Summary
Interactions between the endoderm and mesoderm that mediate myocardial induction are difficult to study in vivo because of the small size of mammalian embryos at relevant stages. However, we and others have demonstrated that signals from endodermal cell lines can influence myocardial differentiation from both mouse and human embryoid bodies (EBs), and because of this, assays that utilize embryonic stem (ES) cells and endodermal cell lines provide excellent in vitro models to study early cardiac differentiation. Extraembryonic endoderm (XEN) stem cells have a particular advantage over other heart-inducing cell lines in that they can easily be derived from both wild type and mutant mouse blastocysts. Here we describe the first isolation of a Nodal mutant XEN stem cell line. Nodal−/− XEN cell lines were not isolated at expected Mendelian ratios, and those that were successfully established, showed an increase in markers for the anterior visceral endoderm (AVE). Since AVE represents the heart-inducing endoderm in the mouse, cardiac differentiation was compared in EBs treated with conditioned medium (CM) collected from wild type or Nodal−/− XEN cells. EBs treated with CM from Nodal−/− cells began beating earlier and showed early activation of myocardial genes, but this early cardiac differentiation did not cause an overall increase in cardiomyocyte yield. By comparison, CM from wild type XEN cells both delayed cardiac differentiation and caused a concomitant increase in overall cardiomyocyte formation. Detailed marker analysis suggested that early activation of cardiac differentiation by Nodal−/− XEN CM caused premature differentiation and subsequent depletion of cardiac progenitors.
Keywords: Nodal, Extraembryonic endoderm (XEN) stem cells, Cardiomyocyte differentiation, Mouse ES cells
Introduction
Inductive interactions between endoderm and mesoderm mediate the initial phases of cardiac differentiation in all vertebrates by activating expression of early cardiac markers such as Nkx2.5 and Tbx5 (Jacobson, 1960; Orts-Llorca and Gil, 1965; Fullilove, 1970; Sugi and Lough, 1994; Tonegawa et al., 1996; Arai et al., 1997; Schneider and Mercola, 1999; Lough and Sugi, 2000; David and Rosa, 2001; Marvin et al., 2001; Mummery et al., 2003; Yuasa et al., 2005; Brown et al., 2010a; Holtzinger et al., 2010). However, because of the small size and relative inaccessibility of mammalian embryos at pertinent developmental stages, little headway has been made in identifying specific cardiogenic signals in the endoderm. A major advance came with the discovery that visceral endoderm (VE)-like cell lines, such as the embryonal carcinoma-derived END2 cells, can activate and/or increase cardiac differentiation from both mouse and human embryonic stem (ES) cells (Mummery et al., 2003). However, since END2 cells cannot be re-derived from mutant embryos, they cannot be used to study specific inducing factors in the endoderm. We recently showed that XEN cells express markers for the AVE (Brown et al., 2010b) and possess cardiogenic potential similar to END2 cells (Brown et al., 2010a). XEN cells can easily be isolated from both wild type and mutant mouse embryos. To accomplish this, 3.5 dpc blastocysts are grown in vitro for several days until they attach to feeder cells. At this point, outgrowths of trophoblast cells can be dissected away and the remaining cells dissociated and replated. Ultimately, cell morphology and marker analysis (Brown et al., 2010b) can be used to distinguish primitive endoderm stem cells (XEN cells) from embryonic stem cells (ES cells). We previously demonstrated that XEN cells comprise a heterogeneous population of cells expressing markers for all primitive endoderm derivatives including the parietal endoderm (PE), visceral endoderm (VE) and anterior visceral endoderm (AVE) (Brown et al., 2010b). More recently it has been demonstrated that XEN cells can be directed toward specific endodermal lineages by the addition of growth factors, with BMP directing them to the VE lineage (Artus et al., 2011), and with NODAL directing cells to the AVE lineage (Julio et al., 2011). Because of this, and because primitive endoderm and epiblast remain in contact for at least two days during XEN cell isolation, we hypothesized that signals present in either the epiblast cells or within the primitive endoderm of the isolated blastocyst could influence the specific subtypes of cells that develop in any given XEN cell line. At preimplantation stages in the mouse, Nodal is expressed throughout the epiblast, and both a Nodal transgene (Varlet et al., 1997; Brennan et al., 2001; Camus et al., 2006; Mesnard et al., 2006; Granier et al., 2011) and Nodal mRNA (Norris et al., 2002; Yamamoto et al., 2004) are transiently expressed in the VE. Moreover, NODAL has been shown to affect AVE marker expression in embryos (Brennan et al., 2001) and in XEN cells (Julio et al., 2011). Therefore, loss of Nodal should also impact the specific lineages that become established in XEN cell lines.
To test this, Nodal−/− XEN cells were isolated and characterized by immunocytochemistry, Western blot and QRT-PCR for expression of primitive endoderm (PrE), VE and AVE markers. XEN cells isolated from Nodal mutant blastocysts showed a surprising upregulation of AVE markers. EBs that were treated with Nodal−/− XEN CM began to beat earlier than those treated with Nodal+/+ XEN CM, suggesting that factors regulated by Nodal play a role in regulating temporal aspects of heart development. Finally, we demonstrate that this early activation of cardiac differentiation correlated with an early activation of Islet-1 expression, which in the embryo, marks cardiac progenitors in the second heart field (SHF). By comparison, expression of Tbx5 and Nkx2.5, markers for the first heart field (FHF), were either unchanged or slightly delayed.
Results
Nodal mutant XEN cell lines were not derived at expected Mendelian ratios
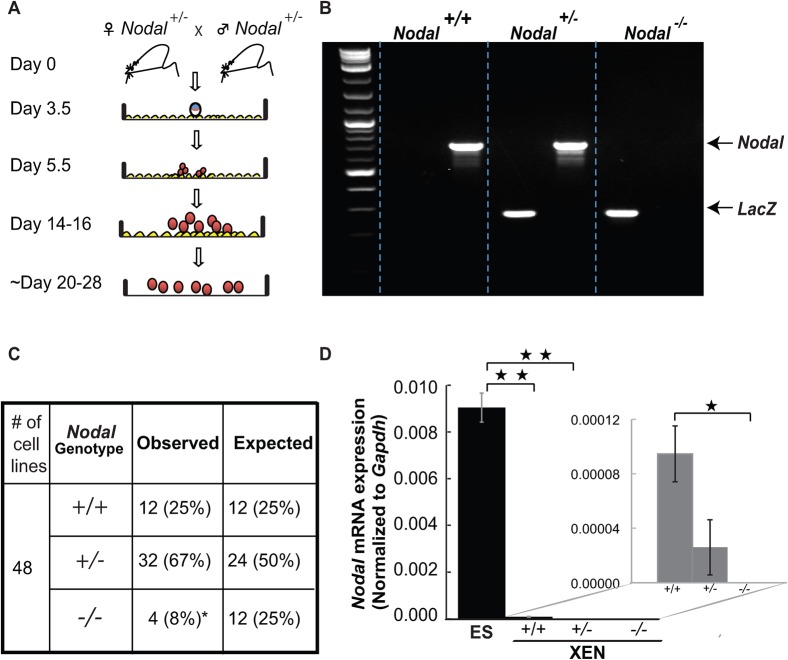
To test the hypothesis that Nodal expression in the blastocyst stage embryo impacts the patterning of XEN cells that are isolated from them, XEN cell lines were established from Nodal mutant embryos. Because Nodal−/− embryos are not viable, XEN cell lines were established by crossing mice that possessed a heterozygous deletion of Nodal (Nodal+/−). Blastocysts of unknown genotype were isolated at 3.5 dpc, and XEN cell lines were derived according to a previously published protocol (Artus et al., 2010) (Fig. 1A). The genotype of individual XEN cell lines was confirmed by PCR after isolation and expansion, using primers specific for either Nodal or LacZ, which was knocked into the Nodal locus as part of the deletion strategy (Collignon et al., 1996) (Fig. 1B).
Fig. 1. Derivation of murine blastocyst-derived eXtraembryonic ENdoderm (XEN) stem cells is impaired in the absence of Nodal.
