Abstract
Erlotinib and gefitinib, tyrosine kinase inhibitors used to block EGFR (epidermal growth factor receptor) signalling in cancer, are thought to bind only the active conformation of the EGFR-TKD (tyrosine kinase domain). Through parallel computational and crystallographic studies, we show in the present study that erlotinib also binds the inactive EGFR-TKD conformation, which may have significant implications for its use in EGFR-mutated cancers.
Keywords: cancer, epidermal growth factor receptor (EGFR), erlotinib, gefitinib, inhibitor, molecular dynamics, receptor tyrosine kinase (RTK), X-ray crystallography
Abbreviations: DFG, Asp-Phe-Gly; DTT, dithiothreitol; EGFR, epidermal growth factor receptor; FEP, free energy perturbation; IFD, Induced Fit Docking; MD, molecular dynamics; MM/PBSA, molecular mechanics/Poisson–Boltzmann surface area; NSCLC, non-small-cell lung cancer; RMSD, root mean square deviation; SASA, surface-accessible surface area; TKD, tyrosine kinase domain; TKI, tyrosine kinase inhibitor
INTRODUCTION
Activating mutations in the EGFR (epidermal growth factor receptor) are now well accepted as oncogenic drivers in such cancers as NSCLC (non-small-cell lung cancer) [1] and glioblastoma [2,3]. Several drugs that inhibit EGFR signalling are now in clinical use, including antibody therapeutics and ATP-competitive small molecule TKIs (tyrosine kinase inhibitors) [4]. Approved EGFR-targeted TKIs include erlotinib and gefitinib (which are EGFR-specific), lapatinib (which inhibits both EGFR and HER2/ErbB2) and vandetanib [which inhibits EGFR, VEGFR (vascular endothelial growth factor receptor) and Ret]. Many more are currently being developed and/or tested.
Not all EGFR-driven cancers are sensitive to EGFR-targeted TKIs [4]. In EGFR-mutated NSCLC, for example, primary resistance is seen in approximately 25% of cases. Where responses are seen, acquired resistance to first-generation TKIs invariably occurs [5], often through secondary EGFR mutations [6,7]. Intriguingly, the selectivity of response to different EGFR-targeted TKIs appears to depend on how the receptor is activated. For example, EGFR activated by TKD (tyrosine kinase domain) mutations in NSCLC can be inhibited by erlotinib and CI-1033 (canertinib) but not by lapatinib or HKI-272 (neratinib), whereas the same receptor activated by extracellular mutations in glioblastoma is conversely inhibited by lapatinib and HKI-272 (neratinib) but not by erlotinib or CI-1033 (canertinib) [3].
Previous structural studies of EGFR-TKD bound to different TKIs [8] suggest that individual drugs differ in their preference for the ‘active’ compared with ‘inactive’ conformation of the kinase, although they are all strictly type I TKIs [9] in that they bind EGFR-TKD in the ‘DFG (Asp-Phe-Gly)-in’ conformation. Crystal structures of erlotinib-bound [10] or gefitinib-bound [11] EGFR-TKD show the kinase in the same active conformation, allosterically stabilized by a characteristic asymmetric dimer formed in the crystals [12]. By contrast, crystals of EGFR-TKD bound to lapatinib [13] or HKI-272 (neratinib) [7] show a distinct Src-like inactive conformation in which a short activation loop α-helix makes autoinhibitory interactions with the displaced αC helix [12]. On the basis of these studies, erlotinib and gefitinib are thought to bind only the active EGFR-TKD conformation, whereas lapatinib and neratinib are thought to bind only the inactive conformation.
