Summary
The Notch signalling pathway plays an essential role in the intricate control of cell proliferation and pattern formation in many organs during animal development. In addition, mutations in most members of this pathway are well characterized and frequently lead to tumour formation. The Drosophila imaginal wing discs have provided a suitable model system for the genetic and molecular analysis of the different pathway functions. During disc development, Notch signalling at the presumptive wing margin is necessary for the restricted activation of genes required for pattern formation control and disc proliferation. Interestingly, in different cellular contexts within the wing disc, Notch can either promote cell proliferation or can block the G1-S transition by negatively regulating the expression of dmyc and bantam micro RNA. The target genes of Notch signalling that are required for these functions have not been identified. Here, we show that the Hes vertebrate homolog, deadpan (dpn), and the Enhancer-of-split complex (E(spl)C) genes act redundantly and cooperatively to mediate the Notch signalling function regulating cell proliferation during wing disc development.
Key words: Notch signalling, bHLH factors, deadpan, E(spl)C
Introduction
The diversity of cell types present in multicellular organisms largely depends on developmental decisions that are determined by various intercellular signals during development. The Notch signalling pathway is well-known to influence cell fate decisions. In addition to the classical role of this pathway in binary cell fate decisions, it is also widely employed in inductive cell fate interactions, such as the one that occurs in the dorsal/ventral compartment boundary of the Drosophila imaginal wing disc (Bray, 2006). The Drosophila wing and notum originate from this disc. The wing disc primordium develops from 20–30 ectodermic cells that are specified during embryogenesis and proliferate during the larval stages (Cohen et al., 1993; Garcia-Bellido and Merriam, 1971). During its development, the wing disc is divided into lineage units known as compartments (García-Bellido, 1975). The compartmentalisation of the wing discs allows localised expression of signalling molecules at the compartment's borders, an essential process in controlling disc growth and patterning (Blair, 1995). One of these borders is the dorsal-ventral (d/v) boundary (Diaz-Benjumea and Cohen, 1995; de Celis et al., 1996a). The interaction between the cells of the dorsal and the ventral compartments causes the activation of Notch signalling at this boundary, which, in turn, induces the expression of different genes, such as wingless (wg) and cut (cut), that are required for the control of pattern formation and wing disc cell proliferation (Rulifson and Blair, 1995; Couso et al., 1995; Diaz-Benjumea and Cohen, 1995; de Celis et al., 1996a; Micchelli et al., 1997). Later in development, the cells that constitute this boundary are arrested in G1 and establish the so-called Zone of Non-proliferation Cells (ZNC) (O'Brochta and Bryant, 1985; Phillips and Whittle, 1993). The Notch pathway is necessary for this regulation (Herranz et al., 2008). Thus, at the d/v boundary, Notch signalling autonomously represses expression of the dmyc proto-oncogene and the bantam micro-RNA, which are required to promote cell cycle progression from G1 to S (Herranz et al., 2008). Therefore, Notch activity would be necessary to restrict proliferation by blocking the G1-S transition in these cells. Interestingly, the function of Notch in the regulation of proliferation depends on the developmental context, because in the proximal and hinge regions of the wing discs the ectopic expression of Notch induces overgrowth (Go et al., 1998; Giraldez and Cohen, 2003; Baonza and Garcia-Bellido, 2000). How these differential responses are modulated by Notch signalling is an interesting question that remains to be elucidated.
The activation of the transmembrane receptor Notch, via its ligands Delta (Dl) and Serrate (Ser), causes the intramembrane proteolysis of the receptor and subsequent release of its intracellular domain (NICD) (Bray, 2006; Fortini, 2009). This domain then translocates to the nucleus to participate in concert with the transcription factor Suppressor of Hairless (Su(H)) in the transcriptional regulation of different target genes (Bray, 2006; Fortini, 2009). Among the best-characterised Notch signalling targets are the Enhancer-of-split complex (E(spl)C) genes (Jennings et al., 1994). In Drosophila, this complex comprises seven genes encoding bHLH proteins (Delidakis and Artavanis-Tsakonas, 1992; Knust et al., 1992). During wing disc development, the E(spl) genes are required for the lateral inhibition processes necessary to define vein thickness and to single out sensory organs precursors. In contrast, the function of the E(spl) proteins seems dispensable for the d/v boundary establishment, the ZNC definition, and the promotion of cell proliferation in the wing disc proximal region (de Celis et al., 1996b). Thus, although it is well established which Notch target genes are involved in the specification of different cell fates, little is known about the genes that mediate the ability of Notch signalling to promote cell proliferation in the wing disc proximal regions or the targets of Notch signalling that are required for the ZNC cell-cycle arrest.
We recently identified that deadpan (dpn), the Hes vertebrate homolog, is a direct target of Notch signalling during neuroblast development (San-Juan and Baonza, 2011). This gene encodes for a bHLH transcription factor that shows strong similarities to the Enhancer-of-split bHLH proteins. Here, we investigate the function of dpn and its relationship to Notch signalling and the E(spl) complex during wing disc development. We find that during neuroblast development, dpn expression is regulated by Notch signalling. Our results indicate that dpn acts redundantly and cooperatively with the E(spl)C genes to maintain d/v integrity and to block cell cycle progression at the ZNC. Moreover, we show that both dpn and the E(spl) complex are required to promote cell proliferation in the wing disc proximal region upon ectopic Notch activation. The data presented here demonstrate a hitherto unrecognised function for dpn and E(spl)C in mediating, at least in part, the function of Notch signalling regulating cell proliferation during wing disc development.
Results
Dpn expression is regulated by Notch signalling
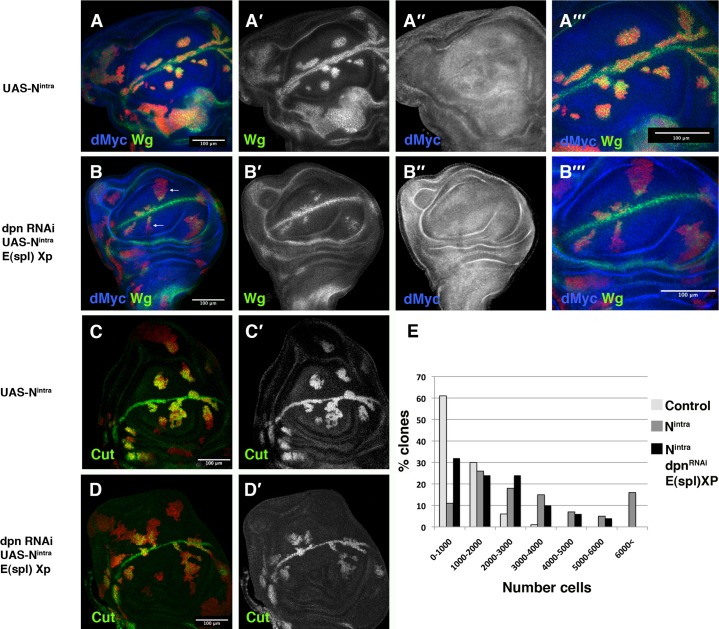
During wing disc development, Notch signalling promotes the localised expression of wg and cut at the d/v boundary (Couso et al., 1995; Rulifson et al., 1996; Neumann and Cohen, 1996; Kim et al., 1995; de Celis et al., 1996a). We found that Dpn was also expressed at high levels in the same d/v boundary cells that express Wg (Fig. 1A,B′). In these cells, Dpn expression co-localised with the reporter of Notch activity E(spl)m4-lacZ (Bailey and Posakony, 1995) (data not shown). These data are compatible with Notch signalling regulating dpn. To analyse whether dpn expression depends on Notch signalling, we induced MARCM clones of the Notch mutant allele N55e11 (see Materials and Methods). We found that Dpn was down-regulated in Notch mutant cells (Fig. 1C,C′). Similarly, in the wing margin, mutant clones of the Notch ligand Delta failed to express Dpn (Fig. 1D,D′).
Fig. 1. Dpn expression is regulated by Notch signalling.
(A–G) Third instar larval wing discs stained with anti-Dpn (in green in A–G and grey in A′–G′) and anti-Wg (in red A,B and grey B′) antibodies. (A–B′) Dpn is expressed at high levels at the d/v boundary in the cells that express Wg. (B,B′) High magnification of panel A. (C,C′) Dpn is not expressed in N55e11 mutant clones (positively marked with GFP in red). (D,D′) In M+ Dlrev10 mutant clones (marked by the absence of GFP in red), Dpn is not expressed, except in some mutant cells at the border of the clone that were non-autonomously rescued by the adjacent wild type cells. (E,E′) Dpn is ectopically expressed in Nintra-expressing cells positively marked with GFP in red. (F–G′) In clones of Delta-expressing cells (marked by the expression of GFP in red), Dpn is expressed at high levels. (G,G′) A longitudinal cross-section at the position of the white line of panel F is shown. Mutant clones are outlined in white.