(A) Schematic of the XEN cell derivation protocol. Nodal+/− mice were crossed on day 0. Blastocysts were collected on day 3.5 in M2 medium and cultured in 4-well gelatin-coated cell culture plates with mitotically inactivated MEFs. Outgrowths were disaggregated at 5.5 days into 2 to 3-cell clumps and replated. Colonies with characteristic XEN cell morphology appeared at day 20–28. (B) PCR genotyping of XEN cell lines. (C) Nodal−/− cell lines were not isolated at expected Mendelian ratios, only 4 of 48 cell lines (8%) were Nodal−/− in genotype as compared to 25% expected (p = 0.018), while heterozygous and wild type XEN cell lines were isolated at or above expected Mendelian ratios. (D) QRT-PCR on cDNA synthesized from XEN cells. Relative Nodal expression was presented after normalization to Gapdh. Insert shows low levels of Nodal expression in Nodal+/+ and Nodal+/− but not in Nodal −/− XEN cell lines. Error bars represent s.e.m. of three samples for each genotype (* indicates a p-value< 0.05, ** indicates a p-value< 0.006).
Wild type and Nodal+/− XEN cell lines were isolated at or above expected Mendelian ratios (12/48 (25%) and 32/48 (67%) respectively). By contrast, Nodal−/− XEN cell lines were isolated well below expected ratios (n = 4/48; 8% as compared to 25% expected, p = 0.018) (Fig. 1C). This suggests that Nodal expression in the embryo is required for optimal XEN cell derivation and/or maintenance.
Nodal mRNA levels were investigated in wild type and Nodal mutant XEN cell lines. Both wild type and Nodal+/− XEN cells expressed very low levels of Nodal when grown in the presence of LIF (Fig. 1D). However, this expression was rapidly lost after the removal of LIF from the medium (data not shown). In addition, since the mouse strain used for XEN cell isolation possessed a knock-in of the LacZ gene into the Nodal locus, XEN cell lines were stained for beta-galactosidase activity. The absence of beta-galactosidase activity corroborated the QRT-PCR data, demonstrating that Nodal is not expressed in these XEN cells (data not shown). These data are consistent with previous studies showing that wild type XEN cells do not express Nodal when expanded in the absence of LIF (Kunath et al., 2005; Brown et al., 2010b). Together, these findings also suggested that Nodal is not normally expressed in cultured XEN cells and is therefore not necessary for their survival, but is probably required for their derivation.
Nodal mutant XEN cells do not express pluripotency markers but do express markers for the primitive (PrE) and visceral endoderm (VE)
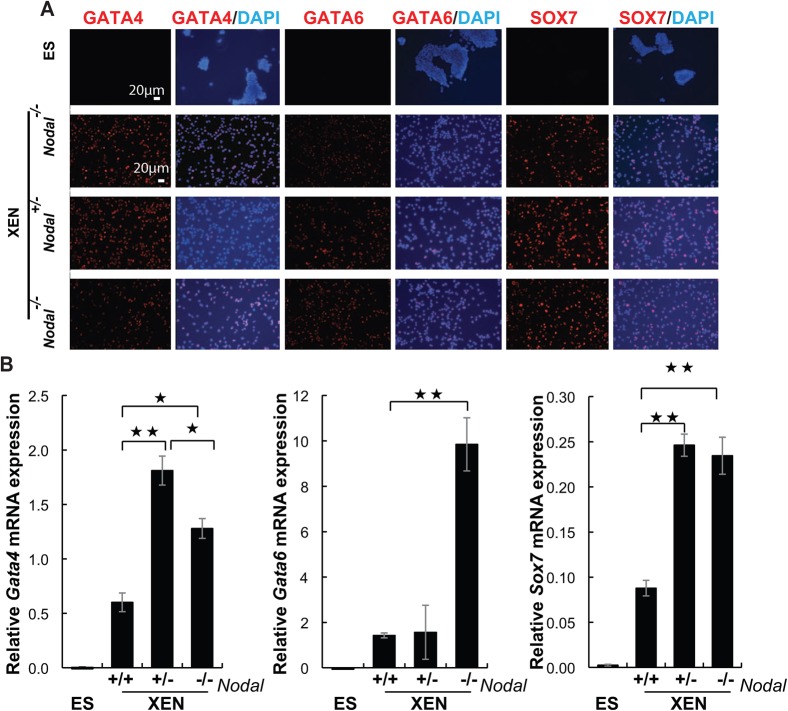
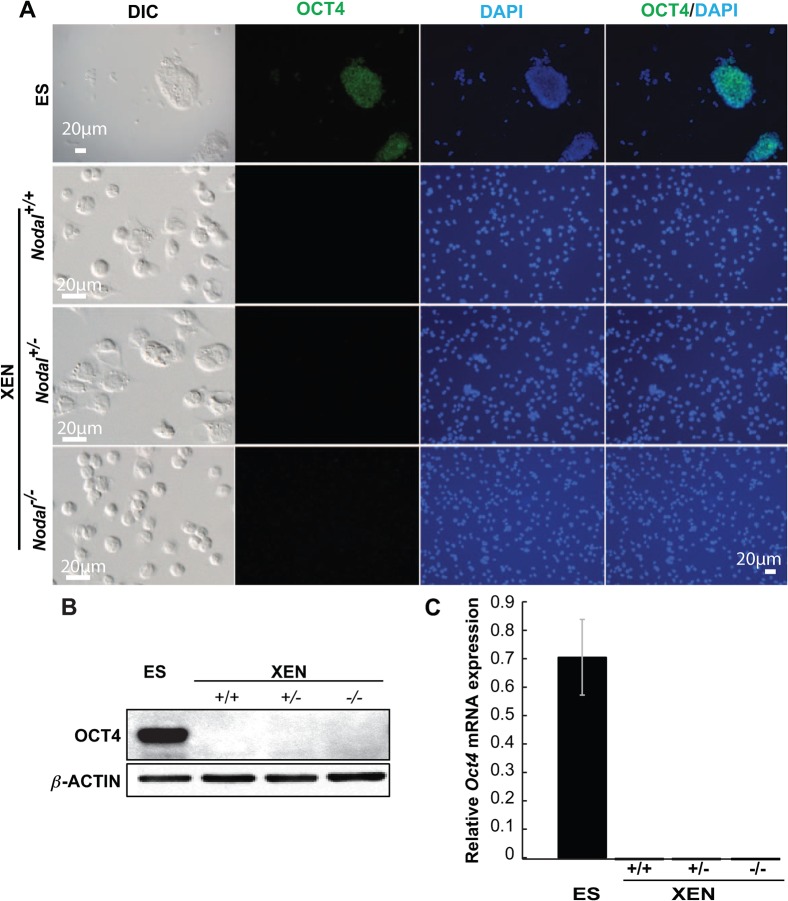
All of the cell lines that were identified, based on their morphology, as putative XEN cell lines, were confirmed as such by expression of the PrE/VE markers GATA4, GATA6, and SOX7. These markers were never detected in ES cells (Fig. 2A,B). Conversely, consistent with previous studies of wild type XEN cells (Kunath et al., 2005), Nodal−/− cell lines did not express the pluripotency marker OCT4, which was strongly expressed in ES cells. This was demonstrated by immunocytochemistry (Fig. 3A), Western blot (Fig. 3B), and QRT-PCR (Fig. 3C). Together, these data confirmed the identity of these cell lines as XEN cells and demonstrated that they were not contaminated with ES cells or any other pluripotent cell type that can be derived from mouse blastocysts.
Fig. 2. XEN cell lines derived from the Nodal mutant embryos express markers for the primitive and visceral endoderm.
Representative images of (A) immunocytochemistry for GATA4 (red), GATA6 (red) and SOX7 (red). Blue reveals DAPI staining of nuclei. Scale bar represents 20 µM. (B) QRT-PCR analysis for these same genes (* indicates a p-value< 0.05, ** indicates a p-value< 0.006).
Fig. 3. XEN cell lines derived from the Nodal mutant embryos do not express the pluripotency marker OCT4.
(A) Representative images of OCT4 (green) immunofluorescence in ES but not in XEN cells. Blue reveals DAPI staining of nuclei. Scale bars represents 20 µM. (B) Western blotting assay showing OCT4 detection in ES but not XEN cells. (C) QRT-PCR data showing Oct4 mRNA expression in ES but not XEN cells.
