Summary
TOR (Target Of Rapamycin) signalling coordinates cell growth and division in response to changes in the nutritional environment of the cell. TOR kinases form two distinct complexes: TORC1 and TORC2. In mammals, the TORC1 controlled S6K1 kinase phosphorylates the ribosomal protein S6 thereby co-ordinating cell size and nutritional status. We show that the Schizosaccharomyces pombe AGC kinase Gad8 co-immunoprecipitates with the ribosomal protein S6 (Rps6) and regulates its phosphorylation status. It has previously been shown that Gad8 is phosphorylated by TORC2. Consistent with this, we find that TORC2 as well as TORC1 modulates Rps6 phosphorylation. Therefore, S6 phosphorylation in fission yeast actually represents a read-out of the combined activities of TORC1 and TORC2. In contrast, we find that the in vivo phosphorylation status of Maf1 (a repressor of RNA polymerase III) specifically correlates with TORC1 activity.
Key words: Gad8, Maf1, S6, S6K, TOR, S. pombe
Introduction
The eukaryotic TOR protein kinases are highly conserved and couple cell growth and cell size with the nutritional context of the cell (Wullschleger et al., 2006). In all cell types, TOR kinases participate in two different complexes; TOR Complex 1 (TORC1) and TOR Complex 2 (TORC2). In TORC1, TOR kinase binds to Raptor, whereas in TORC2, Raptor is replaced by Rictor (Wullschleger et al., 2006). In most eukaryotes, TORC1 is thought to be under nutrient control and inhibited by the drug rapamycin (Jacinto et al., 2004; Loewith et al., 2002; Wullschleger et al., 2006). In contrast to mammals, S. cerevisiae and S. pombe have two TOR kinases. In S. pombe, Tor2 is the main component of TORC1 and Tor1 is the main component of TORC2 (Alvarez and Moreno, 2006; Hayashi et al., 2007; Matsuo et al., 2007). Furthermore, only TORC1 is essential for cell growth in fission yeast (Shinozaki-Yabana et al., 2000; Weisman and Choder, 2001). The Rheb GTPase activates mTORC1 in mammals and Rheb activity, in turn, is regulated by the TSC1-TSC2 complex (Long et al., 2005; Smith et al., 2005). This upstream control of TORC1 is conserved in S. pombe (Matsumoto et al., 2002; Urano et al., 2007; Uritani et al., 2006; van Slegtenhorst et al., 2004). Control of cell growth by mTORC1 involves direct phosphorylation of a number of targets, including Ribosomal S6 kinase (S6K) and MAF1 (repressor of RNA polymerase III) (Michels et al., 2010; Wullschleger et al., 2006). mTORC2 is less well characterized; however, AKT1 and PKC are direct targets of mTORC2, the latter controlling cytoskeletal organisation (Sparks and Guertin, 2010). In S. pombe, TORC2 has been shown to phosphorylate the AGC kinase Gad8, and both Gad8 and TORC2 are required for survival after exposure to stress (Matsuo et al., 2003). Gad8 is believed to be a homologue of the mTORC2 controlled AKT1 kinase.
The conservation of TSC1/2 and Rheb dependent TORC1 regulation makes fission yeast an excellent model organism for studying TOR signalling. Ribosomal protein S6 (Rps6) phosphorylation is believed to be TORC1 specific (Nakashima et al., 2010) and has therefore been used to quantify TORC1 signalling in fission yeast. We previously established that fission yeast Tor1 and Gad8, but not TORC2 components Ste20 (Rictor) or Sin1, are required to control cell size after nutrient stress (Hartmuth and Petersen, 2009). These data indicate that TORC2 independent Gad8 functions exist in S. pombe and that the control of Gad8 kinase activity is not fully understood. We have therefore extended the characterisation of the Gad8 kinase and have identified a role for Gad8 in the phosphorylation of Rps6. Gad8 co-immunoprecipitates with Rps6 and regulates its phosphorylation. We find that both TORC1 and TORC2 modulate Rps6 phosphorylation. Therefore changes in Rps6 phosphorylation represent an integration of both TORC1 and TORC2 signalling. In contrast, we show that the phosphorylation status of Maf1 specifically correlates with TORC1 activity; its phosphorylation therefore offers an excellent alternative to Rps6 phosphorylation in the in vivo assessment of TORC1 signalling in S. pombe.