Next, we investigated whether Notch signalling was also sufficient to induce dpn. Indeed, in the wing blade and hinge regions, clones of cells expressing a constitutively active form of Notch (Nintra) showed an autonomous increase in the expression of Dpn (Fig. 1E,E′). This effect was also observed in clones of Dl-expressing cells, which accumulated high levels of Dpn (Fig. 1F–G′). We further studied the Notch regulation of dpn by using quantitative real-time PCR analysis. Using the Gal4/Gal 80Ts system, we ectopically activated Notch signalling in third instar nub-Gal4 UAS-Nintra/Tub-Gal80Ts wings discs and quantified the levels of dpn mRNA at different times after the induction of Notchintra expression. At all of the times analysed, we found that the dpn mRNA levels were much higher in the mutant discs than in the control discs. Interestingly, dpn transcription appeared to be more sensitive to Notch activity than several E(spl) complex genes (supplementary material Fig. S1). Together, these findings indicate that Delta/Notch signalling was sufficient to activate dpn expression and suggest that dpn might be a Notch signalling target during wing disc development.
Dpn genetically interacts with Notch
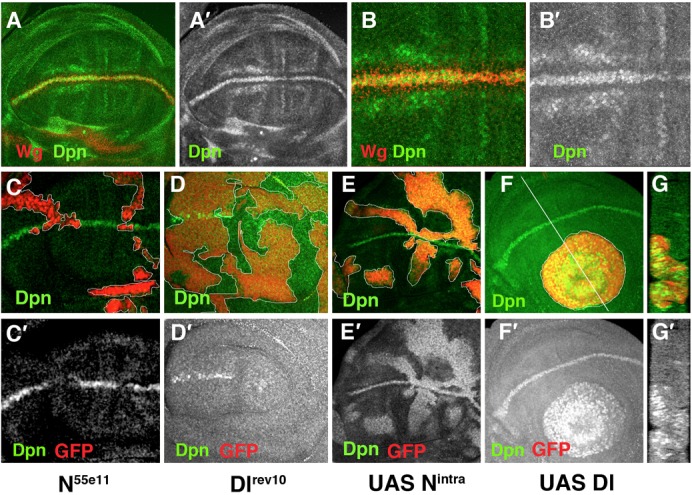
We studied the requirement for dpn during wing disc development by analysing the effects of dpn depletion. We completely eliminated dpn using a heteroallelic combination of dpnDef3D5 and dpn2 with the point allele dpn7 over dpn2. We also examined mutant discs of transgenic flies expressing a dpnRNAi construct in different domains of the wing blade. The phenotypes displayed by the dpn2/dpnDef3D5 and the dpn2/dpn7 mutants were very similar. In both combinations, we found a few adult flies that came out of the puparium and that occasionally presented nicks in the wing margin. This latter phenotype is characteristic of a Notch haploinsufficiency (Fig. 2A). We also occasionally found this phenotype in flies in which dpn was specifically down-regulated throughout the wing blade, namely the UAS-dpnRNAi/nub-Gal4 flies (Fig. 2B). In contrast, the ectopic expression of dpn in the presumptive wing blade (UAS-dpn/nub-Gal4) prevented the differentiation of veins (Fig. 2C), a phenotype also caused by the ectopic activation of Notch signalling. To further test the functional relevance of dpn as a target gene of Notch signalling, we checked possible genetic interactions between dpn and Notch. We found that the wing margin phenotype characteristic of N54/9/+ mutant wings (Fig. 2A) was strongly enhanced when dpn expression was reduced in the wing blade (Fig. 2D). This genetic interaction indicates a functional relationship between dpn and Notch, and supports a role for dpn in the Notch pathway.
Fig. 2. The genetic interactions between dpn and Notch are examined.
(A) N54/9/+; nub-Gal4/+ adult wing. (B) nub-Gal4/nub-Gal4; UAS-dpnRNAi/UAS-dpnRNAi wings occasionally displayed nicks in the margin. In nub-Gal4/+; UAS-dpnRNAi/+, we did not find nicks in the wing margin. (C) The ectopic expression of UAS-dpn in the presumptive wing blade (nub-Gal4/+; UAS-dpn/+) blocked the differentiation of wing veins. (D) N54/9/+; nub-Gal4/+; UAS-dpnRNAi/+ wing. Note the enhancement of the Notch haploinsufficiency scalloping wing phenotype.
dpn and the control of cell proliferation
To better understand dpn function during wing disc development, we studied the proliferation of dpn mutant cells using a twin analysis. In this assay, control clones were generated at the same time as mutant clones, allowing comparison of dpn mutant clones with control clones. The average size of dpn mutant clones, induced 60 h AEL and analysed 48 h later, was similar to that of control clones in all wing disc regions analysed. The ratio of the number of mutant to control cells was 0.99±0.2 (n = 15) for the wing, 0.95±0.1 (n = 5) for the notum, and 0.93±0.2 (n = 16) in the hinge region (supplementary material Fig. S2). This result suggests that dpn function was not essential for the control of cell proliferation during wing disc development. Because our results suggest that dpn functions down-stream of Notch signalling, we analysed whether dpn was required for the expression of the Notch-regulated genes cut and wg at the wing margin. We found that in dpn7 and dpnDef1D6 clone cells that cross the d/v boundary, the expression of Wg was slightly but consistently reduced, while the expression of Cut was not affected. In other wing disc regions, such as the hinge and notum, the expression of Wg was not altered (supplementary material Fig. S2; data not shown).
Next, we studied the effects of ectopic expression of dpn in the wing discs. Expression of dpn in the dorsal compartment (ap-Gal4/+; UAS-dpn) caused an enlargement of the dorsal hinge of third instar wing discs and adult flies (Fig. 3B,B′; supplementary material Fig. S3). Thus, the average size of the hinge regions of control third instar discs was 137,000±30,000 µm3 compared with 393,000±130,000 µm3 of ap-Gal4/+;UAS-dpn wing discs (n = 5 p = 0.001). This enlargement was correlated with a significant increase of proliferation in this region, as assayed by staining of the mitotic marker phospho-histone H3, PH3. The ratio of PH3-positive-cells in the hinge to PH3-positive-cells in the dorsal wing blade control was 1.26±0.026; by contrast, the UAS-dpn flies had a ratio of 1.5±0.1 (n = 5 and P = 0.01). The increased number of mitoses observed in these discs was not solely a consequence of the increase in the size of the hinge, as we observed that the density of PH3 positive cells in the hinge of ap-Gal4/+; UAS-dpn discs was higher than that in the discs of controls (a mitotic index of 2.5×10−4±3×10−5 cells per µm3, compared with 1.5×10−4±3×10−5 for control discs; P = 0.002, n = 5).
Fig. 3. The ectopic expression of dpn, m3 and mγ induce overgrowth of the hinge.
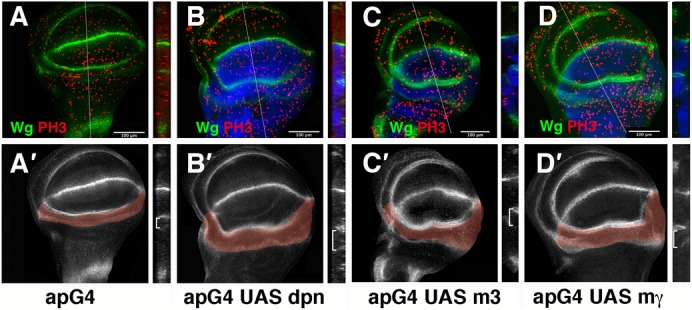
(A–D′) Third instar larval wing discs stained with anti-Wg (in green in A–D, and grey in A′–D′), and anti-PH3 (in red A–D) antibodies. Longitudinal cross-sections at the positions of the white lines of panels A–D are shown in the right panels. In all panels, the expression of GFP driven by ap-Gal4 is shown in blue. (A′–D′) The hinge regions are highlighted in light red. (A,A′) A control wing disc is shown. The ectopic expression of ap-Gal4/+; UAS-dpn/+ (B,B′), ap-Gal4/+;UAS-m3/+ (C,C′), and ap-Gal4/+;UAS-mγ/+ (D,D′) results in a substantial increase in the number of mitotic cells (red) (positive for anti-phospho-histone H3) in the hinge region. See the text for quantification. Note that the over-expression of these genes did not induce Wg ectopic expression.