Despite these structural views, biochemical and binding studies argue that EGFR-targeted TKIs may not in fact be so conformationally selective. Extensive direct binding studies suggested that erlotinib, gefitinib and lapatinib all bind similarly (within 3-fold) to both wild-type inactive and mutationally activated forms of EGFR-TKD [14]. Erlotinib and gefitinib affinities were also reported [15] not to be affected by a C-lobe mutation (V924R) that prevents formation of the activated asymmetric dimer [12], again suggesting that these inhibitors bind similarly to inactive and active EGFR-TKD. Moreover, studies of near full-length EGFR [16] indicated that the apparent Ki value for inhibition by lapatinib is increased only 1.8-fold upon receptor activation. It should be noted, however, that some other published data do suggest affinity differences between active and inactive EGFR-TKD for some of these inhibitors. For example, the apparent Ki for lapatinib inhibition of near full-length EGFR was increased >25-fold when activated by the oncogenic L834R mutation [16]. Blocking EGFR activation with cetuximab also increased the apparent Ki for erlotinib [16], suggesting that inactive EGFR has a reduced affinity for erlotinib. Moreover, one set of direct gefitinib-binding studies suggested that this TKI binds more tightly to the activated (L834R) EGFR-TKD than to the inactive conformation [11]. On balance, it therefore remains unclear whether EGFR inhibitors are truly conformationally selective. It is clear, however, that a full understanding of which TKIs bind more tightly to which mutated EGFR variants found in patients could have significant potential impact on inhibitor choice in the clinic.
To investigate conformational selectivity of EGFR inhibitors further, we undertook computational and crystallographic studies of erlotinib binding to EGFR-TKD in its active and inactive states. Our computational studies predicted that erlotinib binds both inactive and active TKD conformations with similar affinities. We used X-ray crystallography to test this prediction, and describe a structure of inactive EGFR-TKD with bound erlotinib. The findings of the present study cast further doubt on the conformational selectivity of EGFR inhibitors, and also suggest that other characteristics, such as differences in inhibitor dissociation [13] or cycling [17] rates, may underlie the distinct effects of erlotinib and lapatinib on NSCLC and glioblastoma cell lines [3], and will need to be understood to counter TKI resistance.
EXPERIMENTAL
Plasmid construction
DNA encoding kinase domain residues 672–998 (mature protein numbering), equivalent to residues 696–1022 (precursor protein numbering) of human EGFR (NCBI sequence NP_005219.2) was amplified by PCR using primers that included an N-terminal His6 tag and SpeI/XhoI restriction sites. The PCR product was subcloned into pFastBac1(Invitrogen) for generating recombinant baculovirus with the Bac-to-Bac system (Invitrogen) for protein expression in Spodoptera frugiperda Sf9 cells. To generate the V924R mutation, the codon for Val924 (mature protein numbering) was substituted with an arginine codon using the QuikChange® method (Agilent Technologies).
Protein production and purification
Sf9 cells at (1.5–2)×106/ml were infected with recombinant baculovirus, and harvested by centrifugation after 3 days. Cells expressing histidine-tagged EGFR672–998/V924R (~3 litres of medium) were lysed by sonication in 100 ml of lysis buffer [20 mM Tris/HCl (pH 8.0), containing 500 mM NaCl, 5 mM 2-mercaptoethanol and protease inhibitor cocktail (Roche)]. After centrifugation at 40000 g for 30 min to remove cell debris, the supernatant was passed through a 0.45 μm filter and incubated with Ni-NTA (Ni2+-nitrilotriacetate) agarose beads (Qiagen) for 1 h at 4°C. Beads were washed in 50 column volumes of lysis buffer, and bound EGFR-TKD protein was eluted in lysis buffer containing increasing concentrations of imidazole. Eluted protein was then further purified using an UnoQ anion-exchange column (Bio-Rad Laboratories) equilibrated with 20 mM Tris/HCl (pH 8.0), containing 5% glycerol and 2 mM DTT (dithiothreitol), and eluting with a gradient from 75 mM to 1 M NaCl over 20 column volumes. EGFR-TKD protein was then subjected to a final step of size-exclusion chromatography using a Superdex 200 column (GE Healthcare) equilibrated in 20 mM Tris/HCl (pH 8.0), containing 150 mM NaCl and 2 mM DTT. In total 1–2 mg of purified EGFR672–998/V924R protein was typically obtained per litre of Sf9 cell culture.