Results and Discussion
Gad8 co-immunoprecipitates with the ribosomal protein S6
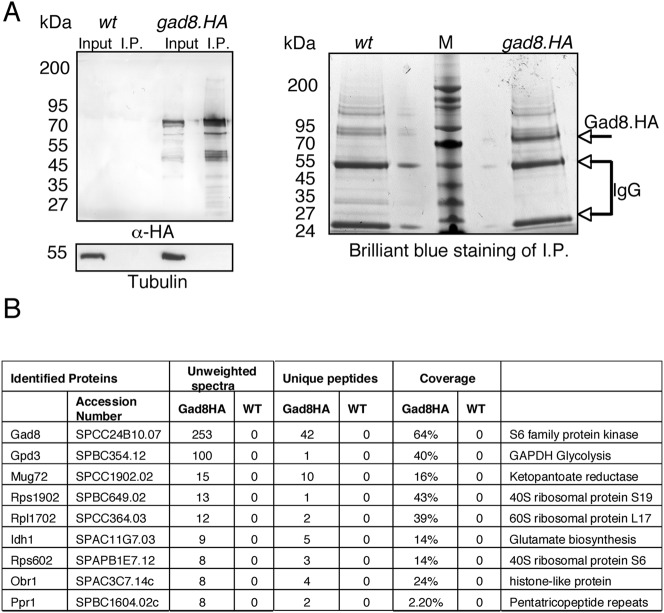
Gad8 is phosphorylated by TORC2 and both are required for survival after stress (Matsuo et al., 2003). However, because we previously established that Gad8 has TORC2 independent roles (Hartmuth and Petersen, 2009), we decided to characterise it further. To identify Gad8 interacting proteins, the kinase was immunoprecipitated from large-scale cultures and subjected to mass spectrometric analysis to identify interacting molecules. The immunocomplexes were subjected to mild washing conditions (0.2% Tween 20) in an attempt to preserve weak interactions (Fig. 1A). A wild type untagged strain was used as a control. Co-purifying candidates that were found only in the Gad8 immunoprecipitates were considered good candidates to be Gad8 specific interacting proteins (Fig. 1B). Interestingly, proteins involved in metabolic pathways, such as Gpd3 and Idh1, specifically co-purified with Gad8. In addition, several ribosomal proteins including the fission yeast homologue of the S6 protein (Rps602) immunoprecipitated with Gad8 (Fig. 1B).
Fig. 1. Rps6 co-immunoprecipitates with Gad8.
(A) Immunoprecipitation of Gad8.HA and wild type control cultures. A western blot probed with anti-HA antibody shows the specific Gad8.HA immunoprecipitation (left). The Brilliant blue stained NUPAGE gel used for analysis by mass spectrometry (right). Intense band indicated by arrow shows Gad8.HA immunoprecipitate. (B) Mass spectrometry data analysed by ScaffoldTM 3. The number of unweighted spectra and unique peptides for each protein are shown.