Interestingly, the ectopic activation of Notch also induces extra cell proliferation in the hinge region of the wing discs (Fig. 4; supplementary material Fig. S4) (Go et al., 1998; Baonza and Garcia-Bellido, 2000; Giraldez and Cohen, 2003). We checked whether higher levels of dpn expression were sufficient to up-regulate Cut and Wg, as is seen upon ectopic activation of Notch (Fig. 4). However, in contrast to Notch, the over-expression of dpn in different regions of the wing discs (using ap-Gal4, nub-Gal4 and dpp-Gal4 in combination with UAS-dpn) was not sufficient to induce the expression of these genes (data not shown).
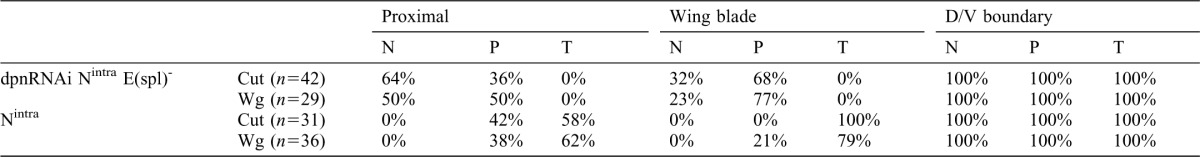
Fig. 4. The down-regulation of dpn partially suppressed the effects caused by the ectopic expression of Nintra.
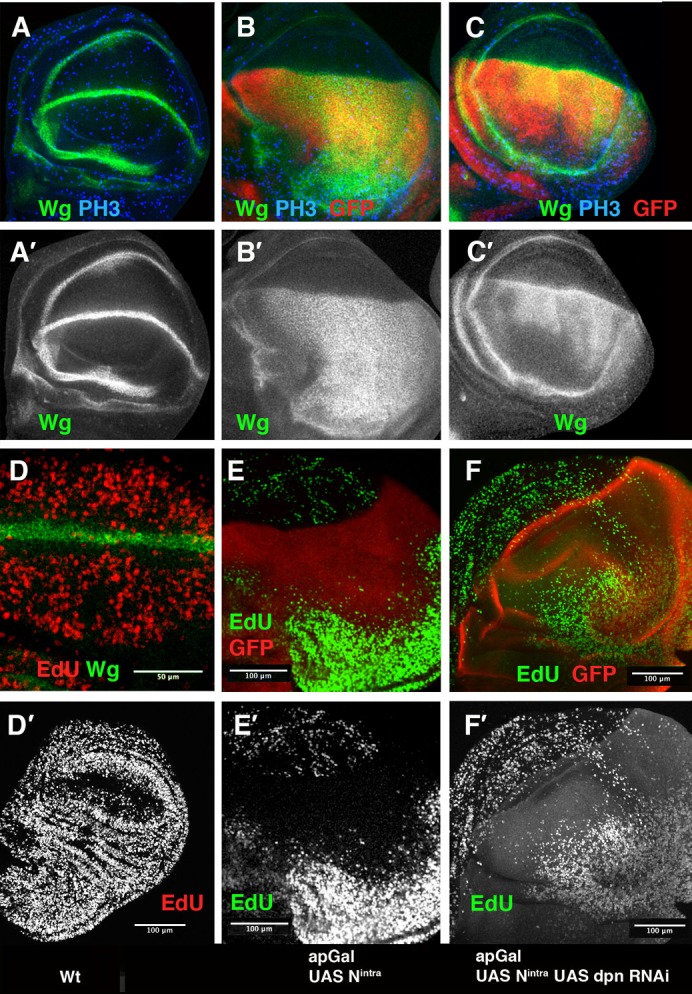
(A–F) Third instar larval wing discs stained with anti-Wg (in green in A–D, and grey in A′–C′), anti-PH3 (in blue A–C) and EdU incorporation (red in D, green E,F and grey in D′–F′). (A,A′,D,D′) Wild type discs. (B,B′,E,E′) ap-Gal4/UAS-Nintra; UAS-GFP/+ discs. (C,C′,F,F′) ap-Gal4/UAS-Nintra; UAS-dpnRNAi/UAS-GFP discs. In all panels, the expression of GFP driven by ap-Gal4 is shown in red. The ectopic expression of Nintra, under the regulation of ap, induces high levels of Wg (green) throughout the dorsal compartment (B,B′) compared with the control (A,A′). In ap-Gal4/UAS-Nintra; UAS-dpnRNAi/UAS -GFP wing discs, this effect was partially suppressed (C,C′ compared with B,B′). (D) High magnification of the ZNC in a wild type disc, the expression of Wg is shown in green. The incorporation of EdU (red) is reduced in the wing margin. (E,E′) The ZNC, monitored by EdU incorporation, was expanded in ap-Gal4/UAS-Nintra; UAS-GFP/+ discs (E compared with the control D), although in the hinge region the number of cells that incorporate EdU was increased (F,F′) The down-regulation of dpn in ap-Gal4/UAS-Nintra; UAS-dpnRNAi/UAS-GFP discs partially suppressed this effect and the extension of the ZNC was reduced (F,F′).
Loss of dpn partially suppressed the effects caused by ectopic activation of Notch
To further test the dpn role downstream of Notch, we checked whether the dpn elimination suppresses the effects caused by ectopically activating Notch signalling. Late third instar wing discs (140 h AEL) expressing UAS-Nintra under ap control (ap-Gal4/+; UAS-Nintra) are much larger than the wild-type discs (Fig. 4B,B′; supplementary material Fig. S4). In accordance with previous reports (Herranz et al., 2008), we found that the ZNC in these discs was expanded in the dorsal compartment, as monitored by EdU incorporation (Fig. 4E,E′). In contrast to the wing pouch region, the number of dividing cells in the hinge was strongly increased when compared with control discs (supplementary material Fig. S4). These mutant larvae displayed a delay in pupation, likely caused by the continuous growth of the disc proximal regions. Thus, at 220 h (AEL), we found ap-Gal4/+; UAS-Nintra larvae containing very large wing discs with overgrown hinges (Fig. 4; supplementary material Fig. S4). In agreement with previous results (Couso et al., 1995; Rulifson et al., 1996; Neumann and Cohen, 1996; Kim et al., 1995; de Celis et al., 1996a; Herranz et al., 2008), the expression of Wg and Cut in these discs was expanded throughout the dorsal compartment, whereas dMyc was down-regulated (Fig. 4B,B′; supplementary material Fig. S4). All these effects were partially suppressed when dpn was depleted by over-expressing UAS-dpnRNAi. Thus, at 220 hs AEL, ap-Gal4/UAS-dpnRNAi; UAS-Nintra wing discs were significantly smaller than the ap-Gal4/+; UAS-Nintra discs (supplementary material Fig. S4). Furthermore, the expression of dMyc was partially restored in the dorsal compartment (supplementary material Fig. S4), whereas the ectopic expression of Cut and Wg was reduced; thus Cut and Wg were only ectopically expressed in the most distal regions of the presumptive wing blade (Fig. 4B–C′; supplementary material Fig. S4). Consequently, the ZNC extension was reduced, as monitored by EdU incorporation (Fig. 4E–F′).
The effects of the Nintra-expressing clones are dependent on the region of the wing discs where they appear. Thus, clones located in the wing pouch proximal region and in the hinge region induce extra cell proliferation, both autonomously and non-autonomously (Go et al., 1998; Baonza and Garcia-Bellido, 2000; Giraldez and Cohen, 2003). On the contrary, clones located in the distal regions of the wing blade blocked cell cycle progression and expanded the ZNC (Johnston and Edgar, 1998; Herranz et al., 2008). To further analyse the requirement for dpn in the different Notch pathway functions, we induced dpn7 mutant clones that simultaneously expressed Nintra. dpn depletion partially suppressed the ectopic expression of Wg and Cut, which is caused by Notch activation (supplementary material Fig. S5; data not shown). In agreement with previous results, the mutant clone cells located at the proximal region of the wing discs were smaller than the clones of Nintra-expressing cells in the same region (supplementary material Fig. S5). In addition to this autonomous rescue, we found that the proportion of dpn7 UAS-Nintra clones that caused non-autonomous overgrowth of the hinge and proximal regions of the wing discs was reduced compared to the that of Nintra-expressing clone cells (43%, n = 46 vs 61%, n = 32). Taken together, our data suggest that dpn is not only transcriptionally activated by Notch signalling but also mediates, at least in part, the function of this pathway during wing disc development.