Our previous studies demonstrated that XEN cells express markers for primitive, visceral and parietal endoderm. This study also described XEN cell morphology in some detail and noted that several distinct cellular morphologies were discernible in wild type XEN cells (Brown et al., 2010b). Interestingly, XEN cells isolated from Nodal+/− and Nodal−/− embryos were more homogeneous in morphology than wild type XEN cells, and based on these morphological differences, cell lines of each of the three genotypes were readily distinguishable from the others (Fig. 3A and data not shown). Other studies have recently demonstrated that XEN cells become lineage restricted after the addition of specific growth factors (Artus et al., 2011; Julio et al., 2011), and one hallmark of this lineage restriction was a distinct change in the morphology of the XEN cells (Artus et al., 2011). To determine if the loss of Nodal during XEN cell isolation similarly impacted gene expression, the XEN cell lines were analyzed for quantitative changes in gene expression by QRT-PCR (Figs 2B, 4). Gata4 and Sox7 expression were highly sensitive to NODAL dosage since their expression was altered in both heterozygous and homozygous mutant cell lines. However, Gata6 expression was only significantly increased in Nodal−/− cell lines. Based on our previous work, these data are consistent with an increase in VE fates within the Nodal−/− XEN cell lines (Brown et al., 2010b).
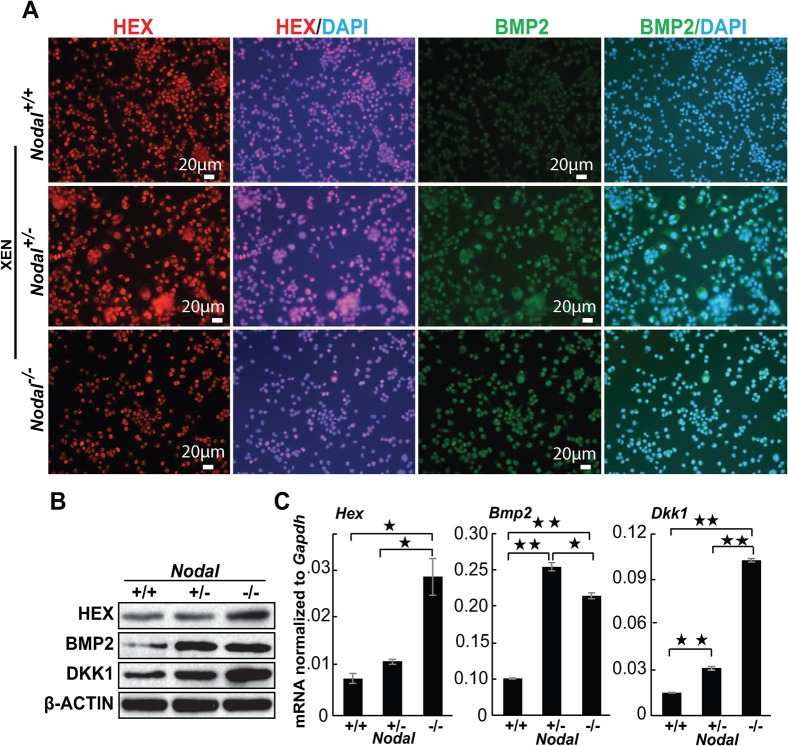
Fig. 4. Nodal regulates anterior visceral endoderm gene expression in XEN cells.
(A) Representative images of immunocytochemistry analysis. HEX (red) and BMP2 (green) expression in XEN cells. Blue reveals DAPI staining of nuclei. XEN cells of all genotypes expressed HEX and BMP2. Scale bar represents 20 µM. (B) Western blot of HEX, BMP2, and DKK1 in XEN cells. (C) QRT-PCR analysis of Hex, Bmp2, and Dkk1 mRNA expression in XEN cells. mRNA expression levels were presented after normalization to Gapdh. Error bars indicate s.e.m. (* indicates a p-value< 0.05, ** indicates a p-value< 0.006).
Nodal regulates anterior visceral endoderm (AVE) gene expression in XEN cells
Because previous studies had suggested a specific role for Nodal in regulating marker expression in the AVE (Brennan et al., 2001), we analyzed the AVE specific markers HEX, DKK1 and BMP2 (Madabhushi and Lacy, 2011), by immunocytochemistry, Western blot, and QRT-PCR. Both Nodal−/− and Nodal+/− XEN cells expressed higher levels of the AVE markers BMP2 and DKK1, while only Nodal−/− (but not Nodal+/−) XEN cells expressed higher levels of the AVE marker HEX (Fig. 4). We also analyzed the expression of Cerberus-like and Lefty1; known targets of NODAL signaling in the AVE. Both markers were absent from XEN cells of all genotype (data not shown). These data demonstrated that Nodal negatively regulates the expression of a subset of AVE markers in XEN cells.
Medium conditioned by Nodal mutant XEN cells impacts the timing of cardiogenesis in EBs
AVE and AVE-like cell lines, including XEN cells, have been shown to increase cardiac differentiation from both nascent mesoderm and differentiating EBs (Arai et al., 1997; Mummery et al., 2003; Stary et al., 2005; Nijmeijer et al., 2009; Brown et al., 2010a). Since our data showed an upregulation of at least a subset of AVE markers in the Nodal−/− XEN cells, we hypothesized that these cells might possess enhanced cardiogenic potential as compared to wild type XEN cells. We previously showed that adding wild type XEN CM from days 4–6 of differentiation, increased cardiac formation within EBs (Brown et al., 2010a). Therefore, XEN CM collected from the different genotypes (Nodal+/+ XEN CM, Nodal+/− XEN CM or Nodal−/− XEN CM) was added to EB culture medium between days 4 and 6 of differentiation. The ES cells used in these studies possessed a MHCα::GFP reporter (Takahashi et al., 2003) that we previously validated as a faithful reporter for cardiomyocyte formation (Brown et al., 2010a). Cardiomyocyte differentiation was first evaluated by assessing the percentage of EBs that contained beating areas at each day of differentiation (Fig. 5A). EBs treated with Nodal−/− XEN CM or Nodal+/− XEN CM began beating on day 5 (with 18% and 2% beating, respectively). EBs treated with Nodal+/+ XEN CM started beating on day 6. Finally EBs treated with control medium began beating on day 7. By day 14, there were no statistically significant differences among the four samples. Together these data suggest that the dosage of Nodal altered the characteristics XEN cells, which in turn, changed the way that XEN cells impacted the timing of cardiomyocyte differentiation.
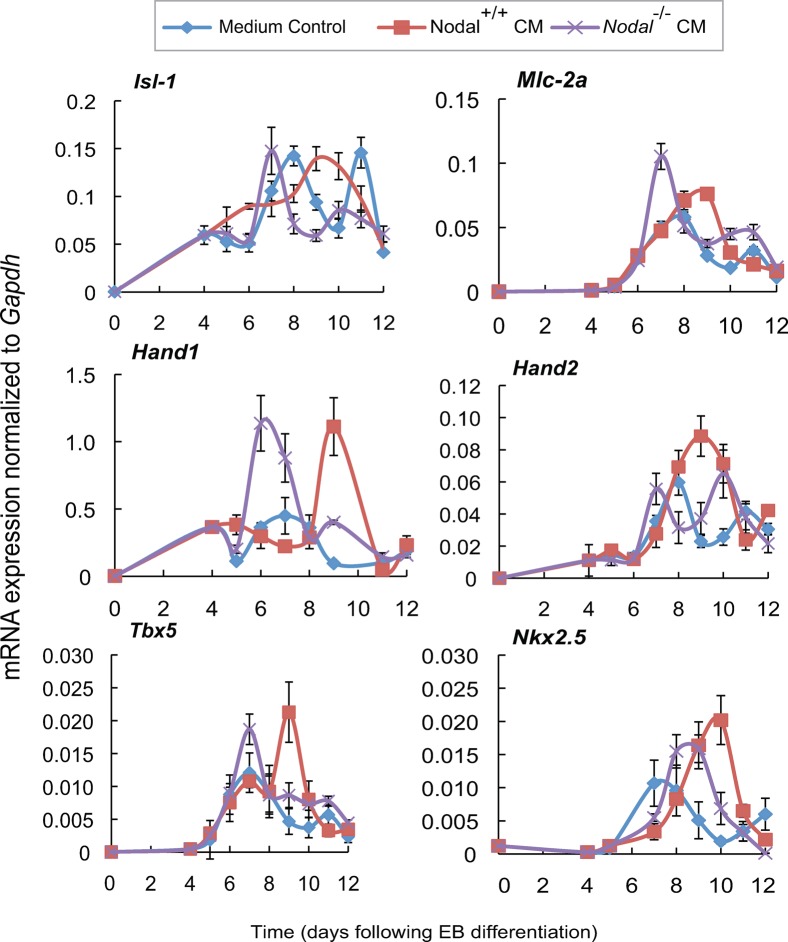
Fig. 5. Nodal impacts the initiation of cardiogenesis in embryoid bodies.