Ribosomal S6 phosphorylation is Gad8 and Tor1 dependent
Since phosphorylation of ribosomal S6 is thought to be specific to TORC1 signalling, we decided to look further into its phosphorylation. It has previously been demonstrated that the anti-PAS antibody, which recognises phosphorylation of the [R/K]X[R/K]XX[S/T] consensus motif in mammalian S6K1 and other AGC kinase substrates (Manning et al., 2002; Pearson and Kemp, 1991) (Cell Signalling Technology), detects phosphorylation of fission yeast Rps6 serine 235 (Nakashima et al., 2010). Ribosomal S6 is encoded by two genes in fission yeast: rps601 and rps602. It is Rps6 expressed from rps602 (co-purifying with Gad8) (Fig. 1B) that is responsible for the majority of the Rps6 phosphorylation signal (Nakashima et al., 2010). Ribosomal proteins are very abundant in the cell; therefore to improve linearity and avoid saturation when detecting Rps6 phosphorylation, gels were loaded with very low levels of total protein. Furthermore, secondary alkaline phosphatase coupled antibodies were used, which have a wider linear range compared to their horseradish peroxidase ECL detected counterparts. In both a gad8 deletion strain (gad8Δ) and a mutant in which gad8 has been mutated to generate a catalytically inactive Gad8 kinase (gad8.KD), the level of Rps6 phosphorylation was reduced in glutamate grown cells (Fig. 2A). This Gad8 dependent phosphorylation of Rps6 was seen in cells grown in minimal medium with either ammonium or glutamate as the nitrogen source (Fig. 2A). As previously shown, we found that Gad8 is phosphorylated by TORC2 (Matsuo et al., 2003) (Fig. 2B). The main TOR kinase found in S. pombe TORC2 is Tor1 (Alvarez and Moreno, 2006; Hayashi et al., 2007; Matsuo et al., 2007). We therefore asked whether Rps6 phosphorylation was reliant upon Tor1 activity. In contrast to previous reports (Nakashima et al., 2010), we find that under conditions in which the detection of Rps6 phosphorylation is linear, there is a major reduction in phosphorylation of Rps6 in the tor1Δ strain (Fig. 2C). Importantly, the signal is not completely abolished by the removal of Tor1, accounting for the previous reports of a persistence of Rps6 phosphorylation in tor1Δ cells (Nakashima et al., 2010). Furthermore, a very weak anti-PAS signal was still detected in gad8 deficient strains. It is therefore likely that one or more kinases act alongside Gad8 to control Rps6 phosphorylation levels. The S. pombe genome contains three kinases that are closely related to Gad8; Psk1, Sck1 and Sck2 (Bimbó et al., 2005). We find that, like Gad8, Psk1 also contributes to Rps6 anti-PAS phosphorylation levels (Fig. 2E). Thus S6 phosphorylation in fission yeast depends on two AGC kinase homologues Gad8 and Psk1. When comparing wild type cells grown in minimal medium with either glutamate or ammonium as the nitrogen source, we detect increased Rps6 phosphorylation in cells relying upon glutamate (Fig. 2D), possibly reflecting increased TOR activity in this environment. In ammonium, Rps6 phosphorylation is slightly elevated when gad8 is deleted, whereas in psk1Δ cells it is slightly elevated in glutamate (Fig. 2F). We conclude that Gad8, Psk1 and Tor1 all regulate Rps6 phosphorylation and that the particular nutrient environment of the cell alters TOR activity and the regulation of Rps6 phosphorylation.