Ectopic expression of E(spl)m3 and mγ reproduces some of the effects caused by the over-expression of dpn
The mild phenotype displayed by dpn mutant cells in the wing disc when compared to the Notch or the Su(H) mutant cells (Schweisguth and Posakony, 1992; de Celis and García-Bellido, 1994) and the partial rescue of the phenotypes produced by the ectopic activation of Notch signalling by dpn alleles, suggests the existence of additional Notch signalling targets. We have confirmed the results of previous studies (Klein et al., 2000) that indicated that during wing development, all functions of Notch signalling occurred through Su(H). We found that in the Su(H)O47 mutant clones that simultaneously express UAS-Nintra, all of the effects caused by the ectopic expression of Notch are completely suppressed (data not shown). These data indicate that the target genes of Su(H) must be enacting the different Notch signalling functions during wing disc development. The best-characterised Notch/Su(H) targets genes are the members of the Enhancer-of-split complex (E(spl)C) (Delidakis and Artavanis-Tsakonas, 1992; Knust et al., 1992; Jennings et al., 1994). During wing disc development, the mutant clone cells, containing a deficiency that eliminates all the members of this complex, did not cause loss of wing margin; however, the differentiation of sensory structures and the pattern of veins was affected (de Celis et al., 1996b). Because the E(spl)C proteins show strong sequence similarities to Dpn, we wondered whether those genes could act redundantly with dpn. To study the function of different E(spl)C components, we induced the ectopic expression of the complex members in the dorsal compartment. Most of the genes studied gave rise to adults with vein loss but normally sized wings. However, the ectopic expression of two E(spl) proteins, E(spl)m3 and E(spl)mγ, resulted in adults with small wings that had large hinge regions (supplementary material Fig. S3). The third instar wing discs in these conditions displayed overgrowth in the wing hinge (241,300±44,000 and 224,000±59,000 µm3 for E(spl)mγ and E(spl)m3, respectively, compared with 137,000±30,000 µm3 in the control) that was associated with increased cell proliferation and confirmed by anti-PH3 staining (the ratios of hinge PH3-positive-cells/dorsal wing blade PH3-positive-cells were as follows: control 1.26±0.026, m3 1.96±0.12, and mγ 2.03±0.11 (n = 10 for each genotype)) (Fig. 3C,D). The mitotic index in these disc hinge regions was also increased (2.6×10−4±5×10−5 and 2.2×10−4±2×10−5 cells per µm3 for mγ and m3, respectively, compared with 1.5×10−4±3×10−5 for the control discs). This phenotype appeared similar to the hinge region enlargement caused by dpn over-expression (Fig. 3B,B′). These results suggest that at least some of the E(spl) complex members might also participate downstream of Notch to promote cell proliferation in the hinge.
The E(spl)C genes and dpn act redundantly downstream of Notch signalling
To further analyse the relationships between dpn and the members of the E(spl) complex, we induced MARCM mutant clones containing the deficiency E(spl)Xp (Bardin et al., 2010) and simultaneously depleted dpn by means of a dpnRNAi. Double E(spl)Xp UAS-dpnRNAi mutant clones in the wing blade were comparably sized with control clones (72±44 cells (n = 24) and 79±51 cells (n = 25), respectively), suggesting that in contrast to Notch signalling, their function was not essential for cell proliferation control during wing disc development. However, when these clones crossed the d/v boundary, they caused a strong down-regulation of the expression of Wg and Cut (Fig. 5A–D″). Neither clones of E(spl)Xp nor UAS-dpnRNAi caused these strong effects (supplementary material Fig. S2; data not shown). We next examined the contribution of E(spl)Xp and dpn in the G1 arrest of the d/v boundary cells. While wild-type cells at the d/v boundary were arrested in G1, the UAS-dpnRNAi; E(spl) XP mutant cells located at the ZNC progressed in the cell cycle, as monitored by EdU incorporation and by the expression of the G2/M specific marker Cyclin-B (Fig. 5E–H). Furthermore, in contrast to the wild-type cells, the UAS-dpnRNAi; E(spl) XP mutant cells express dMyc (Fig. 5A–A″). This effect is similar to the phenotype seen with a decrease in Notch activity (Duman-Scheel et al., 2004; Herranz et al., 2008) (data not shown). Taken together, our results suggest that the functions of Notch signalling that promote a cell-cycle arrest and the induction of Wg and Cut at the d/v boundary are, at least in part, mediated by dpn and some of the E(spl) complex members.
Fig. 5. The down-regulation of E(spl) and dpn partially suppressed the definition of the d/v boundary and the establishment of the ZNC.
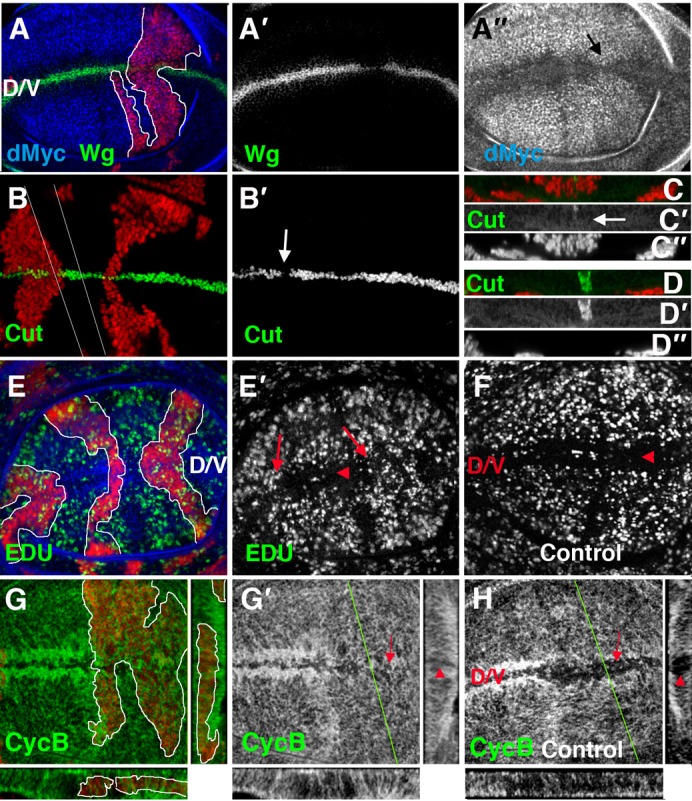
(A–E′,G,G′) Clones of E(spl)Xp UAS-dpnRNAi mutant cells. Clones were positively marked with GFP (A–D, E,G), shown in red, and outlined by a white line. (A–A″) In E(spl)Xp UAS-dpnRNAi mutant cells, Wg (green in A and grey in A′) was strongly down-regulated. In these mutant cells, dMyc is up-regulated at the d/v boundary (black arrow) (in blue in A and grey in A″). (B–D″) In E(spl)Xp UAS-dpnRNAi clones, Cut (green in B,C,D, and grey in B′,C′,D′) was almost absent, and we found a gap where it was totally eliminated (white arrows in B,C′, compare with D,D′). Optical Z-sections of panel B are shown in C and D. White lines indicate the position of the sections. (E–F) EdU (green in E and grey in E′,F) was incorporated in the E(spl)Xp UAS-dpnRNAi mutant cells that crossed the d/v margin (red arrows in E′) but not in wild type d/v boundary cells (red arrowheads in E′,F). (G–H) The expression of CycB (green in G, and grey in G′,H) in wing discs containing clones of E(spl)Xp UAS-dpnRNAi mutant cells (G,G′) and in control discs (H). The mutant cells that crossed the ZNC expressed CycB (compare the red arrow in G′ with the expression of CycB in control cells (the red arrow in H)). Optical Z-sections (right) were taken of panels G and H at the position of the green line, whereas the X-Z projections below the panels show a cross-section along the d/v boundary. In the control cells, CycB was not expressed in the ZNC (red arrowheads in H), but it was expressed in the mutant cells, seen by a comparison of the expression of CycB in the X-Z projections of mutant cells (red arrowhead in G′) with the wild-type expression (red arrowheads in H).