Crystallization and structure determination
Crystals were obtained using the hanging-drop vapour diffusion method, by mixing equal volumes of protein and reservoir solutions and equilibrating over the reservoir solution at 21°C. EGFR-TKD protein was concentrated to 6 mg/ml in 20 mM Tris/HCl (pH 8.0), containing 150 mM NaCl and 2 mM DTT. Crystals were obtained with a reservoir solution of 0.25 M sodium thiocyanate (pH 6.9) and 27% (w/v) PEG [poly(ethylene glycol)] 3350, and when 10 mM taurine had been included as ‘additive’ in the hanging drop. Crystals were soaked for 2 h at 21°C in mother liquor containing 1 mM erlotinib. Crystals were cryo-protected in reservoir solution with 20% (w/v) glycerol added and flash frozen in liquid nitrogen. Diffraction data were collected at beamline 23ID-D of GM/CA@APS (Advanced Photon Source), where crystals diffracted to 2.75 Å (1 Å=0.1 nm), and were processed using HKL2000 [18] (see Table 1). The structure was solved by molecular replacement using Phaser [19] with the inactive EGFR (V924R)-TKD structure (PDB code 3GT8 [20]) as the search model. Repeated cycles of manual building/rebuilding using Coot [21] were alternated with rounds of refinement using REFMAC [19,22], plus composite omit maps calculated using CNS [23]. PHENIX [24] and TLS refinement [25] were used in the later stages. Co-ordinates, parameter files and molecular topology of erlotinib were generated by PRODRG [26]. Data collection and refinement statistics are shown in Table 1. One molecule of EGFR672–998/V924R is present in the asymmetric unit, and the model of its structure complexed with erlotinib includes amino acids 679–709 and 714–960 (mature EGFR numbering). Structural figures were generated with PyMOL (http://www.pymol.org).
Table 1. Data collection and refinement statistics (molecular replacement).
Each dataset was collected from a single crystal. Values for highest resolution shell are shown in parentheses.
| Parameter | EGFR/erlotinib |
|---|---|
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 78.0, 114.3, 84.9 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50–2.75 |
| Rsym | 0.159 (0.494) |
| I/σ | 12.8 (2.1) |
| Completeness (%) | 96.6 (82.1) |
| Redundancy | 4.8 (2.8) |
| Refinement | |
| Resolution (Å) | 50–2.75 |
| Number of reflections | 9413 |
| Rwork/Rfree | 0.23/0.25 |
| Number of atoms | |
| Protein | 2201 |
| Ligand | 29 |
| Water | 34 |
| B-factors | |
| Protein | 47.2 |
| Ligand | 47.5 |
| Water | 43.3 |
| RMSDs | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.081 |
System preparation and molecular docking
Active EGFR-TKD was modelled on the basis of PDB entries 1M17 (which also provided the initial erlotinib conformation) [10] and 2ITX [11], and the L834R mutant was modelled on the basis of PDB entry 2ITV [11]. Inactive EGFR-TKD was modelled based on PDB entries 2GS7 [12] and 1XKK [13]. Protein and ligand conformations were prepared using the Protein Preparation Wizard and LigPrep protocols from Schrödinger Software. All docking simulations used the OPLS (Optimized Potentials for Liquid Simulations) force field [30], and used Schrödinger's IFD (Induced Fit Docking) package [31]. Ligand was first docked to rigid protein using Glide XP [32]. For the resulting top 20 complex conformations, the protein side chains within 5.0 Å of the ligand in that pose were subjected to conformational search and minimized using Prime [33] and the ligand was redocked to the 20 new receptor conformations.
Parameterization of erlotinib for MD (molecular dynamics)
For MD-based analysis of EGFR-TKD–inhibitor interactions, we first generated a CHARMM format force field for erlotinib by following the procedure detailed in the Supplementary Online Data (at http://www.BiochemJ.org/bj/448/bj4480417add.htm), adding nine new atom types to the CHARMM27 [34] topology file to represent new atom types in erlotinib (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/448/bj4480417add.htm). Tests of erlotinib parameterization are shown in Supplementary Figure S2 and Supplementary Table S1 (at http://www.BiochemJ.org/bj/448/bj4480417add.htm).