Fig. 2. Phosphorylation of Rps6 is Gad8 and Tor1 dependent.
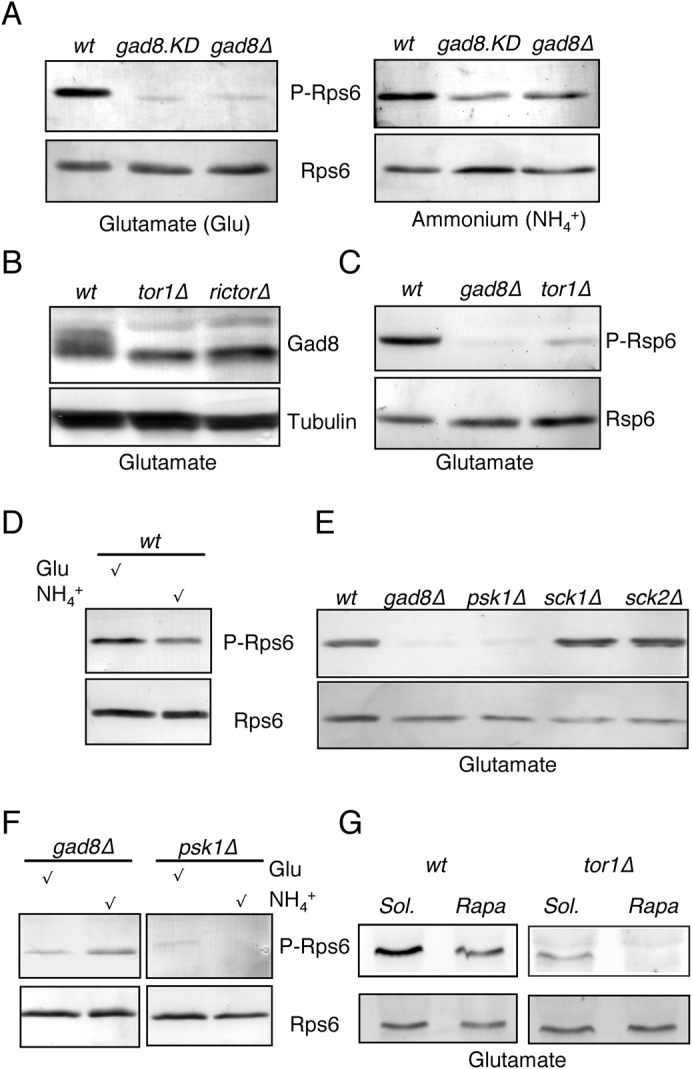
(A) WT, gad8.KD and gad8Δ were harvested at early exponential phase in minimal medium using glutamate or ammonium as the nitrogen source. The phosphorylated Rps6 was probed by anti-PAS antibody. Total Rps6 was detected by anti-S6 antibody, as a loading control. (B) Deletion of tor1 or the Rictor homologue ste20 diminished the phosphorylation of Gad8. WT, tor1Δ and ste20Δ strains were cultured in EMMG to early exponential phase. Gad8 was probed with anti-Gad8 antibodies and tubulin was used as loading control. (C) gad8Δ and tor1Δ down-regulates Rps6 phosphorylation. Cells were cultured in EMMG medium and the blots were probed as in (A). (D) Rps6 phosphorylation levels in WT cells were compared in glutamate and ammonium containing medium. (E) Psk1, but not Sck1 or Sck2 regulate Rps6 phosphorylation. (F) gad8Δ and psk1Δ were grown in EMMG and EMM2 and the Rps6 phosphorylation levels were compared. (G) Rapamycin inhibits Rps6 phosphorylation. WT and tor1Δ were grown in EMMG to early exponential phase and treated with 0.3 µg/ml rapamycin for 30 min.
Rps6 phosphorylation is both TORC1 and TORC2 dependent
TOR kinases are the target of the macrolide Rapamycin, which efficiently inhibits the TORC1 complex and after prolonged exposure disrupts the formation of TORC2 and therefore its activity (Lamming et al., 2012; Wullschleger et al., 2006). When Rapamycin was added to wild type cells, we observed a reduced level of Rps6 phosphorylation (Fig. 2G). In addition, when Rapamycin was added to tor1Δ cells, the low level of Rsp6 phosphorylation was completely abolished (Fig. 2G). This dependency on TORC1 is in agreement with Nakashima et al., who reported that Rps6 phosphorylation is regulated by TORC1 (Nakashima et al., 2010). We previously found that amino-terminal myc-tagged Tor1 can be incorporated into both TORC1 and TORC2 (Hartmuth and Petersen, 2009). Therefore, to address whether regulation of Rps6 is by either TORC1 or TORC2 or both, we examined its phosphorylation in mutants that impact upon the function of each complex. Increasing the temperature to 37°C in cultures of the temperature sensitive mutants of tor2.52 and mip1.310 (Raptor) inactivates TORC1, whereas TORC2 signalling is ablated in the deletion mutants of sin1 and ste20 (Rictor) (Alvarez and Moreno, 2006; Shinozaki-Yabana et al., 2000; Tatebe et al., 2010). Rps6 phosphorylation was reduced in all mutant backgrounds (Fig. 3A). Loss of the TORC2 specific Rictor clearly has an impact upon Rps6 phosphorylation, indicating that ribosomal S6 phosphorylation in S. pombe is not a measure of TORC1 specific signalling as previously suggested; rather it is a read-out of both TORC1 and TORC2 activities. Interestingly, S6 phosphorylation is reduced but not absent in cells derived from S6K1(−/−)/S6K2(−/−) mice (Pende et al., 2004), raising the possibility that S6 phosphorylation in mammalian cells is non-specific to TORC1 as well. Significantly, it is the phosphorylation of the S6 kinase itself, and not its substrate the S6 ribosomal protein, that is routinely used as a TORC1 specific read-out in mammalian cells (Magnuson et al., 2012).