Embryoid bodies (EBs) were treated between days 4 and 6 of differentiation with CM from XEN cells of all three genotypes (Nodal+/+ XEN CM, Nodal+/− XEN CM, Nodal−/− XEN CM), or medium control. (A) Beating EBs were counted and represented as a percentage of the total number of EBs counted. (B,C) Stills from supplementary material Movies 1 and 2, showing representative beating areas in EBs treated with Nodal+/− XEN CM and control EBs, respectively. EBs express the MHCα::GFP reporter that indicates cardiomyocyte differentiation. Scale bar represent 100 µM. (D) Myosin heavy chain alpha (MHCα) mRNA expression (normalized to Gapdh) was examined in EBs by QRT-PCR. (E) Flow cytometry analysis on day 14 of cardiac differentiation comparing untreated EBs to those treated on days 4–6 with Nodal mutant XEN CM. EBs were made from CGR8 ES cells in which the MHCα promoter drives GFP protein expression upon cardiomyocyte formation. The percentage of MHCα expressing (GFP positive) cells were significantly increased in EBs treated with wild type XEN CM as compared to both controls and EBs treated with Nodal−/− XEN CM. Error bars indicate s.e.m.(*indicates a p-value< 0.05).
By day14, EBs developed large areas of beating cardiomyocytes expressing GFP. Interestingly, the contracting loci that formed in response to Nodal−/− CM (Fig. 5B, supplementary material Movie 1) appeared to be well organized as compared to untreated EBs (Fig. 5C, supplementary material Movie 2), with cardiomyocytes aligned in highly-organized, synchronously contracting sheets (supplementary material Movie 1). In contrast, contracting loci that formed in control EBs and those treated with wild type CM, appeared more disorganized with many areas within a single contractile region appearing to initiate contractions independently of one another (supplementary material Movie 2). These findings suggest that there is a higher degree of electrophysiological coupling in cardiomyocytes that develop from EBs treated with Nodal mutant CM and may suggest that these cells differentiate simultaneously. On the other hand, our data suggest that cardiomyocytes derived from either untreated EBs or EBs treated with wild type XEN CM may develop in several waves of differentiation.
To quantitatively compare overall cardiomyocyte differentiation between treated and untreated EBs, MHCα levels were investigated by QRT-PCR. MHCα expression peaked on day 7 in all samples, and mRNA levels were significantly increased in EBs treated with Nodal−/− XEN CM. No change in MHCα expression was observed in response to Nodal+/+ XEN CM or Nodal+/− XEN CM as compared to control EBs (Fig. 5D).
To quantify total myocardial differentiation after treatment with CM, EBs possessing the MHCα::GFP reporter (Takahashi et al., 2003) were treated as described above and analyzed by flow cytometry on day 14 for overall cardiac formation. Unexpectedly, although EBs treated with Nodal−/− XEN CM initiated cardiogenesis earlier and showed increased expression of MHCα on day 7 of differentiation, this treatment did not increase the total number of cardiomyocytes that formed as compared to controls. Instead, consistent with our previous findings (Brown et al., 2010a), EBs treated with wild type XEN CM showed an approximate 2-fold increase in overall cardiomyocyte differentiation (Fig. 5E).
Cardiac gene expression is delayed in EBs treated with Nodal +/+ XEN CM
Treatment of EBs with Nodal−/− XEN CM caused earlier initiation of cardiogenesis, the formation of large, well-ordered beating areas and an increase in MHCα expression at day 7 of differentiation but with no subsequent increase in the total percentage of cardiomyocytes that formed. Based on these seemingly contradictory results, we hypothesized that Nodal−/− XEN CM might cause a premature differentiation of cardiac progenitors with a subsequent depletion of the progenitor pool. The vertebrate heart develops from two sources of multipotent cardiac progenitors, the first heart field (FHF) and the second heart field (SHF) (Kelly et al., 2001; Cai et al., 2003; Meilhac et al., 2004; Zaffran et al., 2004; Waldo et al., 2005; Moretti et al., 2006; Abu-Issa and Kirby, 2007; Black, 2007; Qyang et al., 2007; Dyer and Kirby, 2009). FHF cells in the cardiac crescent express Nkx2.5 and Tbx5, the latter of which is considered to be a more specific marker for the FHF, whereas Nkx2.5 is expressed in both the FHF and SHF. At the cardiac crescent and linear heart tube stages, SHF cells, identified by the expression of Islet-1 (Cai et al., 2003; Moretti et al., 2006; Qyang et al., 2007; Bu et al., 2009) and Fgf10 (Kelly et al., 2001), reside adjacent to the FHF (Cai et al., 2003). Differentiation of SHF cells into myocardial cells is delayed for several days by a mechanism that is not well understood. The atria contain derivatives from both the FHF and the SHF. By comparison, the left ventricle develops primarily from the FHF and the right ventricle and outflow tract (OFT) primarily from SHF progenitors (Stennard et al., 2005; Vincent and Buckingham, 2010).
To assess whether endodermal signals impact one or both of these cardiac progenitor populations we assessed the expression of FHF and SHF markers by QRT-PCR over the course of EB differentiation with or without the addition of XEN CM. Untreated EBs typically showed two pulses of Islet-1 transcription over the first 12 days of EB differentiation (blue line, Fig. 6). By comparison, EBs treated with CM from wild type XEN cells had only a single, broader peak of Islet-1 expression, the timing of which was delayed as compared to controls (red line, Fig. 6). Similarly, EBs treated with Nodal−/− XEN CM also showed a single large peak of Islet-1 expression, but in this case the peak was narrower and occurred earlier than the first peak in control EBs (purple line, Fig. 6). This finding suggested that CM from wild type XEN cells delayed the differentiation of Islet-1 expressing cells, whereas CM from Nodal−/− XEN cells caused Islet-1 expressing cells to differentiate prematurely. Consistent with this, markers for differentiated cardiomyocytes including Mlc2a, Hand1 and Hand2 were all delayed in EBs treated with wild type CM (red lines, Fig. 6) but were initiated early in EBs treated with Nodal−/− XEN CM (purple line, Fig. 6).
Fig. 6. Conditioned media from Nodal mutant XEN cells shifts the timing of cardiac gene expression in embryoid bodies (EBs).
EBs were treated with Nodal+/+ XEN CM, Nodal−/− XEN CM, or medium control from days 4 to 6 of differentiation. Cardiac genes were examined over the course of differentiation by QRT-PCR for expression of the cardiac progenitor marker, Islet-1, markers for differentiated cardiomyocytes, Mlc2a, Hand1 and Hand2, and the early cardiac markers Tbx5 and Nkx2.5. Relative mRNA expression was presented after normalization to Gapdh. Comparatively, EBs treated with Nodal+/+ XEN CM were shown to have a significant temporal shift in the onset of cardiac gene expression.