MD simulations
Conformations generated from IFD were energy-minimized and subsequently equilibrated by performing MD using the CHARMM27 force field [34]. Each system was subjected to constant temperature and constant pressure MD runs at 300 K and 101.325 kPa, followed by constant temperature equilibrium at 300 K with periodic boundaries enforced and long-range electrostatics taken into consideration for 10 ns.
MM/PBSA (molecular mechanics/Poisson–Boltzmann surface area) calculation
We used MM/PBSA [35,36] to calculate the absolute binding free energy and compared it with the Glide [32] score based on the 10 ns MD simulation. MM/PBSA energies of each modelled complex conformation were calculated as the average of the single-point MM/PBSA energy of 500 snapshots taken from the 10 ns simulation. Water molecules and salt ions were removed from the trajectory before calculation. The molecular mechanics energy, UMM, was evaluated by averaging energies over all structures using an infinite cut-off for non-bonded interactions. The electrostatic contribution to the solvation free energy, WPB, was calculated using Poisson–Boltzmann Solver in CHARMM [37]. The reference system had a solvent dielectric constant of 1 and salt concentration of 0 M, and the solvated system had a dielectric constant of 80 and salt concentration of 100 mM. Non-polar contribution to the solvation free energy, WSA, was approximated with the surface area model ΔWSA=[0.00542 kcal/mol/Å2]×SASA+0.92 kcal/mol [38], where the SASA (surface-accessible surface area) was estimated using a 1.4 Å solvent probe radius. Terms for entropy changes in the MM/PBSA score were neglected.
RESULTS AND DISCUSSION
Computational analysis of erlotinib binding to EGFR-TKD
To investigate the possible basis for preferential binding of erlotinib to the active conformation of EGFR-TKD, we used computational approaches. Docking erlotinib on to wild-type EGFR-TKD in its active conformation, as described in the Experimental section, closely reproduced the binding mode observed crystallographically in PDB entry 1M17 [10] (see Supplementary Online Data and Supplementary Figure S4 at http://www.BiochemJ.org/bj/448/bj4480417add.htm). Erlotinib was stable in this bound conformation (cyan in Figure 1) throughout a 10 ns MD run (Supplementary Figure S5 at http://www.BiochemJ.org/bj/448/bj4480417add.htm). Notably, two water molecules (W1 and W2 in Figure 1) were also found to remain in the binding pocket, contributing to a stable network of hydrogen bonds involving residues Thr766, Gln767 and Thr830 of EGFR-TKD. Docking erlotinib into L834R-mutated EGFR-TKD gave very similar results (see Supplementary Online Data). Moreover, estimates of binding energy using Glide [32], MM/PBSA calculations (see the Experimental section) or FEP (free energy perturbation) calculations (see Supplementary Online Data) indicated no increase in erlotinib binding affinity to the L834R mutant (Table 2).
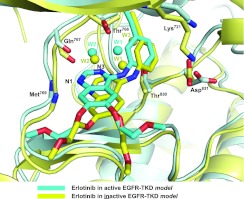
Figure 1. Comparison of erlotinib binding to active and inactive EGFR-TKD models.
On the basis of the computational models described in the present study, erlotinib is shown bound to active EGFR-TKD (cyan) and inactive EGFR-TKD (yellow). The protein structure in the background is similarly coloured (cyan for active, yellow for inactive). Functional groups in several EGFR-TKD residues that interact directly or indirectly with the bound erlotinib are shown. The backbone amide of Met769 donates a hydrogen bond to N1 of the erlotinib quinazoline moiety. The backbone carbonyl of Gln767 and side chains of Thr766 and Thr830 participate in a hydrogen-bonding network to which water molecules (W1 and W2) also contribute. The Lys721 and Asp831 side chains are shown for reference. Polypeptide in the foreground has been removed for clarity.