To further study this requirement, we analysed whether the elimination of the dpn and the E(spl) genes was sufficient to suppress the effects caused by the ectopic expression of Nintra. We induced mutant E(spl)Xp clones that simultaneously expressed UAS-Nintra and UAS-dpnRNAi. The effects on Wg, Cut and dMyc expression that are seen with ectopic expression of Notch were strongly suppressed by depletion of E(spl)Xp and dpn. Thus, in contrast to clones of Nintra-expressing cells that up-regulate Wg and Cut in all wing blade regions, clones of UAS-dpnRNAi; E(spl)Xp UAS-Nintra located in the proximal regions of the wing pouch either did not express Cut and Wg or did so at very low levels (Table 1). In the case of clones generated in distal regions, we found that only the mutant cells closest to the d/v boundary ectopically expressed high levels of Wg and Cut (Fig. 6B,D). Moreover, a down-regulation of dMyc was observed only in these mutant clones, contrasting with its general down-regulation in all clones of the Nintra-expressing cells in the wing pouch (Fig. 6A–D). Interestingly, the simultaneous down-regulation of dpn and the genes of the E(spl) complex partially suppressed the autonomous and non-autonomous excessive growth caused by ectopically activating Notch in the proximal and hinge regions of the wing discs. Thus, the average size of UAS-dpnRNAi; E(spl)Xp UAS-Nintra mutant clones in these regions was smaller than Nintra-expressing clone cells, such that 35% of the mutant clones were smaller than 1,000 cells, compared to 10% of the Nintra-expressing cells. Moreover, we never found UAS-dpnRNAi; E(spl)Xp UAS-Nintra clones larger than 5,000-6,000 cells, whereas 15% of the UAS-Nintra clones were much larger (Fig. 6E). The proportion of the UAS-dpnRNAi; E(spl)Xp UAS-Nintra clones that caused non-autonomous overgrowth of the wing disc hinge and proximal regions was also reduced; only 36% (n = 22) of these clones produced this effect, contrasting with 64% (n = 28) of the Nintra-expressing clone cells. In all cases, we found that the depletion of dpn in conjunction with E(spl)C produced stronger suppression of the phenotype displayed by the NotchIntra expressing clone cells than when only dpn function was eliminated.
Table 1. Quantification of the effects of clones of dpnRNAi Nintra E(spl)XP and Nintra in the expression of Cut or Wg.
We distinguished between clones in proximal regions (clones adjacent to the hinge regions), D/V boundary (clones that are in the D/V boundary) and clones in the wing blade (clones not included in the other categories that are in the wing blade region). N stands for No (clones with no expression of Cut or Wg), P stands for partial (clones in which some cells expressed Wg or Cut), and T stands for total (clones in which all the cells expressed these proteins).
Fig. 6. E(spl)C and dpn mediate the functions of Notch signalling in regulating cell proliferation and the defining the d/v boundary.
(A–D′) Third instar larval wing discs stained with anti-Wg (in green in A,B,A‴,B‴, and grey in A′–B′), anti-Cut (green C,D, and grey C′,D′) and anti-dMyc (in blue A,B,A‴ ,B‴, and grey A″,B″) antibodies. Clones were positively marked with GFP in red (A,A‴,B,B‴,C,D). (A–A‴) In UAS-Nintra-expressing mutant cells, Wg was expressed at high levels throughout the wing blade. In these mutant cells, dMyc was down-regulated. (A‴) A high magnification view of panel A. (B–B‴) The up-regulation of Wg, caused by the ectopic activation of Nintra, was strongly suppressed in clones of UAS-dpnRNAi UAS-Nintra E(spl)Xp cells. In most of these clones, the ectopic expression of Wg was restricted to regions close to the wing margin (arrows), compared with the discs in A. (B‴) A high magnification view of panel B. (C,C′) The expression of Cut in discs contained clones of Nintra-expressing cells. (D,D′) In UAS-dpnRNAi UAS-Nintra E(spl)Xp mutant cells, Cut was ectopically expressed near the d/v boundary compared with C. (E) Quantitative analysis of the size of wild type (n = 59), UAS-Nintra (n = 79), and UAS-dpnRNAi UAS-Nintra E(spl)Xp (n = 50) mutant clones. Clone sizes were analysed using ImageJ (see Materials and Methods). The sizes of UAS-dpnRNAi and E(spl)Xp clones were similar to those of controls. Clones were induced at 60±12 h AEL and were analysed 96 h after induction.
Our results indicate that the elimination of dpn in conjunction with E(spl)C was sufficient to notably suppress the effects of the ectopic activation of Notch signalling. However, the rescue was not complete, as UAS-dpnRNAi; E(spl)Xp UAS-Nintra mutant clones were still larger than the controls (Fig. 6E) and the ectopic expression of Cut and Wg was not completely eliminated (Fig. 6). These results suggest that Notch signalling must be regulating other genes during this process that may act redundantly or in parallel with dpn and E(spl)C.
Ectopic co-expression of dpn and m3 or mγ induces the ectopic expression of Wg and extra-growth
Our data are consistent with a requirement for dpn and some of the E(spl)C genes in mediating multiple Notch signalling functions during wing disc development. However, when we induced clones over-expressing dpn or E(spl)C genes, we found that they never induced the expression of Wg or Cut seen upon ectopic activation of Notch. Moreover, their over-expression caused neither the arrest of the cell cycle at the d/v boundary nor the large autonomous and non-autonomous extra-growth produced by Nintra-expressing clone cells in the proximal regions of the wing pouch (data not shown). We then questioned whether dpn and the genes of the E(spl) complex, in addition to acting redundantly, could also cooperate. To this end, we examined clones that ectopically co-express dpn and different members of the E(spl)C. We found that the expression of dpn together with most of the complex genes did not reproduce the effects found in Nintra-expressing clone cells (Table 2). However, some clones co-expressing dpn and either E(spl)m3 or E(spl)mγ induced weak ectopic expression of Wg and down-regulated the dMyc expression when they are located close to the wing margin (Table 2; Fig. 7). Interestingly, we found that in the wing disc proximal and hinge regions, some of these clones could induce large overgrowths (Table 2; Fig. 7A). As seen in clones ectopically expressing Nintra, these areas of overgrowth contained mutant cells as well as wild-type cells (Fig. 7). A characteristic feature of the Nintra expressing clone cells is that, within the wing pouch, clones closer to the endogenous d/v boundary proliferated less and their average size was smaller than control clones (53±48 cells, n = 75 compared to 79±51 cells, n = 25 in controls p = 0.02) (Go et al., 1998; Baonza and Garcia-Bellido, 2000; Giraldez and Cohen, 2003) (Fig. 6A). The dpn and E(spl)mγ or E(spl)m3-expressing clone cells produced comparable results, as the average sizes of these mutant clones in distal regions of the wing pouch were 42±25 cells (n = 28) and 46±45 cells (n = 31), respectively.
Table 2. Effects caused by clones of over-expression of dpn together with different members of the E(spl)C.
We included the percentage of clones that caused the effects. n indicates the number of clones analysed.
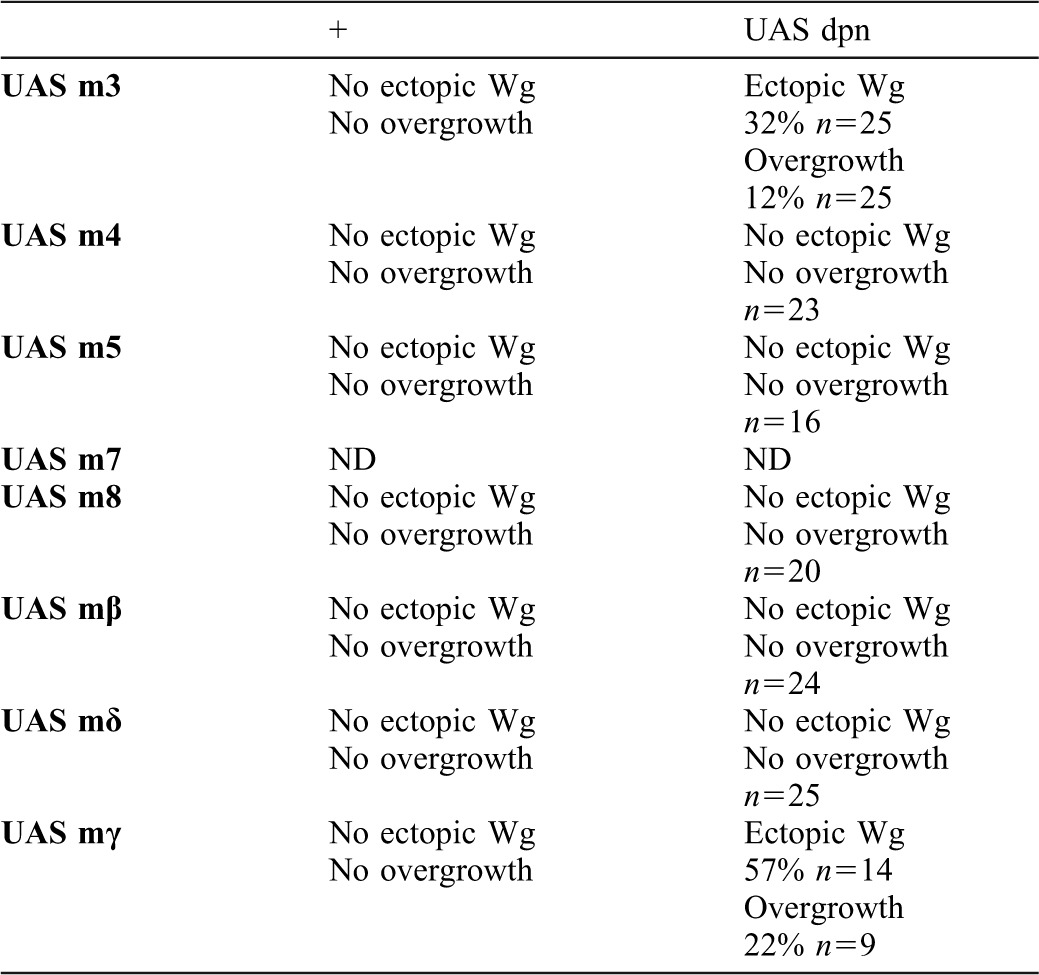
Fig. 7. The co-expression of dpn and m3 or mγ can mimic the effects caused by the ectopic expression of Notch signalling.