Fig. 3. Both TORC1 and TORC2 regulate Rps6 phosphorylation while Maf1 phosphorylation is TORC1 specific.
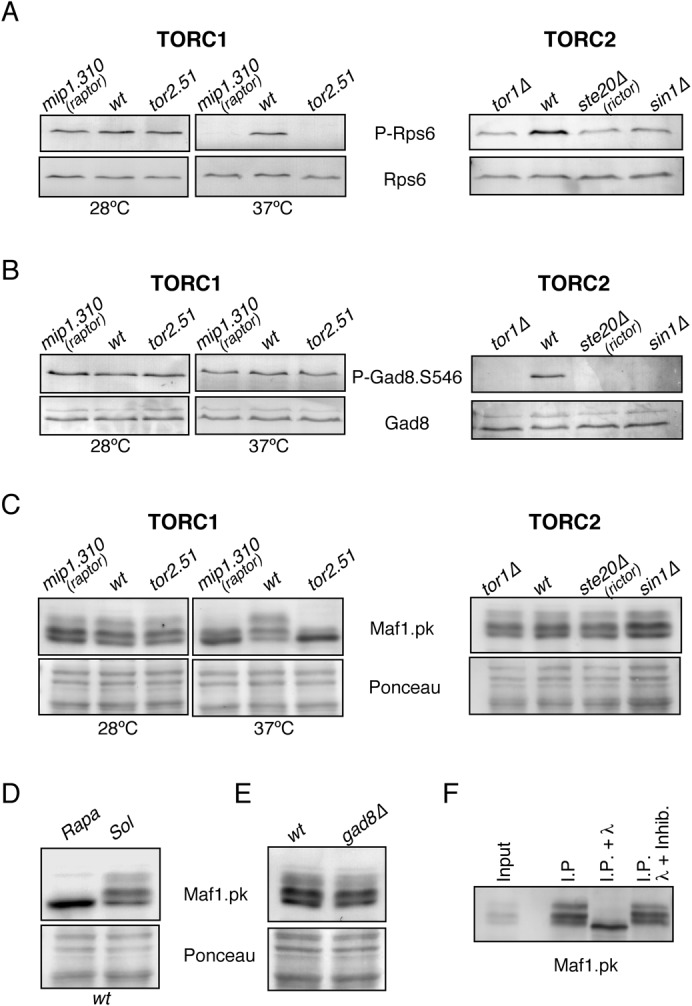
All cells were grown in EMMG (A) Phosphorylation of Rps6 is TORC1 and TORC2 dependent. WT and temperature sensitive TORC1 mutants mip1.310 (Raptor) and tor2.51 were cultured to exponential phase at permissive temperature (28°C). The cells were heat shocked at 37°C for 2 hr (left). Deletion of tor1, ste20 (Rictor) or sin1 reduced the phosphorylation level of Rps6 (right). (B) Gad8 S546 phosphorylation is TORC2 specific. All strains were treated as in (A) and the phosphorylated protein was detected by P-Gad8.S546 specific antibodies and total Gad8 by anti-Gad8 antibodies. (C) Maf1 phosphorylation is TORC1 but not TORC2 dependent. TORC1 and TORC2 mutants expressing Maf1.pk were treated as in (A). Maf1.pk was detected by anti-pk antibodies. (D) Rapamycin inhibits Maf1.pk phosphorylation. Cells were grown and treated with rapamycin, and Maf1.pk was detected as previously described. (E) Gad8 is not required for Maf1 phosphorylation. (F) Slower migrating Maf1.pk bands represent phosphorylation. When λ phosphatase was added to immunoprecipitated Maf1.pk, only the faster migrating non-phosphorylated Maf1.pk was observed.