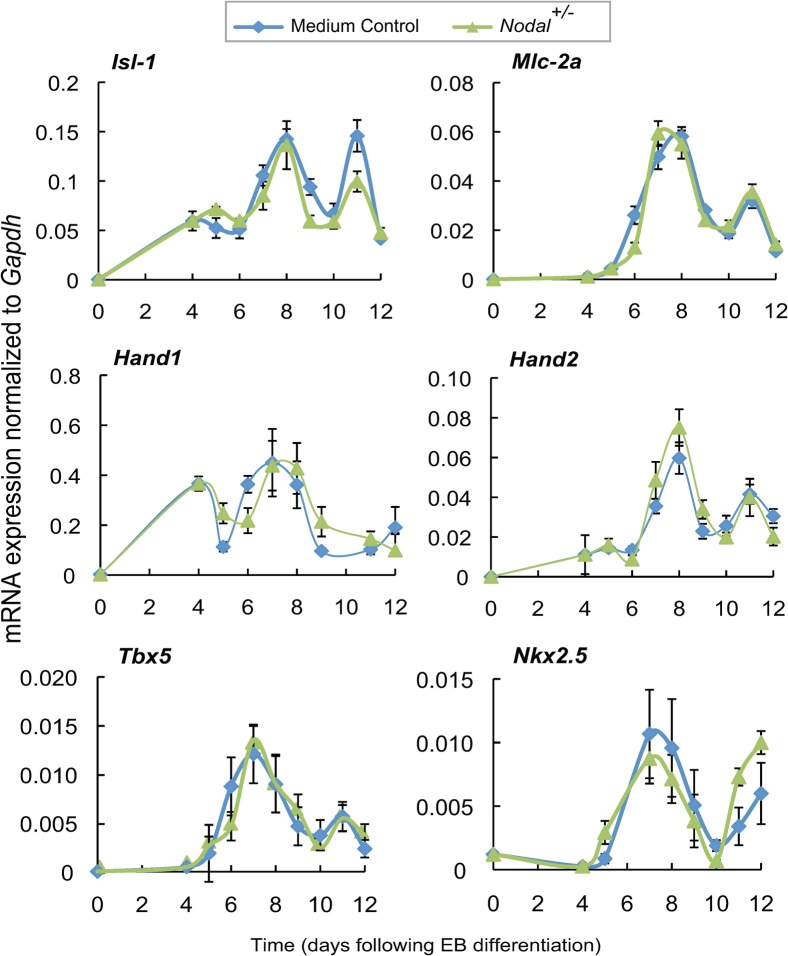
In contrast, Tbx5 and Nkx2.5 were also delayed in EBs treated with wild type XEN CM, but addition of Nodal−/− XEN CM did not cause premature activation of these markers. Rather, Nodal−/− XEN CM had no effect on the timing of Tbx5 and delayed the expression of Nkx2.5 (purple line, Fig. 6). Therefore, the premature differentiation of cardiomyocytes observed in response to Nodal−/− XEN CM is more likely to be due to a specific effect on Islet-1 expressing cells rather than premature differentiation of cardiac progenitors in general. All of these markers were also assessed in EBs treated with Nodal+/− XEN CM, but no differences were observed when compared to controls (Fig. 7). Together these findings suggest that a premature differentiation of Islet-1 positive progenitors could account for the early differentiation of cardiomyocytes that we observed in EBs treated with Nodal−/− XEN CM. Likewise, the increased number of cardiomyocytes observed when EBs were treated with wild type XEN CM could be accounted for by the delay discovered in the differentiation of cardiac progenitors. Furthermore, this delay may have allowed for increased proliferation of the progenitor pool.
Fig. 7. Conditioned media from Nodal+/− XEN cells did not shift the timing of cardiac gene expression in EBs.
EBs were treated with Nodal+/− XEN CM or medium control from days 4 to 6. Cardiac genes were examined over the course of differentiation by QRT-PCR for expression of the cardiac progenitor marker Islet-1, which marks the secondary heart field, the chamber specific markers Mlc2a, Hand1 and Hand2, and the early cardiac markers Tbx5 and Nkx2.5. Relative mRNA expression levels were presented after normalization to Gapdh. Comparatively, EBs treated with Nodal+/− XEN CM showed no significant changes in gene expression as compared to controls.
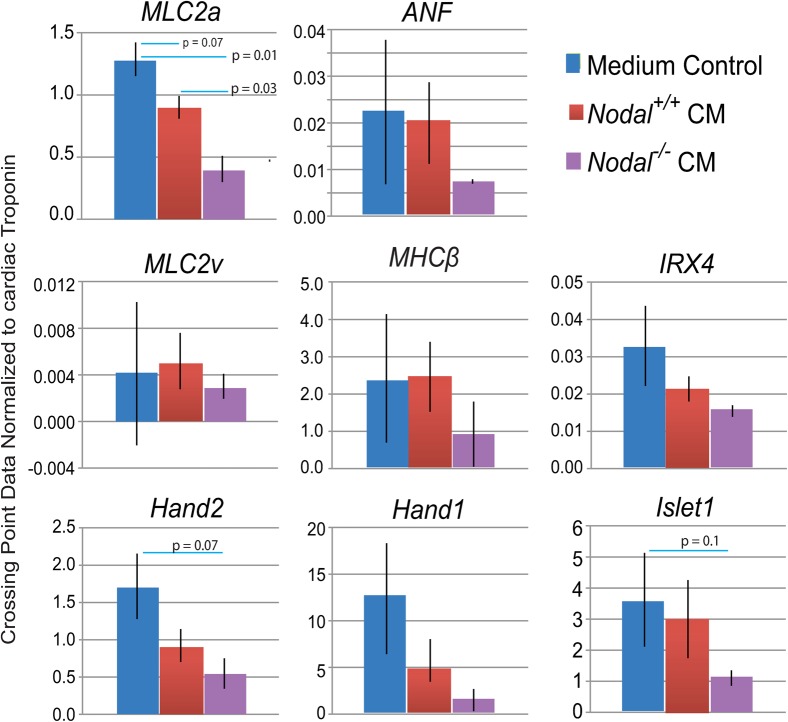
In the embryo, the FHF contributes primarily to the left ventricle with a smaller contribution to the atria, whereas the SHF contributes to all lineages of the heart (but with a relatively small contribution to the left ventricle). If cardiac differentiation in EBs is equivalent to cardiac differentiation in the mouse embryo, we would expect that an early depletion of Islet-1expressing cells would lead to a general depletion of SHF derivatives and this should impact the ratio of atrial to ventricular cell types. Previous studies suggest that ES-derived cardiomyocytes begin to show electrophysiological maturation toward specific cardiac phenotypes 5–7 days after the onset of beating (Kolossov et al., 2005). To address whether premature differentiation of Islet-1 progenitors might lead to a depletion of markers for the SHF and its derivatives, day 14 EBs (7+ days after the onset of beating) were analyzed for the expression of cardiac markers that become regionalized to specific chambers during development. To ensure that differences are due to changes in the regional expression of cardiac markers and not to differences in overall cardiac induction, QRT-PCR data were normalized first to Gapdh and then to the general cardiac marker Cardiac Troponin. At day 14, Mlc2a, which becomes restricted to the atria, was expressed at significantly higher levels in untreated EBs and EBs treated with wild type CM as compared to those treated with Nodal−/− XEN CM. The average expression of Anf was also lower in EBs treated with Nodal−/− XEN CM. However, this marker was not informative because its expression was highly variable in untreated EBs. Islet-1 and Hand2, which mark the SHF and SHF-derivatives respectively, were also significantly decreased in EBs treated with Nodal−/− XEN CM. There were no statistically significant differences in the expression of ventricular markers between EBs treated with wild type XEN CM and those treated with Nodal mutant XEN CM (Fig. 8). Taken together, these data are consistent with the idea that premature differentiation of cardiac progenitors leads to a subsequent depletion of markers characteristic of the SHF and its derivatives.
Fig. 8. Markers that become localized to specific cardiac chambers begin to be differentially expressed in day 14 EBs that were previously treated with Nodal−/− XEN CM.
QRT-PCR data on day 14 of differentiation for markers that become regionalized to atrial or ventricular cardiomyocytes. Markers that become restricted to the ventricles (Mlc2v, MHCβ, Irx4) or which mark derivatives of the FHF in the embryo (Hand1) were not different between the two populations whereas those that become atrial-restricted (Mlc2a), mark the SHF (Islet-1) or mark derivatives of the SHF (Hand2) were expressed at lower levels in cells treated with Nodal−/− XEN CM. Error bars indicated s.e.m. Data considered significant had a p-value< 0.1.