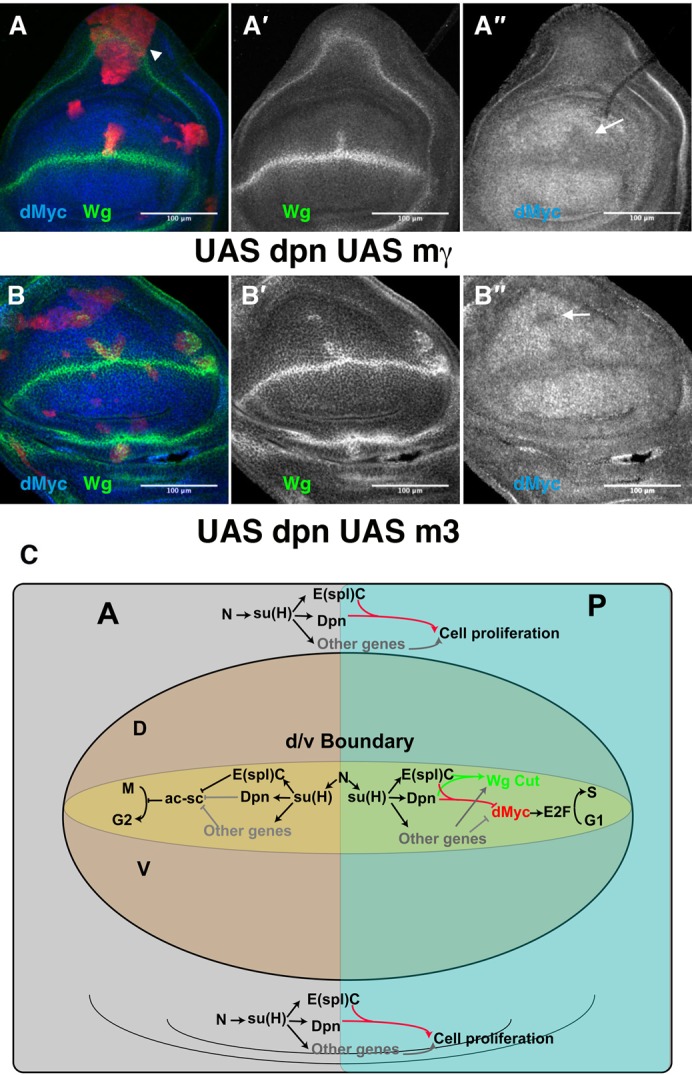
(A–B″) Third instar larval brains containing clones of cells (positively marked with GFP in red) expressing dpn with mγ (A–A″) or m3 (B–B″). In both mutant conditions, we found some clones that ectopically expressed Wg (in green in A,B, and grey in A′,B′) and displayed decreased levels of dMyc (arrows) (in blue in A,B, and grey in A″,B″). In the proximal region of the wing discs, we found large clones that induced non-autonomous overgrowth (arrowhead in A). (C) An illustration describing the role of Notch signalling in controlling the cell cycle in the anterior and posterior compartments of the wing margin, through the dpn and E(spl)C genes.
These data indicate that the co-expression of dpn and E(spl)m3 or E(spl)mγ was sufficient to reproduce most of the effects of the ectopic activation of Notch signalling during wing disc development, although at lower penetrance and expressivity. Our data suggest that dpn, E(spl)m3, and E(spl)mγ might cooperatively act to define the wing margin and the ZNC, as well as act in controlling cell proliferation of the wing disc proximal region.
Discussion
Notch signalling is required for various processes during wing disc development. In addition to its well-known function in defining the d/v boundary (Rulifson and Blair, 1995; Couso et al., 1995; Diaz-Benjumea and Cohen, 1995; de Celis et al., 1996a; Micchelli et al., 1997), this pathway also regulates cell proliferation in a regional-dependent manner. Thus, whereas Notch is necessary to autonomously arrest the cell cycle in G1 at the wing margin, in order to establish the ZNC (Herranz et al., 2008), the Notch activity in the proximal regions of the wing pouch and in the wing hinge stimulates cell proliferation both autonomously and non-autonomously (Go et al., 1998; Baonza and Garcia-Bellido, 2000; Giraldez and Cohen, 2003). The different cellular regulatory responses to Notch activity and the genes that enact its different functions during wing disc development are still largely unknown. Previous studies have shown that the Su(H) target genes of the E(spl)C were only required for a subset of the processes involving Notch signalling. Thus, although some members of this complex are transcribed at the wing margin in a Notch-dependent manner, their functions seem to be unnecessary for the definition of the wing margin and the establishment of the ZNC (de Celis et al., 1996b). In addition, this complex appears to be dispensable for controlling proliferation (de Celis et al., 1996b). These results could be obscured due to genes acting cooperatively and redundantly with the E(spl)-C genes downstream of Notch/Su(H). Recently, we have identified the bHLH repressor Dpn as a target of Notch during neuroblast division (San-Juan and Baonza, 2011). Dpn shows strong similarities to the E(spl) bHLH proteins. In this study, we have shown that the expression of dpn depends on Notch signalling during wing disc development. Furthermore, we show that the ectopic co-expression of dpn with E(spl)m3 or E(spl)mγ could reproduce, although at a weaker level, many of the Notch over-expression effects, whereas the simultaneous loss of function of dpn and E(spl)C mimics most of the effects produced by the Notch signalling insufficiency. Moreover, we have found that the concomitant elimination of dpn and E(spl) strongly suppresses the phenotypes caused by the ectopically active Notch. Taken together, our results indicate that dpn and members of the E(spl) complex partially mediate the function of Notch signalling in defining the wing margin and the ZNC and in controlling cell proliferation of the wing disc proximal region.
This provides evidence of a surprisingly linear pathway, because Notch, through Su(H), would transcriptionally activate the expression of dpn and E(spl)C, which might in turn regulate the expression of other genes required for the control of cell proliferation in the proximal region of the wing (Fig. 7C). In the wing margin, dpn and E(spl)C could contribute to the repression of dMyc (Fig. 7C) to block cell cycle progression and arrest the cells in the G1 phase. In addition, E(spl)C is a well characterised repressor of Achaete/Scute Complex protein activity (Giagtzoglou et al., 2005). The genes of this complex are activated by Wg and negatively regulate the expression of string, the cdc25 phosphatase, in the cells adjacent to the wing margin (Johnston and Edgar, 1998; Duman-Scheel et al., 2004; Herranz et al., 2008), arresting the cells in G2. In the wing margin cells, Notch transcriptional activation of the E(spl)C genes, and likely dpn, would prevent Wg from promoting the expression of the ac/sc complex genes in these cells, allowing them to proliferate when the G1/S transition repression is released (Fig. 7C). Our results show that the elimination of the E(spl)C genes and dpn neither completely reproduces the loss of Notch phenotype nor fully suppressed the ectopic expression pathway effects. These data indicate that Notch signalling must be regulating other gene(s) during this process that may act redundantly with dpn and E(spl)C genes (Fig. 7C).