Table 2. Erlotinib-binding energies calculated by Glide, MM/PBSA and FEP for wild-type and L834R EGFR-TKD in the active conformation.
| Molecule | Glide [32] score (kcal/mol) | MM/PBSA (kcal/mol) | FEP (kcal/mol) |
|---|---|---|---|
| Wild-type | −9.34 | −28.2 | |
| L834R | −9.35 | −30.7 | |
| ΔΔG | −0.01 | −2.5 | 0.83±1.16 |
Computational studies of erlotinib binding to inactive EGFR-TKD
To challenge the assumption that erlotinib, gefitinib and other related inhibitors only bind to (and stabilize) the active EGFR-TKD conformation, we used the same computational approaches to ask whether erlotinib can bind to the inactive conformation of the kinase. Surprisingly, docking erlotinib into a crystal structure of the inactive EGFR-TKD (PDB entry 1XKK [13]), after removing lapatinib from the model, yielded a very similar Glide [32] score (−9.72 kcal/mol) to that seen for active EGFR-TKD (−9.34 kcal/mol). Moreover, when the two models were overlaid (Figure 1), the orientation of erlotinib and the conformation of its binding site were very similar in active (cyan) and inactive (yellow) EGFR-TKD models. N1 of the erlotinib quinazoline moiety receives a hydrogen bond from the amide nitrogen of Met769 in both cases. The two water molecules (W1 and W2) mentioned above form essentially the same hydrogen-bonding network in each model, which also involves the Gln767 backbone carbonyl, the Thr766 side chain, nitrogen N3 of the erlotinib quinazoline moiety and the Thr830 side chain (Figure 1). An additional third strongly bound water molecule (W3 in Figure 1) was seen in the model of erlotinib-bound inactive EGFR-TKD. Distances between erlotinib and protein (and between bound waters and erlotinib) were stable throughout a 10 ns MD simulation, as plotted in Supplementary Figure S7 (at http://www.BiochemJ.org/bj/448/bj4480417add.htm). Thus our computational studies failed to provide any explanation for why erlotinib might bind preferentially to the active conformation of EGFR-TKD. Yun et al. [11] suggested that favourable van der Waals interactions between gefitinib and the back of the ATP-binding cleft in active EGFR-TKD may be lost in the inactive state, but this was not seen in our modelling.
Crystal structure of erlotinib bound to inactive EGFR-TKD
Following the unexpected suggestion from our computational studies that erlotinib binds similarly to active and inactive EGFR-TKD, we sought to crystallize the kinase domain in its inactive conformation with erlotinib bound. Previous crystal structures of active EGFR-TKD with erlotinib [10] or gefitinib [11] bound were obtained by soaking the drug into pre-formed crystals. However, it is now clear that wild-type EGFR-TKD (or variants harbouring NSCLC mutations) always adopt the active conformation in crystals [10,12], because of EGFR-TKD's intrinsic tendency to form asymmetric dimers in which the C-lobe of one molecule associates with the N-lobe of its neighbour to stabilize the active conformation. Although EGFR-TKD co-crystallized with lapatinib [13], HKI-272 [7] or related inhibitors adopt the inactive conformation, wild-type EGFR-TKD only crystallizes in the inactive conformation when the asymmetric dimer is disrupted by a mutation in its interface (such as V924R in the C-lobe) [12,20] or by a Mig6-derived peptide that binds to the C-lobe dimerization site [39].