Phosphorylation of S. pombe Maf1 is TORC1 specific
Both S. cerevisiae and S. pombe are popular model organisms for characterising TOR signalling. In contrast to S. cerevisiae, the Rheb and TSC1/2 dependent TORC1 regulation that is characteristic of control of mammalian TORC1 is conserved in S. pombe (Matsumoto et al., 2002; Urano et al., 2007; Uritani et al., 2006; van Slegtenhorst et al., 2004), making fission yeast an attractive model for particular aspects of TOR signalling. The use of a specific substrate to differentiate between the in vivo activity of either TORC1 or TORC2 in this organism is therefore essential. For TORC2, the previously shown phosphorylation of Gad8 on serine 546 is a good specific marker of its activity (Fig. 3B) (Tatebe et al., 2010). Mammalian MAF1 (a repressor of RNA polymerase III) was recently shown to be a direct mTORC1 specific substrate (Michels et al., 2010). Extensive phosphorylation by mTORC1 on three conserved sites inhibits MAF1. We therefore asked whether Maf1 would be a potential candidate for a specific read-out of TORC1 activity in fission yeast. We fused sequences encoding an epitope tag to the 3′ end of the maf1 open reading frame at the native locus to generate maf1.pk. In a wild type background several forms of Maf1.pk were observed (Fig. 3C). Interestingly, inhibition of TORC1 for 60 min with Rapamycin condensed all of the slower migrating bands into one faster migrating form (Fig. 3D). This observation suggested that fission yeast mimics mammals in relying upon TORC1 for Maf1 control. To assess whether Maf1 phosphorylation was TORC1 specific and had no input from TORC2, we looked at Maf1 migration in mutants that compromise the functions of both TORC1 and TORC2 (Alvarez and Moreno, 2006; Shinozaki-Yabana et al., 2000; Tatebe et al., 2010) (Fig. 3C). Only TORC1 specific mutants influenced Maf1 migration. In addition, no change in Maf1.pk migration was observed in gad8Δ cells (Fig. 3E). When lambda phosphatase was added to immunoprecipitated Maf1.pk the slower migrating bands were absent and an increase in the faster migrating non-phosphorylated Maf1.pk was observed (Fig. 3F). Together this indicates that Maf1 phosphorylation may represent a more specific read-out of TORC1 activity in fission yeast.
Conclusions
We find that several ribosomal proteins co-purify with the AGC kinase Gad8. Given that Gad8 is phosphorylated by TORC2 (Matsuo et al., 2003) (Fig. 3B), it may be significant that active mTORC2 co-purifies with ribosomes (Zinzalla et al., 2011), raising the possibility that Gad8 may also be phosphorylated by TORC2 on ribosomes. The TORC2 dependent regulation of Rps6 phosphorylation that we observed here was unexpected because mTORC1 specifically phosphorylates the S6 kinase in mammalian cells. However, because S6 phosphorylation is reduced but not absent in cells derived from S6K1(−/−)/S6K2(−/−) mice (Pende et al., 2004), this TORC2 dependent regulation of S6 phosphorylation may be conserved in mammalian cells. The conservation of TSC1/2 and Rheb dependent TORC1 regulation has made fission yeast an attractive model for particular aspects of TOR signalling. In this organism S6 phosphorylation was considered to be TORC1 specific (Nakashima et al., 2010). However, as summarised in Fig. 4, we find that TORC1, TORC2 and Gad8 all regulate Rps6 phosphorylation. This highlights the need for an alternative read-out of TORC1 specific signalling in S. pombe. Maf1 appears to represent an ideal candidate as its phosphorylation status shows a direct correlation with in vivo TORC1 activity. In addition, as previously shown (Tatebe et al., 2010), Gad8.S546 phosphorylation represents a TORC2 specific substrate in fission yeast.
Fig. 4. TORC dependent substrates.
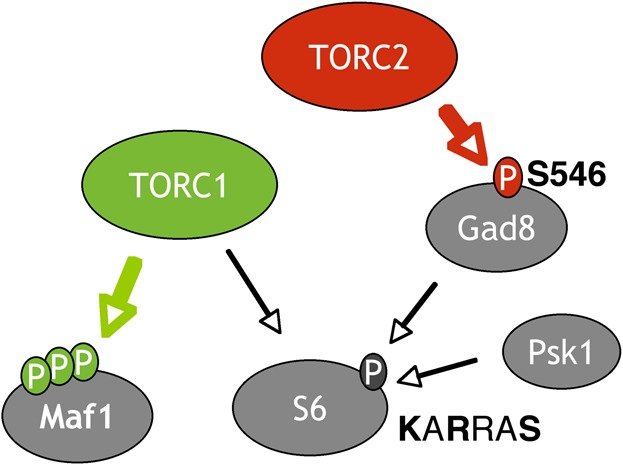
A schematic model of TOR signalling in fission yeast. TORC1 specific control of Maf1 phosphorylation is shown in green. TORC2 specific phosphorylation of Gad8 is shown in red. The ribosomal protein S6 is regulated by both TORC1 and TORC2 and the AGC kinases Gad8 and Psk1.