Control of cell proliferation by Dpn and E(spl)C
Dpn and the E(spl)-C genes encode bHLH family transcription factors. This family of proteins is characterised by the presence of three conserved domains that confer a transcriptional function, the bHLH, Orange and WRPW domains. The bHLH domain includes the basic region for DNA binding and the HLH motif for dimerisation. Most bHLH factors bind to a consensus sequence that is contained in the promoter region of their target genes. Through the WRPW domain, these factors can interact with the co-repressor Groucho and can actively repress transcription of these target genes by chromatin-inactivation (reviewed by Kageyama et al., 2007). We and others have shown that dMyc is negatively regulated by Notch signalling during wing disc development (Duman-Scheel et al., 2004; Herranz et al., 2008). Interestingly, it has been shown that in human T-cell acute lymphoblastic leukaemia/lymphoma, the cmyc proto-oncogene acts as a target of Notch1 (Sharma et al., 2006). The fact that an activated form of Su(H) was able to repress dMyc (Herranz et al., 2008) suggests that Notch regulates dMyc through a Su(H) target gene. Our results indicate that dpn in combination with the E(spl)C genes could mediate this transcriptional regulation. Additionally, the promoter region of dmyc contains putative binding sites for Dpn (Southall and Brand, 2009). Dpn and E(spl)C may act simultaneously to modulate the expression of a set of genes that is required to control proliferation, which is likely to block differentiation and allow cell cycle progression. Interestingly, it has been proposed that Notch activity can maintain cells in a proliferative state by antagonising the cyclin dependent kinase inhibitor P21/P27 homolog Dacapo in the developing Drosophila CNS (Griffiths and Hidalgo, 2004). Moreover, dpn has been proposed as a possible regulator of dacapo expression (Wallace et al., 2000). Therefore, it is conceivable that Notch signalling, acting through Dpn, might transcriptionally regulate dacapo, as well as other cell cycle regulators. In addition to this active repression, bHLH factors can also passively repress gene expression. The bHLH repressor factors can form heterodimers with bHLH activators, preventing them from binding to DNA or blocking their transcriptional activation domains. Thus, Dpn could display a dominant-negative effect on different bHLH transcriptional factors.
Materials and Methods
Genetic strains
The following alleles were used: E(spl)XP (Bardin et al., 2010), Dlrev10, N55e11 and dpn7. The following UAS lines were used: UAS-NintrLH50 (II), UAS-Nintra(III), UAS-dpn (San-Juán and Baonza, 2011), UAS-E(spl)m8, UAS-E(spl)mδ 1−2 and h8 (Ligoxygakis et al., 1998), UAS-E(spl)m5.2 and R4, UAS-E(spl)m4, and UAS-E(spl)β (de Celis et al., 1996b), UAS-E(spl)m3 and UAS-E(spl)mγ (Ligoxygakis et al., 1999; Ligoxygakis et al., 1998). The following Gal4 lines were used: ap-Gal4 and nub-Gal4. We also used the reporter line P (E(spl)m4-lac-Z)-96A (Bailey and Posakony, 1995). All of these stocks are described in FlyBase (http://flybase.bio.indiana.edu). The transgenic dpnRNAi strain was obtained from the VDRC (Vienna Drosophila Research Centre) (line 106181).
Generation of mosaics
Mitotic clones were generated by FLP-mediated mitotic recombination (Xu and Rubin, 1993). Clones lacking dpn were obtained by crossing FRTG13 dpn7 to hsp-FLP122 tub-Gal4 UAS-nucGFP; tub-Gal80 FRTG13/Cyo flies. For the twin analysis, the clones were marked by the absence of GFP using hsp-FLP122; FRTG13 UB-GFP/Cyo. Control clones were generated using the FRTG13 UAS-mCD8-GFP chromosome.
FRT82B DlRev10 and FRT82B E(spl)XP clones were marked by the absence of GFP, using y w hsp-FLP122; FRT82B UB-GFP M(3)/TM6B and y w hsp-FLP122; FRT82B UB-GFP/TM6B stocks, respectively.
N55e11 mutant clones were positively marked with GFP in progeny from the cross between male tub-Gal80 FRT18A; hsp-FLP122 tub-Gal4 UAS-GFP and N55e11 FRT18A/FM7 females.
Clones of dpn7 that simultaneously express Nintra, and E(spl)XP mutant cells expressing UAS-dpnRNAi or UAS-Nintra were generated using the Gal4/Gal80 system (Lee and Luo, 1999). FRTG13 dpn7/+; UAS-Nintra/TM6B and FRTG13 UAS-mCD8-GFP/; UAS-Nintra/SM6a-TM6B were crossed to hsp-FLP122 tub-Gal4 UAS-GFP; tub-Gal80 FRTG13/Cyo.
Females of y w hsp-FLP122 tub-Gal4 UAS-GFP; tub-Gal80 FRT82B/TM6 genotype were crossed to males of the following genotypes:
UAS-dpnRNAi; FRT82B UB-GFP/TM6B
UAS-Nintra FRT82B/TM6B
UAS-dpnRNAi; FRT82B E(spl)XP/SM6a-TM6B
UAS-dpnRNAi; UAS-Nintra FRT82B E(spl)XP/SM6a-TM6B
FRT82B E(spl)XP/TM3
The progeny of these crosses were heat shocked at 37°C for 1 hour at 48–72 hours after egg-laying. Wing discs were dissected and analysed 3 days after the induction of the clones.
Clones of cells expressing Gal4 (Ito et al., 1997) were induced 48–72 hours after egg laying by heat shocking at 37°C for 12 minutes in larvae of the genotype FLP1.22; Act5C<FRTyellow+FRT>Gal4 UAS-GFP/+ UAS-dpn/+. Clones of the cells co-expressing UAS-dpn and different members of E(spl) complex were induced in larvae of FLP1.22; Act5C<FRT yellow+ FRT> Gal4 UAS-GFP/UAS-X;UAS-dpn/TM6B, where X represents the different genes of the E(spl)C tested.
The MARCM system is a method to positively mark the off-spring of mutant or wild-type cells. This system combines the Gal80 repressor protein with the Drosophila Gal4 transcription factor-upstream activator sequence (UAS) binary expression system and the FLP/FRT system to genetically label clones. Using the Gal 4-UAS system, it is possible to over-express a specific gene under the regulation of the UAS promoter. The clones of cells that ectopically expressed this gene can be labelled using different markers under UAS regulation, such as UAS-GFP. In the MARCM system, the activity of Gal4 is repressed by the Gal80 factor. After FLP/FRT-dependent mitotic recombination, homozygous mutant cells lack Gal80, allowing active Gal4 to activate marker genes and any gene under the regulation of UAS (Wu and Luo, 2007).
Hinge and clone size quantifications
Quantitative analyses of the size of the clones and hinge regions were performed by measuring the volume of the clones and hinges using the Volumest tool in the ImageJ application. The estimated cell size used for calculating the number of cells was 30 µm3.
EdU staining protocol
Larvae were dissected in Schneider Medium at room temperature and incubated for 1 h at 25°C in 200 µl of 1× EdU working solution supplemented with 1% goat serum. After the EdU incorporation, the tissue was washed with PBS and fixed in 500 µl of 4% PFA for 1 h. Permeabilisation required 3 washes (20 minutes) in 0.1% Triton X-100 in PBS. The tissue was blocked in 3% BSA-PBS (3 times for 20 minutes each). The primary antibody staining was done according to standard protocols before the EdU reaction cocktail was added. We used anti-GFP antibody to retain the fluorescent signal. The Click IT Buffer for the EdU visualisation was prepared just before addition, and the incubation was carried out for 30 minutes at room temperature in the dark. After this step, the samples were washed in 0.1% Triton X-100 in PBS for 40 minutes. Kit Invitrogen (Cat No. A10044, E10187).
Immunohistochemistry
Immunostaining of wing discs was performed according to standard protocols. The following antibodies were used: Guinea pig anti-dMyc (1:200) (Herranz et al., 2008) rabbit anti-Dpn (diluted 1:1000 and 1:500) (Gift from Yan); rabbit anti-phospho-Histone 3 (Upstate) (1:1000); mouse anti-β-galactosidase (Promega Z3778A) (1:200); mouse anti-Dl (C594.9B) (1:50); mouse anti-CycB (F2F4) (1:10); mouse anti-Cut (2B10) (1:100), and mouse anti-Wg (4D4) (1:100), were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. Secondary antibodies (Molecular Probes) were used at dilutions of 1:200.