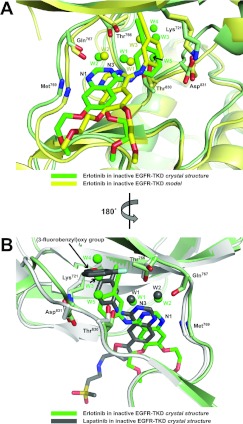
By soaking erlotinib into crystals formed by a V924R-mutated EGFR-TKD variant, we were able to determine the structure of erlotinib bound to inactive EGFR-TKD at a resolution of 2.75 Å (Table 1). As shown in Figure 2, our structure of inactive EGFR-TKD closely resembles that seen in PDB entry 1XKK [13] (in complex with lapatinib). The RMSD (root mean square deviation) between α carbon positions in these two structures was 1.30 Å, and the α carbon RMSD for overlay of our inactive structure with PDB entry 3GT8 [20] was 1.42 Å. As illustrated in Figure 3, the crystal structure shows erlotinib (green in Figure 3A) to be slightly shifted in the binding site compared with its predicted position in the model (yellow in Figure 3A); to the left in the aspect of the Figure by approximately 1 Å. The distance between the amide nitrogen of Met769 in EGFR-TKD and N1 of the erlotinib quinazoline moiety is maintained at 2.9 Å in the crystal structure and 3.2 Å in the model. The slight shift of erlotinib is associated with a similar shift in the polypeptide backbone in the region of residues 767–770. Other key side chains (Thr766, Thr830, Asp831 and Lys721) shown in Figure 3(A) are located very similarly in the model and crystal structure. Importantly, the pattern of water molecules predicted by our model was also observed crystallographically (compare yellow and green waters in Figure 3A). W1 and W2 are in very similar positions in the crystal structure and model, providing crystallographic evidence that the predicted water-mediated hydrogen-bonding network involving the quinazoline moiety of erlotinib, and Thr766/Gln767/Thr830 of EGFR-TKD is maintained in the inactive conformation. Interestingly, equivalent waters are also seen in the published crystal structure of lapatinib-bound inactive EGFR-TKD (Figure 3B) [13]. Moreover, a crystallographic water was seen in our erlotinib/inactive EGFR-TKD structure close to the position of W3 in the computational model, and near the side chains of Thr766 and Thr830. Additional waters labelled W4 and W5 in Figure 3(A) are seen in our erlotinib/EGFR-TKD crystal structure and not the model. Interestingly, as shown in Figure 3(B), waters W3–W5 in our structure lie in the same pocket within inactive EGFR-TKD that is occupied by the (3-fluorobenzyl)oxy group of lapatinib in PDB entry 1XKK [13]; these waters also interact with the EGFR-TKD backbone in parts of the αC helix (Met742) and Phe832 of the DFG motif.
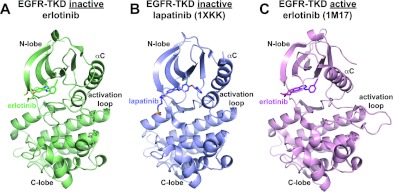
Figure 2. Crystal structures of TKI-bound EGFR-TKD.
(A) The 2.75 Å resolution structure of V924R-mutated EGFR-TKD in its inactive form bound to erlotinib (green) as described in the text and Table 1. (B) Crystal structure from PDB entry 1XKK [13] showing lapatinib (blue) bound to inactive EGFR-TKD. (C) Crystal structure from PDB entry 1M17 [10] showing erlotinib (magenta) bound to active EGFR-TKD. Note that the αC helix is in the ‘in’ or ‘active’ position in (C), but in the ‘out’ or ‘inactive’ position in (A) and (B), and that the characteristic short α-helix seen in the inactive EGFR-TKD activation loop is present in (A) and (B), but not (C). As described in the text, the crystal structure shown in (A) confirms our computational findings that erlotinib can bind to the inactive EGFR-TKD conformation.
Figure 3. Erlotinib binding to inactive EGFR-TKD.
(A) The crystallographically observed mode of erlotinib binding to inactive EGFR-TKD (green) is compared with the computational model (yellow). Protein structure in the background coloured green for the crystal structure and yellow for the model. (B) The mode of erlotinib binding to inactive EGFR-TKD observed crystallographically (green) is compared with the mode of lapatinib binding to inactive EGFR-TKD in PDB entry 1XKK [13] (grey), in a view rotated by 180° about a vertical axis compared with that seen in (A). Waters W1–W5 are labelled for the erlotinib/inactive EGFR-TKD structure. Waters W1 and W2 are also found in the lapatinib complex [13]. W3–W5 in the erlotinib complex structure lie in a pocket occupied by the (3-fluorobenzyl)oxy group of lapatinib in PDB entry 1XKK. Functional groups, side chains and bound waters are shown as described in Figure 1. Polypeptide in the foreground has been removed for clarity.