Discussion
XEN cells allow for the molecular dissection of endodermal signals during heart induction
Signals from the AVE and its avian equivalent, the hypoblast, are required for the initial specification of cardiac cells in amniotes and mammals. In frogs, the DAE, that expresses the same markers as the AVE, is also required for early heart formation (Sater and Jacobson, 1989; Nascone and Mercola, 1995; Schneider and Mercola, 1999). In addition, signals from endodermal cell lines such as END2, PYS2 and XEN can mimic the effect of the AVE in vitro, enhancing cardiac differentiation from both ES cells and explants of undifferentiated mesoderm (Mummery et al., 2003; Stary et al., 2005; Nijmeijer et al., 2009; Brown et al., 2010a). Despite the importance of endodermal signals in myocardial differentiation, minimal progress has been made toward the identification of specific cardiogenic factors in the endoderm. In vivo, studies of endodermal signaling have been hampered by the small size and inaccessibility of gastrula stage mouse embryos. In vitro studies have also been slow because previous works have relied almost exclusively on embryonal carcinoma-derived cell lines that do not allow for the genetic ablation of candidate factors. Cell lines representing the primitive endoderm lineage (XEN cells) have recently been derived directly from blastocyst stage embryos (Kunath et al., 2005). Since XEN cells can be generated from both wild type and mutant strains, our recent finding that XEN CM promotes cardiogenesis (Brown et al., 2010a) suggests that they should provide a useful tool to study the role of endodermal signals in myocardial differentiation. To this end, we produced a Nodal mutant XEN cell line and assessed its ability to activate cardiogenesis as compared to wild type XEN cells.
Loss of Nodal results in the upregulation of a subset of AVE markers in XEN cells
A major finding of these studies is that loss of Nodal upregulated the expression of several AVE markers including HEX, DKK1 and BMP2 in XEN cells. This is surprising given that Nodal activates the expression of the AVE marker Cerberus in Xenopus laevis embryos (Foley et al., 2007) and is required for expression of the AVE marker Lefty1 in mouse embryos (Brennan et al., 2001). In addition, treatment of XEN cells with either recombinant NODAL or its co-factor CRIPTO resulted in an upregulation of Cerberus-like but not Dkk1 or Otx2 (Julio et al., 2011). These data suggest that redundant or parallel pathways pattern subsets of markers within the AVE such that NODAL regulates one set of markers (i.e. Cerberus-like and Lefty), while another set (i.e. Bmp2, Hex, Otx2 and Dkk1) is repressed by NODAL and activated by a different pathway(s). Alternatively, recent studies suggest that cells expressing Lefty and Cerberus-like and those marked by Hex, may represent two distinct subgroups of cells within the AVE (Torres-Padilla et al., 2007; Takaoka et al., 2011).
While the mechanism for AVE marker upregulation in Nodal−/− XEN cells is still unclear, this finding suggested that they might possess superior cardiogenic potential as compared to wild type XEN cells. We had previously shown that conditioned medium from wild type XEN cells enhanced cardiac differentiation when it was added to growth medium from day 4 to 6 of EB differentiation (Brown et al., 2010a). By comparison to both control EBs and those treated with wild type XEN CM, Nodal−/− XEN CM did not enhance overall cardiac differentiation but did result in both earlier beating and increased expression of MHCα mRNA on day 7 of differentiation. To clarify these conflicting findings, we investigated the effect of CM on the expression of cardiac progenitor markers. In EBs treated with wild type XEN CM, Islet-1 expression was delayed as compared to controls, whereas EBs treated with Nodal−/− XEN CM showed a premature increase in the expression of Islet-1 as compared to controls. Two other cardiac progenitor markers, Nkx2.5 and Tbx5, were either unaffected or slightly delayed in response to Nodal−/− XEN CM. While there is no clear definition of FHF and SHF within EBs, our observation demonstrate that markers associated with the FHF (Nkx2.5 and Tbx5) and SHF (Islet-1) respond differently to endodermal signals. Prall and colleagues proposed a model to explain how the embryo regulates the switch from FHF and SHF differentiation involving a negative feed back loop comprised of Nkx2.5, Bmp2 and SMAD-1. In the absence of Nkx2.5, they saw an upregulation of Bmp2 and SMAD phosphorylation, and this resulted in a premature differentiation of Islet-1 positive cells, with a subsequent failure of SHF derivatives to proliferate. Similarly, in these studies we saw an upregulation of BMP2, premature expression of Islet-1 and a failure of the cardiac pool to expand. By day 14, we also observed a significant decrease in markers for the SHF and its derivatives within EBs.
Notably, Prall and colleagues observed that the failure of SHF progenitors to proliferate in Nkx2.5 null embryos led to the formation of a malformed heart containing a single ventricle and a severely truncated OFT (Prall et al., 2007). Consistent with this, mice in which an enhancer of early Nodal expression is knocked out, had normal cardiac specification but subsequent heart defects including ventricular septal defects and misplacement of the outflow tract (Norris et al., 2002). In addition, many human congenital heart defects including Tetralogy of Fallot, that are thought to be related to defects in SHF development, have been linked to specific mutations at the Nodal locus (Roessler et al., 2009). Our data suggests that endodermal signals that are regulated by Nodal may mediate the time of the embryos switch from FHF to SHF development.
Nodal is not the direct cardiogenic signal
Both wild type and Nodal heterozygous mutant XEN cells expressed very low levels of Nodal mRNA when grown in the presence of LIF. However, this expression was rapidly lost after the removal of LIF from the media. In addition, we previously showed that wild type XEN cells, grown in the absence of LIF possessed cardiogenic potential despite the fact that they expressed no Nodal (Brown et al., 2010b). These findings suggest that the cardiogenic potential of XEN cells is not due to Nodal signaling from the XEN cells themselves, but is an indirect consequence of Nodal's ability to pattern XEN cells. These XEN cells are most likely patterned during the initial phases of isolation when the primitive endoderm and epiblast cells remain in culture. More specifically, these data suggest that genes regulated by Nodal in the endoderm may be required to maintain SHF progenitors in pre-cardiac state.
Materials and Methods
Derivation and culture of Nodal mutant XEN cell lines
Mice carrying a NodalLacZ allele (Collignon et al., 1996) were maintained on an ICR genetic background. NodalLacZ/+ mice were crossed to obtain NodalLacZ/LacZ embryos (referred to in the text as Nodal−/−). XEN cell lines were derived from these embryos according to a previously described method (Kunath et al., 2005; Artus et al., 2010) (Fig. 1A). After 2–3 weeks, colonies with XEN cell morphology were observed in over half the cultures. The colonies were expanded onto gelatinized 10 cm plates with mouse embryonic fibroblasts (MEFs). After passaging off MEFs, genomic DNA was purified from cell pellets using DNeasy Blood & Tissue Kit (QIAGEN) and putative XEN cells lines were genotyped using previously described primer sequences (Collignon et al., 1996) (Table 1).
Table 1. QRT-PCR primers used in this study. Primers were designed for detecting murine gene expression.
XEN cells were maintained on MEFs in DMEM supplemented with 20% fetal bovine serum (FBS; GIBCO), 1 mM sodium pyruvate (Mediatech), 2 mM L-glutamine (Mediatech), 50 mg/ml penicillin/streptomycin (Cellgro), 55 mM β-mercaptoethanol (Sigma), 100 mM non-essential amino acids (Mediatech), and 1000 units/ml of ESGRO (LIF) (Millipore).
To collect CM, XEN cells (no MEFs), were cultured in media without LIF. Media were conditioned with Nodal+/+ XEN cells (Nodal+/+ XEN CM), Nodal+/− XEN cells (Nodal +/− XEN CM), or Nodal −/− XEN cells (Nodal−/− XEN CM) and incubated at 37°C, 5% CO2. Medium control consisted of medium that was not conditioned by XEN cells. After 24-hour incubation, media were sterile filtered and stored at −20°C for later use.
Culture and differentiation of ES cells
Mouse R1 or CGR8 embryonic stem (ES) cells were cultured and differentiated as previously described (Brown et al., 2010a). For in vitro differentiation, ES cells were passaged off MEFs then plated as hanging drops at a concentration of 15,000 cells/mL (300 cells per hanging drop) in Iscove's Modified Dulbecco Medium (IMDM; Gibco), supplemented with 20% serum (Gibco), 50 mg/mL each of penicillin/streptomycin (100 units/mL and 100 µg/mL, respectively) (Cellgro), 200 mg/mL apo-transferrin (Sigma), 5% protein-free hybridoma medium (PFHMII; Gibco), 0.5 mM 1-thioglycerol (Sigma), and 0.5 mM ascorbic acid (Sigma).