Supplementary Material
Acknowledgments
We thank L. A. Baena, S. Campuzano, J. de Celis and S. Diaz for their helpful comments and constructive criticism. We are very grateful to J. Culi, Y. N. Jan, J. Skeath, T. Cline, A. Martinez-Arias, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for providing fly strains and antibodies. This work was supported by grants from the MICINN (BFU2008-03664/BMC) and Consolider (20072D9110) and by an institutional grant from the F. Ramón Areces to the CBM-SO. B.P.S. was supported by a FPI fellowship from the MICINN, and I.A. was supported by a JAE-PRE fellowship from CSIC.
Footnotes
Competing interests: The authors declare that there are no competing interests.
References
- Bailey A. M., Posakony J. W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609–2622 10.1101/gad.9.21.2609 [DOI] [PubMed] [Google Scholar]
- Baonza A., Garcia-Bellido A. (2000). Notch signaling directly controls cell proliferation in the Drosophila wing disc. Proc. Natl. Acad. Sci. USA 97, 2609–2614 10.1073/pnas.040576497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Perdigoto C. N., Southall T. D., Brand A. H., Schweisguth F. (2010). Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137, 705–714 10.1242/dev.039404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair S. S. (1995). Compartments and appendage development in Drosophila. Bioessays 17, 299–309 10.1002/bies.950170406 [DOI] [PubMed] [Google Scholar]
- Bray S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Cohen B., Simcox A. A., Cohen S. M. (1993). Allocation of the thoracic imaginal primordia in the Drosophila embryo. Development 117, 597–608. [DOI] [PubMed] [Google Scholar]
- Couso J. P., Knust E., Martínez Arias A. (1995). Serrate and wingless cooperate to induce vestigial gene expression and wing formation in Drosophila. Curr. Biol 5, 1437–1448 10.1016/S0960-9822(95)00281-8 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., García-Bellido A. (1994). Roles of the Notch gene in Drosophila wing morphogenesis. Mech. Dev. 46, 109–122 10.1016/0925-4773(94)90080-9 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., García-Bellido A., Bray S. J. (1996a). Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122, 359–369. [DOI] [PubMed] [Google Scholar]
- de Celis J. F., de Celis J., Ligoxygakis P., Preiss A., Delidakis C., Bray S. (1996b). Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122, 2719–2728. [DOI] [PubMed] [Google Scholar]
- Delidakis C., Artavanis-Tsakonas S. (1992). The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 89, 8731–8735 10.1073/pnas.89.18.8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Benjumea F. J., Cohen S. M. (1995). Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development 121, 4215–4225. [DOI] [PubMed] [Google Scholar]
- Duman-Scheel M., Johnston L. A., Du W. (2004). Repression of dMyc expression by Wingless promotes Rbf-induced G1 arrest in the presumptive Drosophila wing margin. Proc. Natl. Acad. Sci. USA 101, 3857–3862 10.1073/pnas.0400526101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M. E. (2009). Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647 10.1016/j.devcel.2009.03.010 [DOI] [PubMed] [Google Scholar]
- García-Bellido A. (1975). Genetic control of imaginal disc morphogenesis in Drosophila. Developmental Biology: Pattern Formation, Gene Regulation Vol. 2, (ed. McMahon D, Fox C F.), pp. 40–59 Menlo Park, CA, USA: W. A. Benjamin. [Google Scholar]
- Garcia-Bellido A., Merriam J. R. (1971). Genetic analysis of cell heredity in imaginal discs of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68, 2222–2226 10.1073/pnas.68.9.2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagtzoglou N., Koumbanakis K. A., Fullard J., Zarifi I., Delidakis C. (2005). Role of the Sc C terminus in transcriptional activation and E(spl) repressor recruitment. J. Biol. Chem. 280, 1299–1305 10.1074/jbc.M408949200 [DOI] [PubMed] [Google Scholar]
- Giraldez A. J., Cohen S. M. (2003). Wingless and Notch signaling provide cell survival cues and control cell proliferation during wing development. Development 130, 6533–6543 10.1242/dev.00904 [DOI] [PubMed] [Google Scholar]
- Go M. J., Eastman D. S., Artavanis-Tsakonas S. (1998). Cell proliferation control by Notch signaling in Drosophila development. Development 125, 2031–2040. [DOI] [PubMed] [Google Scholar]
- Griffiths R. L., Hidalgo A. (2004). Prospero maintains the mitotic potential of glial precursors enabling them to respond to neurons. EMBO J. 23, 2440–2450 10.1038/sj.emboj.7600258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H., Pérez L., Martín F. A., Milán M. (2008). A Wingless and Notch double-repression mechanism regulates G1-S transition in the Drosophila wing. EMBO J. 27, 1633–1645 10.1038/emboj.2008.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Awano W., Suzuki K., Hiromi Y., Yamamoto D. (1997). The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–771. [DOI] [PubMed] [Google Scholar]
- Jennings B., Preiss A., Delidakis C., Bray S. (1994). The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120, 3537–3548. [DOI] [PubMed] [Google Scholar]
- Johnston L. A., Edgar B. A. (1998). Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature 394, 82–84 10.1038/27925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. (2007). The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 10.1242/dev.000786 [DOI] [PubMed] [Google Scholar]
- Kim J., Irvine K. D., Carroll S. B. (1995). Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell 82, 795–802 10.1016/0092-8674(95)90476-X [DOI] [PubMed] [Google Scholar]
- Klein T., Seugnet L., Haenlin M., Martinez Arias A. (2000). Two different activities of Suppressor of Hairless during wing development in Drosophila. Development 127, 3553–3566. [DOI] [PubMed] [Google Scholar]
- Knust E., Schrons H., Grawe F., Campos-Ortega J. A. (1992). Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 132, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P., Yu S. Y., Delidakis C., Baker N. E. (1998). A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development 125, 2893–2900. [DOI] [PubMed] [Google Scholar]
- Ligoxygakis P., Bray S. J., Apidianakis Y., Delidakis C. (1999). Ectopic expression of individual E(spl) genes has differential effects on different cell fate decisions and underscores the biphasic requirement for notch activity in wing margin establishment in Drosophila. Development 126, 2205–2214. [DOI] [PubMed] [Google Scholar]
- Micchelli C. A., Rulifson E. J., Blair S. S. (1997). The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development 124, 1485–1495. [DOI] [PubMed] [Google Scholar]
- Neumann C. J., Cohen S. M. (1996). A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development 122, 3477–3485. [DOI] [PubMed] [Google Scholar]
- O'Brochta D. A., Bryant P. J. (1985). A zone of non-proliferating cells at a lineage restriction boundary in Drosophila. Nature 313, 138–141 10.1038/313138a0 [DOI] [PubMed] [Google Scholar]
- Phillips R. G., Whittle J. R. S. (1993). wingless expression mediates determination of peripheral nervous system elements in late stages of Drosophila wing disc development. Development 118, 427–438. [DOI] [PubMed] [Google Scholar]
- Rulifson E. J., Blair S. S. (1995). Notch regulates wingless expression and is not required for reception of the paracrine wingless signal during wing margin neurogenesis in Drosophila. Development 121, 2813–2824. [DOI] [PubMed] [Google Scholar]
- Rulifson E. J., Micchelli C. A., Axelrod J. D., Perrimon N., Blair S. S. (1996). wingless refines its own expression domain on the Drosophila wing margin. Nature 384, 72–74 10.1038/384072a0 [DOI] [PubMed] [Google Scholar]
- San-Juan B. P., Baonza A. (2011). The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev. Biol. 352, 70–82 10.1016/j.ydbio.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., Posakony J. W. (1992). Suppressor of Hairless the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell 69, 1199–1212 10.1016/0092-8674(92)90641-O [DOI] [PubMed] [Google Scholar]
- Sharma V. M., Calvo J. A., Draheim K. M., Cunningham L. A., Hermance N., Beverly L., Krishnamoorthy V., Bhasin M., Capobianco A. J., Kelliher M. A. (2006). Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol. Cell. Biol. 26, 8022–8031 10.1128/MCB.01091-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall T. D., Brand A. H. (2009). Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 28, 3799–3807 10.1038/emboj.2009.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K., Liu T. H., Vaessin H. (2000). The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis 26, 77–85 [DOI] [PubMed] [Google Scholar]
- Wu J. S., Luo L. (2007). A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat. Protoc. 1, 2583–2589 10.1038/nprot.2006.320 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G. M. (1993). Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.