Conclusions
It is generally assumed that the EGFR inhibitors gefitinib and erlotinib bind selectively to the active conformation of EGFR-TKD, whereas lapatinib selectively binds the inactive configuration. A preference of lapatinib for inactive EGFR-TKD can be rationalized, since it has a bulky [(3-fluorobenzyl)oxy] substituent on its aniline ring that projects into the space opened up upon displacement of the αC helix in the inactive kinase (as shown in Figure 2B). For erlotinib, however, no significant difference in the binding affinity for inactive and active EGFR-TKD conformations could be detected using a range of computational approaches. This led us to confirm crystallographically that erlotinib can indeed bind to EGFR-TKD in its inactive conformation (Figures 2A and 3). These findings have several important implications.
First, our findings complicate suggestions in the literature that erlotinib and gefitinib drive EGFR dimerization by stabilizing the active configuration. Since asymmetric dimerization of EGFR-TKD promotes its acquisition of the active conformation, the converse should also be true: that stabilization of the active TKD conformation drives dimerization. Thus if erlotinib and gefitinib selectively bind and stabilize the active EGFR-TKD conformation, their binding should drive asymmetric dimer formation. Indeed, it has been reported that gefitinib stabilizes formation of EGFR-containing heterodimers in cells [40]. Moreover, Springer and colleagues have reported in electron microscopy studies [15,41] that gefitinib promotes dimerization of near full-length EGFR by inducing structures that resemble asymmetric EGFR-TKD dimers. The finding in the present study that erlotinib also binds to inactive (monomeric) EGFR-TKD is difficult to reconcile with the simple interpretation of these reports, as are other published data indicating that neither stabilizing the active conformation of EGFR-TKD with oncogenic mutations nor disrupting the asymmetric dimer interface with a V924R mutation (and thus stabilizing the inactive conformation) alters the affinity of EGFR-TKD for gefitinib or erlotinib [14,15].
Secondly, the findings of the present study argue that the relative abilities of erlotinib/gefitinib-type EGFR TKIs and lapatinib/neratinib TKIs to inhibit different mutationally activated EGFR variants is more complicated than previously thought. Given the results of the present study, the fact that NSCLC mutants and glioblastoma mutants are selectively inhibited by erlotinib and lapatinib respectively [3] seems unlikely simply to reflect stabilization of different conformational states of EGFR-TKD in the two cancers (active for NSCLC, inactive for glioblastoma). Rather, the different sensitivities of the EGFR mutants are likely to reflect more complicated conformational, and dynamic, characteristics, as suggested by a previous study [17]. It is clear, for example, that lapatinib dissociates from EGFR-TKD much more slowly than erlotinib or gefitinib [13] (although covalent inhibitors dissociate even more slowly, yet still show differential effects). More extensive analysis is required to identify the differences between these groups of inhibitors, which ultimately should aid in tailoring the type of inhibitor used clinically to the mode in which EGFR has been aberrantly activated.
Online data
AUTHOR CONTRIBUTION
Jin Park performed all of the experimental and crystallographic work, guided by Mark Lemmon. Yingting Liu performed all computational modelling work guided by Ravi Radhakrishnan. Jin Park, Yingting Liu, Mark Lemmon and Ravi Radhakrishnan all interpreted results and prepared the paper.
ACKNOWLEDGEMENTS
We thank Kathryn Ferguson and members of the Lemmon, Ferguson and Radhakrishnan laboratories for advice and reading of the paper before submission.
FUNDING
Crystallographic data were collected at beamline 23ID-D of GM/CA@APS, funded by the National Cancer Institute [grant number Y1-CO-1020], the National Institute of General Medical Sciences [grant number Y1-GM-1104] and the U.S. Department of Energy [contract DE-AC02-06CH11357]. This work was supported, in part, by the National Institutes of Health [grant numbers R01-CA079992 (to M.A.L.) and R01-GM084959 (to Graham Carpenter and M.A.L.)], the National Science Foundation [grant numbers 0835539 and 0835389 (to R.R.)] and a Predoctoral Fellowship from the Great Rivers Affiliate of the American Heart Association [grant number 11PRE7670020 (to J.H.P.)].