Materials and Methods
Strains and cell cultures
Strains used in this study are listed in supplementary material Table S1. Cells were cultured at 28°C in yeast extract medium (YES) or in EMM2 minimal medium – using ammonium or glutamic acid (EMMG) as the nitrogen source (Fantes and Nurse, 1977). Rapamycin was added to early exponential cultures at a final concentration of 0.3 µg/ml.
Molecular manipulations
To generate the gad8 kinase dead allele, the gad8 ORF was cloned into TOPO TA Cloning vectors (Invitrogen) and mutated by the standard site directed mutagenesis method. The ORF was sequenced and integrated into JP1295 by 5-FOA selection. Strains were then backcrossed to remove pku70::kanMx and selective markers. Generation of the genomic maf1.pk was achieved through PCR-based gene-tagging (Bähler et al., 1998).
Western blotting
Total protein extracts were prepared by TCA precipitation (Caspari et al., 2000). Gad8.HA was detected by anti-HA antibody (1:2000, Santa Cruz Biotechnology). Gad8 was detected with antibodies raised to the first 60 amino acids of the N-terminus of the protein, at 1:200 dilution. Phosphorylated Rps6 was detected by Phospho-(Ser/Thr) Akt Substrate (PAS) Antibody (Cell Signalling) at 1:2000. Total Rps6 was detected by anti-S6 antibody (Abcam) at 1:2000. Maf1.pk was detected by anti-V5 antibody (AbDserotec) at 1:2000. Alkaline phosphatase coupled secondary antibodies were used for all blots followed by direct detection with NBT/BCIP (VWR) substrates on PVDF membranes. Phospho-Gad8.S546 antibodies were generated by Eurogentec.
Large scale native Gad8 IP for mass spectrometry
Cells were grown to 3×106 cells/ml (20 litres) harvested and disrupted using a Spex 6870 freezer mill in liquid nitrogen. The cell powder was thawed with IP buffer (50 mM Tris pH 8.0, 1 mM EDTA, 20 mM sodium glycerol phosphate, 0.1 mM Na3VO4, 50 mM NaF, 1 mM PMSF, 150 mM NaCl, 0.2% Tween 20 and Sigma protease inhibitor cocktail). The supernatant was incubated with Invitrogen protein Dynabeads pre-coupled with anti-HA antibodies for 30 min at 4°C. Beads were then washed three times with IP buffer and heated to 80°C for 10 min to elute the proteins. The samples were loaded on a NUPAGE Bis-Tris 4–12% gel (Invitrogen). The gel was fixed with 7% acetic acid and 25% methanol and stained by Brilliant blue. The entire lane was cut into small bands, before being sent for protein identification. Mass spectrometry data were analysed by ScaffoldTM 3 software.
