., Blazar B. R. (2009). Expression of telomerase and telomere length are unaffected by either age or limb regeneration in Danio rerio. PLoS ONE 4, e7688 10.1371/journal.pone.0007688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E. Y., Rubel E. W., Raible D. W. (2008). Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J. Neurosci. 28, 2261–2273 10.1523/JNEUROSCI.4372-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H. (1900). Regeneration in teleosts. Dev. Genes Evol. 10, 120–134 10.1007/BF02156348 [DOI] [Google Scholar]
- Nachtrab G., Czerwinski M., Poss K. D. (2011). Sexually dimorphic fin regeneration in zebrafish controlled by androgen/GSK3 signaling. Curr. Biol. 21, 1912–1917 10.1016/j.cub.2011.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollycove M., Feinendegen L. E. (2003). Radiation-induced versus endogenous DNA damage: possible effect of inducible protective responses in mitigating endogenous damage. Hum. Exp. Toxicol. 22, 290–306 10.1191/0960327103ht365oa [DOI] [PubMed] [Google Scholar]
- Poss K. D. (2007). Getting to the heart of regeneration in zebrafish. Semin. Cell Dev. Biol. 18, 36–45 10.1016/j.semcdb.2006.11.009 [DOI] [PubMed] [Google Scholar]
- Poss K. D., Wilson L. G., Keating M. T. (2002). Heart regeneration in zebrafish. Science 298, 2188–2190 10.1126/science.1077857 [DOI] [PubMed] [Google Scholar]
- Poss K. D., Keating M. T., Nechiporuk A. (2003). Tales of regeneration in zebrafish. Dev. Dyn. 226, 202–210 10.1002/dvdy.10220 [DOI] [PubMed] [Google Scholar]
- Raya A., Koth C. M., Büscher D., Kawakami Y., Itoh T., Raya R. M., Sternik G., Tsai H. J., Rodríguez-Esteban C., Izpisúa-Belmonte J. C. (2003). Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 100, Suppl 111889–11895 10.1073/pnas.1834204100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A., Consiglio A., Kawakami Y., Rodriguez-Esteban C., Izpisúa-Belmonte J. C. (2004). The zebrafish as a model of heart regeneration. Cloning Stem Cells 6, 345–351 10.1089/clo.2004.6.345 [DOI] [PubMed] [Google Scholar]
- Sadler K. C., Krahn K. N., Gaur N. A., Ukomadu C. (2007). Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc. Natl. Acad. Sci. USA 104, 1570–1575 10.1073/pnas.0610774104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría J. A., Becerra J. (1991). Tail fin regeneration in teleosts: cell-extracellular matrix interaction in blastemal differentiation. J. Anat. 176, 9–21. [PMC free article] [PubMed] [Google Scholar]
- Sawada A., Shinya M., Jiang Y. J., Kawakami A., Kuroiwa A., Takeda H. (2001). Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128, 4873–4880. [DOI] [PubMed] [Google Scholar]
- Schnabel K., Wu C. C., Kurth T., Weidinger G. (2011). Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE 6, e18503 10.1371/journal.pone.0018503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J., Chen D., Ye Q., Cui J., Li Y., Li L. (2011). Tissue regeneration after injury in adult zebrafish: the regenerative potential of the caudal fin. Dev. Dyn. 240, 1271–1277 10.1002/dvdy.22603 [DOI] [PubMed] [Google Scholar]
- Shinya M., Koshida S., Sawada A., Kuroiwa A., Takeda H. (2001). Fgf signalling through MAPK cascade is required for development of the subpallial telencephalon in zebrafish embryos. Development 128, 4153–4164. [DOI] [PubMed] [Google Scholar]
- Tal T. L., Franzosa J. A., Tanguay R. L. (2010). Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish - a mini-review. Gerontology 56, 231–240 10.1159/000259327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. B., Tucci V., Uchiyama J., Fabian N. J., Lin M. C., Bayliss P. E., Neuberg D. S., Zhdanova I. V., Kishi S. (2007). Differential effects of genotoxic stress on both concurrent body growth and gradual senescence in the adult zebrafish. Aging Cell 6, 209–224 10.1111/j.1474-9726.2007.00278.x [DOI] [PubMed] [Google Scholar]
- Tsang M., Maegawa S., Kiang A., Habas R., Weinberg E., Dawid I. B. (2004). A role for MKP3 in axial patterning of the zebrafish embryo. Development 131, 2769–2779 10.1242/dev.01157 [DOI] [PubMed] [Google Scholar]
- Tu S., Johnson S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725–732 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Panáková D., Kikuchi K., Holdway J. E., Gemberling M., Burris J. S., Singh S. P., Dickson A. L., Lin Y. F., Sabeh M. K. et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138, 3421–3430 10.1242/dev.068601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead G. G., Makino S., Lien C. L., Keating M. T. (2005). fgf20 is essential for initiating zebrafish fin regeneration. Science 310, 1957–1960 10.1126/science.1117637 [DOI] [PubMed] [Google Scholar]