References
- 1.Sharma S. V., Bell D. W., Settleman J., Haber D. A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vivanco I., Robins H. I., Rohle D., Campos C., Grommes C., Nghiemphu P. L., Kubek S., Oldrini B., Chheda M. G., Yannuzzi N., et al. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discovery. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pao W., Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jänne P. A., Gray N., Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat. Rev. Drug Discovery. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa S., Kukimoto-Niino M., Parker L., Handa N., Terada T., Fujimoto T., Terazawa Y., Wakiyama M., Sato M., Sano S., et al. Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene. 2012 doi: 10.1038/onc.2012.21. doi:10.1038/onc.2012.21. [DOI] [PubMed] [Google Scholar]
- 7.Yun C. H., Mengwasser K. E., Toms A. V., Woo M. S., Greulich H., Wong K. K., Meyerson M., Eck M. J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eck M. J., Yun C. H. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim. Biophys. Acta. 2010;1804:559–566. doi: 10.1016/j.bbapap.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Gray N. S. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 10.Stamos J., Sliwkowski M. X., Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 11.Yun C. H., Boggon T. J., Li Y., Woo M. S., Greulich H., Meyerson M., Eck M. J. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Wood E. R., Truesdale A. T., McDonald O. B., Yuan D., Hassell A., Dickerson S. H., Ellis B., Pennisi C., Horne E., Lackey K., et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 14.Fabian M. A., Biggs, 3rd W. H., Treiber D. K., Atteridge C. E., Azimioara M. D., Benedetti M. G., Carter T. A., Ciceri P., Edeen P. T., Floyd M., et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 15.Lu C., Mi L. Z., Schürpf T., Walz T., Springer T. A. Mechanisms for kinase-mediated dimerization of the EGF receptor. J. Biol. Chem. 2012 doi: 10.1074/jbc.M112.414391. doi:10.1074/jbc.M112.414391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Longo P. A., Tarrant M. K., Kim K., Head S., Leahy D. J., Cole P. A. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat. Struct. Mol. Biol. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkovich K. J., Hariono S., Garske A. L., Zhang J., Blair J. A., Fan Q. W., Shokat K. M., Nicolaides T., Weiss W. A. Kinetics of inhibitor cycling underlie therapeutic disparities between EGFR-driven lung and brain cancers. Cancer Discovery. 2012;2:450–457. doi: 10.1158/2159-8290.CD-11-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Jura N., Endres N. F., Engel K., Deindl S., Das R., Lamers M. H., Wemmer D. E., Zhang X., Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winn M. D., Isupov M. N., Murshudov G. N. Use of TLS anisotropic displacements in macromolecular refinement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 26.Schüttelkopf A. W., van Aalten D. M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 27. Reference deleted.
- 28. Reference deleted.
- 29. Reference deleted.
- 30.Jorgensen W. L., Tiradorives J. The OPLS potential functions for proteins: energy minimizations for crystals of cyclic-peptides and crambin. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 31.Sherman W., Day T., Jacobson M. P., Friesner R. A., Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 32.Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., Sanschagrin P. C., Mainz D. T. Extra Precision Glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson M. P., Pincus D. L., Rapp C. S., Day T. J. F., Honig B., Shaw D. E., Friesner R. A. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 34.Foloppe N., MacKerell A. D. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21:86–104. [Google Scholar]
- 35.Wang J., Morin P., Wang W., Kollman P. A. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 36.Swanson J. M. J., Henchman R. H., McCammon J. A. Revisiting free energy calculations: a theoretical connection between MM/PBSA and direct calculation of the association free energy. Biophys. J. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Im W., Beglov D., Roux B. Continuum Solvation Model: computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput. Phys. Commun. 1998;111:59–75. [Google Scholar]
- 38.Sanner M. F., Olson A. J., Spehner J. C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anido J., Matar P., Albanell J., Guzmán M., Rojo F., Arribas J., Averbuch S., Baselga J. ZD1839, a specific epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, induces the formation of inactive EGFR/HER2 and EGFR/HER3 heterodimers and prevents heregulin signaling in HER2-overexpressing breast cancer cells. Clin. Cancer Res. 2003;9:1274–1283. [PubMed] [Google Scholar]
- 41.Mi L. Z., Lu C., Li Z., Nishida N., Walz T., Springer T. A. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat. Struct. Mol. Biol. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.