Supplementary Material
Acknowledgments
We thank Masayuki Yamamoto, Kazuhiro Shiozaki, Sergio Moreno and Mohan Balasubramanian for strains; the Biological Mass Spectrometry Facility at Manchester University for protein identification; Iain Hagan and members of the laboratory for stimulating discussions and valuable comments on the manuscript. This work was supported by a Cancer Research UK Senior Fellowship [grant number C10888/A11178] and The University of Manchester.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Alvarez B., Moreno S. (2006). Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119, 4475–4485 10.1242/jcs.03241 [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- Bimbó A., Jia Y., Poh S. L., Karuturi R. K., den Elzen N., Peng X., Zheng L., O'Connell M., Liu E. T., Balasubramanian M. K.et al. (2005). Systematic deletion analysis of fission yeast protein kinases. Eukaryot. Cell 4, 799–813 10.1128/EC.4.4.799-813.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T., Dahlen M., Kanter–Smoler G., Lindsay H. D., Hofmann K., Papadimitriou K., Sunnerhagen P., Carr A. M. (2000). Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20, 1254–1262 10.1128/MCB.20.4.1254-1262.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P., Nurse P. (1977). Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp. Cell Res. 107, 377–386 10.1016/0014-4827(77)90359-7 [DOI] [PubMed] [Google Scholar]
- Hartmuth S., Petersen J. (2009). Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J. Cell Sci. 122, 1737–1746 10.1242/jcs.049387 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., Yanagida M. (2007). Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 12, 1357–1370 10.1111/j.1365-2443.2007.01141.x [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004). Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- Lamming D. W., Ye L., Katajisto P., Goncalves M. D., Saitoh M., Stevens D. M., Davis J. G., Salmon A. B., Richardson A., Ahima R. S.et al. (2012). Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 10.1126/science.1215135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002). Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz–Vega S., Yonezawa K., Avruch J. (2005). Rheb binds and regulates the mTOR kinase. Curr. Biol. 15, 702–713 10.1016/j.cub.2005.02.053 [DOI] [PubMed] [Google Scholar]
- Magnuson B., Ekim B., Fingar D. C. (2012). Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem. J. 441, 1–21 10.1042/BJ20110892 [DOI] [PubMed] [Google Scholar]
- Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. (2002). Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10, 151–162 10.1016/S1097-2765(02)00568-3 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Bandyopadhyay A., Kwiatkowski D. J., Maitra U., Matsumoto T. (2002). Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics 161, 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Kubo Y., Watanabe Y., Yamamoto M. (2003). Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J. 22, 3073–3083 10.1093/emboj/cdg298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Otsubo Y., Urano J., Tamanoi F., Yamamoto M. (2007). Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 27, 3154–3164 10.1128/MCB.01039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels A. A., Robitaille A. M., Buczynski–Ruchonnet D., Hodroj W., Reina J. H., Hall M. N., Hernandez N. (2010). mTORC1 directly phosphorylates and regulates human MAF1. Mol. Cell. Biol. 30, 3749–3757 10.1128/MCB.00319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Sato T., Tamanoi F. (2010). Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J. Cell Sci. 123, 777–786 10.1242/jcs.060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R. B., Kemp B. E. (1991). Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 200, 62–81 10.1016/0076-6879(91)00127-I [DOI] [PubMed] [Google Scholar]
- Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L., Mestan J., Mueller M., Fumagalli S., Kozma S. C., Thomas G. (2004). S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24, 3112–3124 10.1128/MCB.24.8.3112-3124.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki–Yabana S., Watanabe Y., Yamamoto M. (2000). Novel WD-repeat protein Mip1p facilitates function of the meiotic regulator Mei2p in fission yeast. Mol. Cell. Biol. 20, 1234–1242 10.1128/MCB.20.4.1234-1242.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. M., Finn S. G., Tee A. R., Browne G. J., Proud C. G. (2005). The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 280, 18717–18727 10.1074/jbc.M414499200 [DOI] [PubMed] [Google Scholar]
- Sparks C. A., Guertin D. A. (2010). Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene 29, 3733–3744 10.1038/onc.2010.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H., Morigasaki S., Murayama S., Zeng C. T., Shiozaki K. (2010). Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr. Biol. 20, 1975–1982 10.1016/j.cub.2010.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J., Sato T., Matsuo T., Otsubo Y., Yamamoto M., Tamanoi F. (2007). Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. USA 104, 3514–3519 10.1073/pnas.0608510104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani M., Hidaka H., Hotta Y., Ueno M., Ushimaru T., Toda T. (2006). Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells 11, 1367–1379 10.1111/j.1365-2443.2006.01025.x [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M., Carr E., Stoyanova R., Kruger W. D., Henske E. P. (2004). Tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279, 12706–12713 10.1074/jbc.M313874200 [DOI] [PubMed] [Google Scholar]
- Weisman R., Choder M. (2001). The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276, 7027–7032 10.1074/jbc.M010446200 [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Zinzalla V., Stracka D., Oppliger W., Hall M. N. (2011). Activation of mTORC2 by association with the ribosome. Cell 144, 757–768 10.1016/j.cell.2011.02.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.