Immunocytochemistry
Cells were plated onto gelatinized chamber slides. Immunocytochemistry was performed at various degrees of confluence to ensure that there were no changes in gene expression resulting from cell density (data not shown). The cells were fixed in 4% paraformaldehyde at room temperature then permeabilized with 0.25% Triton X-100-PBS at room temperature. Blocking was carried out with 10% heat-inactivated FBS-0.25% Triton X-100 in PBS at room temperature then incubated with primary antibody overnight at 4°C. After this, cells were incubated with secondary antibody and counterstained with DAPI. Primary antibodies used were: OCT4 (1:250; Cell Signaling), GATA4 (1:1000; Santa Cruz), GATA6 (1:500; R&D systems), SOX7 (1:500; R&D systems), BMP2 (1:250; Abcam), and HEX (1:500; Abcam). Staining was detected by Alexa Fluor 555-conjugated anti-goat antibody (1:1000; Invitrogen), Alexa Fluor 555-conjugated anti-rabbit antibody (1:1000; Invitrogen), Alexa Fluor 555-conjugated anti-mouse antibody (1:1000; Invitrogen), or Alexa Fluor 488-conjugated anti-mouse antibody (1:1000; Invitrogen).
Western blot
Whole cell extracts (20 mg) were prepared using RIPA lysis buffer, electrophoresed through 4–20% NuPAGE Bis-Tris gel (Invitrogen), transferred to a PVDF membrane (Pall Life Sciences) and blotted by standard techniques. Primary antibodies used were: OCT4 (1:1000; Cell Signaling), GATA4 (1:1000; Santa Cruz), GATA6 (1:1000; R&D systems), SOX7 (1:1000; R&D systems), BMP2 (1:200; Abcam), DKK1 (1:200; Santa Cruz), and HEX (1:200; Abcam). Secondary antibodies were anti-mouse HRP (1:3000; Cell Signaling), anti-rabbit HRP (1:3000; GE Healthcare). As a loading control, membranes were stripped and probed with anti-ACTIN (1:2000; Santa Cruz) and labeled with anti-goat HRP (1:10,000; Santa Cruz). Chemoluminescent detection (Thermo Scientific) was performed according to the manufacturer's instructions.
RNA isolation, reverse transcription, and QRT-PCR analysis
Total RNA from cells was isolated with Tri-Reagent (Sigma). First strand cDNA synthesis was performed with 1 µg of total RNA using QuantiTect Reverse Transcription kit (QIAGEN). cDNAs were analyzed by QRT-PCR for the expression of Nodal, Oct4, Gata4, Gata6, Sox7, Hex, Dkk1, Bmp2, Tbx5, Islet-1, Nkx2.5, Mlc2a, Hand1, and Hand2 (Table 1). QRT-PCR was performed on a Roche Lightcycler 480, and analysis was done with the Roche Lightcycler 480 software package. Crossing point data was corrected for primer efficiency (as determined by analysis of a standard dilution curve for a positive control) then normalized to expression of Gapdh.
Flow cytometry
CGR8 ES cells expressing the Green Fluorescent Protein (GFP) reporter under the Myosin Heavy Chain-alpha (MHCα) promoter (MHCα::GFP) (Takahashi et al., 2003) were used to form EBs. EBs were dissociated with trypsin. Acquisition and analysis for amount of GFP positive cells was performed on a BD FACSCalibur cytometer (BD Biosciences) with the aid of the BD Cell Quest software.
Statistical Analysis
The statistical significance of differences among treatment groups was assessed with Scheffe multiple-comparison test. Error bars indicate standard error. (s.e.m.). Differences were considered significant at p-value< 0.05 (a p-value< 0.05 is indicated by one star, p-value< 0.006 is indicated by two stars in figures). Significance of regional cardiac markers was determined by unpaired two-tailed t-test. Differences were considered significant at a p-value< 0.1.
Supplementary Material
Acknowledgments
We are grateful to: Elizabeth Robertson and Kat Hadjantonakis for providing Nodal mutant mice; Kat Hadjantonakis for helpful discussion and advice on the manuscript; Stephanie Hausler for assistance with mouse husbandry; Ania Piliszek and Jérôme Artus for teaching preimplantation embryo collection and XEN cell isolation. This work was supported by grants from the American Heart Association as well as funds from Raymond and Beverly Sackler.
Footnotes
Competing Interests: There is no potential competing conflict of interests in connection with this manuscript. All sources of financial support for this study have been disclosed and are indicated in the acknowledgements.
References
- Abu-Issa R., Kirby M. L. (2007). Heart field: from mesoderm to heart tube. Annu. Rev. Cell Dev. Biol. 23, 45–68 10.1146/annurev.cellbio.23.090506.123331 [DOI] [PubMed] [Google Scholar]
- Arai A., Yamamoto K., Toyama J. (1997). Murine cardiac progenitor cells require visceral embryonic endoderm and primitive streak for terminal differentiation. Dev. Dyn. 210, 344–353 [DOI] [PubMed] [Google Scholar]
- Artus J., Panthier J. J., Hadjantonakis A. K. (2010). A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development 137, 3361–3372 10.1242/dev.050864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J., Douvaras P., Piliszek A., Isern J., Baron M. H., Hadjantonakis A. K. (2012). BMP4 signaling directs primitive endoderm-derived XEN cells to an extraembryonic visceral endoderm identity. Dev. Biol. 361, 245–262 10.1016/j.ydbio.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B. L. (2007). Transcriptional pathways in second heart field development. Semin. Cell Dev. Biol. 18, 67–76 10.1016/j.semcdb.2007.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J., Lu C. C., Norris D. P., Rodriguez T. A., Beddington R. S., Robertson E. J. (2001). Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411, 965–969 10.1038/35082103 [DOI] [PubMed] [Google Scholar]
- Brown K., Doss M. X., Legros S., Artus J., Hadjantonakis A. K., Foley A. (2010a). eXtraembryonic ENdoderm (XEN) Cells Produce Factors that Activate Heart Formation. PLoS ONE 5, e13446 10.1371/journal.pone.0013446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Legros S., Artus J., Doss M. X., Khanin R., Hadjantonakis A. K., Foley A. (2010b). A comparative analysis of extra-embryonic endoderm cell lines. PLoS ONE 5, e12016 10.1371/journal.pone.0012016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D. J., Huang P. L., Domian I. J., Chien K. R. (2009). Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460, 113–117 10.1038/nature08191 [DOI] [PubMed] [Google Scholar]
- Cai C. L., Liang X., Shi Y., Chu P. H., Pfaff S. L., Chen J., Evans S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus A., Perea-Gomez A., Moreau A., Collignon J. (2006). Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev. Biol. 295, 743–755 10.1016/j.ydbio.2006.03.047 [DOI] [PubMed] [Google Scholar]
- Collignon J., Varlet I., Robertson E. J. (1996). Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature 381, 155–158 10.1038/381155a0 [DOI] [PubMed] [Google Scholar]
- David N. B., Rosa F. M. (2001). Cell autonomous commitment to an endodermal fate and behaviour by activation of Nodal signalling. Development 128, 3937–3947 [DOI] [PubMed] [Google Scholar]
- Dyer L. A., Kirby M. L. (2009). The role of secondary heart field in cardiac development. Dev. Biol. 336, 137–144 10.1016/j.ydbio.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley A. C., Korol O., Timmer A. M., Mercola M. (2007). Multiple functions of Cerberus cooperate to induce heart downstream of Nodal. Dev. Biol. 303, 57–65 10.1016/j.ydbio.2006.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullilove S. L. (1970). Heart induction: distribution of active factors in newt endoderm. J. Exp. Zool. 175, 323–326 10.1002/jez.1401750306 [DOI] [PubMed] [Google Scholar]
- Granier C., Gurchenkov V., Perea-Gomez A., Camus A., Ott S., Papanayotou C., Iranzo J., Moreau A., Reid J., Koentges G. et al. (2011). Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo. Dev. Biol. 349, 350–362 10.1016/j.ydbio.2010.10.036 [DOI] [PubMed] [Google Scholar]
- Holtzinger A., Rosenfeld G. E., Evans T. (2010). Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev. Biol. 337, 63–73 10.1016/j.ydbio.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. G. (1960). Influences of ectoderm and endoderm on heart differentiation in the newt. Dev. Biol. 2, 138–154 10.1016/0012-1606(60)90003-8 [DOI] [PubMed] [Google Scholar]
- Julio M. K., Alvarez M. J., Galli A., Chu J., Price S. M., Califano A., Shen M. M. (2011). Regulation of extra-embryonic endoderm stem cell differentiation by Nodal and Cripto signaling. Development 138, 3885–3895 10.1242/dev.065656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. G., Brown N. A., Buckingham M. E. (2001). The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell 1, 435–440 10.1016/S1534-5807(01)00040-5 [DOI] [PubMed] [Google Scholar]
- Kolossov E., Lu Z., Drobinskaya I., Gassanov N., Duan Y., Sauer H., Manzke O., Bloch W., Bohlen H., Hescheler J. et al. (2005). Identification and characterization of embryonic stem cell-derived pacemaker and atrial cardiomyocytes. FASEB J. 19, 577–579 10.1016/fj.03-1451fje [DOI] [PubMed] [Google Scholar]
- Kunath T., Arnaud D., Uy G. D., Okamoto I., Chureau C., Yamanaka Y., Heard E., Gardner R. L., Avner P., Rossant J. (2005). Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649–1661 10.1242/dev.01715 [DOI] [PubMed] [Google Scholar]
- Lough J., Sugi Y. (2000). Endoderm and heart development. Dev. Dyn. 217, 327–342 [DOI] [PubMed] [Google Scholar]
- Madabhushi M., Lacy E. (2011). Anterior Visceral Endoderm Directs Ventral Morphogenesis and Placement of Head and Heart via BMP2 Expression. Dev. Cell 21, 907–919 10.1016/j.devcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin M. J., Di Rocco G., Gardiner A., Bush S. M., Lassar A. B. (2001). Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 15, 316–327 10.1101/gad.855501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilhac S. M., Esner M., Kelly R. G., Nicolas J. F., Buckingham M. E. (2004). The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell 6, 685–698 10.1016/S1534-5807(04)00133-9 [DOI] [PubMed] [Google Scholar]
- Mesnard D., Guzman-Ayala M., Constam D. B. (2006). Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133, 2497–2505 10.1242/dev.02413 [DOI] [PubMed] [Google Scholar]
- Moretti A., Caron L., Nakano A., Lam J. T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S. et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165 10.1016/j.cell.2006.10.029 [DOI] [PubMed] [Google Scholar]
- Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R., van der Heyden M., Opthof T., Pera M., de la Riviere A. B. et al. (2003). Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation 107, 2733–2740 10.1161/01.CIR.0000068356.38592.68 [DOI] [PubMed] [Google Scholar]
- Nascone N., Mercola M. (1995). An inductive role for the endoderm in Xenopus cardiogenesis. Development 121, 515–523 [DOI] [PubMed] [Google Scholar]
- Nijmeijer R. M., Leeuwis J. W., DeLisio A., Mummery C. L., Chuva de Sousa Lopes S. M. (2009). Visceral endoderm induces specification of cardiomyocytes in mice. Stem Cell Res. 3, 170–178 10.1016/j.scr.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Norris D. P., Brennan J., Bikoff E. K., Robertson E. J. (2002). The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129, 3455–3468 [DOI] [PubMed] [Google Scholar]
- Orts-Llorca F., Gil D. R. (1965). Influence of the endoderm on heart differentiation. Roux's Arch. EntwMech. Org. 156, 368–370 10.1007/BF02456124 [DOI] [PubMed] [Google Scholar]
- Prall O. W., Menon M. K., Solloway M. J., Watanabe Y., Zaffran S., Bajolle F., Biben C., McBride J. J., Robertson B. R., Chaulet H. et al. (2007). An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 128, 947–959 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y., Martin-Puig S., Chiravuri M., Chen S., Xu H., Bu L., Jiang X., Lin L., Granger A., Moretti A. et al. (2007). The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell 1, 165–179 10.1016/j.stem.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Roessler E., Pei W., Ouspenskaia M. V., Karkera J. D., Velez J. I., Banerjee-Basu S., Gibney G., Lupo P. J., Mitchell L. E., Towbin J. A. et al. (2009). Cumulative ligand activity of NODAL mutations and modifiers are linked to human heart defects and holoprosencephaly. Mol. Genet. Metab. 98, 225–234 10.1016/j.ymgme.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sater A. K., Jacobson A. G. (1989). The specification of heart mesoderm occurs during gastrulation in Xenopus laevis. Development 105, 821–830 [DOI] [PubMed] [Google Scholar]
- Schneider V. A., Mercola M. (1999). Spatially distinct head and heart inducers within the Xenopus organizer region. Curr. Biol. 9, 800–809 10.1016/S0960-9822(99)80363-7 [DOI] [PubMed] [Google Scholar]
- Stary M., Pasteiner W., Summer A., Hrdina A., Eger A., Weitzer G. (2005). Parietal endoderm secreted SPARC promotes early cardiomyogenesis in vitro. Exp. Cell Res. 310, 331–343 10.1016/j.yexcr.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Stennard F. A., Costa M. W., Lai D., Biben C., Furtado M. B., Solloway M. J., McCulley D. J., Leimena C., Preis J. I., Dunwoodie S. L. et al. (2005). Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 132, 2451–2462 10.1242/dev.01799 [DOI] [PubMed] [Google Scholar]
- Sugi Y., Lough J. (1994). Anterior endoderm is a specific effector of terminal cardiac myocyte differentiation of cells from the embryonic heart forming region. Dev. Dyn. 200, 155–162 10.1002/aja.1002000207 [DOI] [PubMed] [Google Scholar]
- Takahashi T., Lord B., Schulze P. C., Fryer R. M., Sarang S. S., Gullans S. R., Lee R. T. (2003). Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 107, 1912–1916 10.1161/01.CIR.0000064899.53876.A3 [DOI] [PubMed] [Google Scholar]
- Takaoka K., Yamamoto M., Hamada H. (2011). Origin and role of distal visceral endoderm, a group of cells that determines anterior-posterior polarity of the mouse embryo. Nat. Cell Biol. 13, 743–752 10.1038/ncb2251 [DOI] [PubMed] [Google Scholar]
- Tonegawa A., Moriya M., Tada M., Nishimatsu S., Katagiri C., Ueno N. (1996). Heart formative factor(s) is localized in the anterior endoderm of early Xenopus neurula. Roux's Arch. Dev. Biol. 205, 282–289 10.1007/BF00365806 [DOI] [PubMed] [Google Scholar]
- Torres-Padilla M. E., Richardson L., Kolasinska P., Meilhac S. M., Luetke-Eversloh M. V., Zernicka-Goetz M. (2007). The anterior visceral endoderm of the mouse embryo is established from both preimplantation precursor cells and by de novo gene expression after implantation. Dev. Biol. 309, 97–112 10.1016/j.ydbio.2007.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet I., Collignon J., Robertson E. J. (1997). nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development 124, 1033–1044 [DOI] [PubMed] [Google Scholar]
- Vincent S. D., Buckingham M. E. (2010). How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1–41 10.1016/S0070-2153(10)90001-X [DOI] [PubMed] [Google Scholar]
- Waldo K. L., Hutson M. R., Ward C. C., Zdanowicz M., Stadt H. A., Kumiski D., Abu-Issa R., Kirby M. L. (2005). Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 281, 78–90 10.1016/j.ydbio.2005.02.012 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Saijoh Y., Perea-Gomez A., Shawlot W., Behringer R. R., Ang S. L., Hamada H., Meno C. (2004). Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature 428, 387–392 10.1038/nature02418 [DOI] [PubMed] [Google Scholar]
- Yuasa S., Itabashi Y., Koshimizu U., Tanaka T., Sugimura K., Kinoshita M., Hattori F., Fukami S., Shimazaki T., Ogawa S. et al. (2005). Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat. Biotechnol. 23, 607–611 10.1038/nbt1093 [DOI] [PubMed] [Google Scholar]
- Zaffran S., Kelly R. G., Meilhac S. M., Buckingham M. E., Brown N. A. (2004). Right ventricular myocardium derives from the anterior heart field. Circ. Res. 95, 261–268 10.1161/01.RES.0000136815.73623.BE [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.