Abstract
Coronatine is a phytotoxin produced by some plant-pathogenic bacteria. It has been shown that coronatine mimics the action of methyl jasmonate (MeJA) in plants. MeJA is a plant-signaling molecule involved in stress responses such as wounding and pathogen attack. In Arabidopsis thaliana, MeJA is essential for pollen grain development. The coi1 (for coronatine-insensitive) mutant of Arabidopsis, which is insensitive to coronatine and MeJA, produces sterile male flowers and shows an altered response to wounding. When the differential display technique was used, a message that was rapidly induced by coronatine in wild-type plants but not in coi1 was identified and the corresponding cDNA was cloned. The coronatine-induced gene ATHCOR1 (for A. thaliana coronatine-induced) is expressed in seedlings, mature leaves, flowers, and siliques but was not detected in roots. The expression of this gene was dramatically reduced in coi1 plants, indicating that COI1 affects its expression. ATHCOR1 was rapidly induced by MeJA and wounding in wild-type plants. The sequence of ATHCOR1 shows no strong homology to known proteins. However, the predicted polypeptide contains a conserved amino acid sequence present in several bacterial, animal, and plant hydrolases and includes a potential ATP/GTP-binding-site motif (P-loop).
JA and its methyl ester, MeJA, and related compounds derived from linolenic acid are recognized as signaling molecules synthesized by plants in response to wounding and herbivore and pathogen attack (Creelman et al., 1992; Farmer and Ryan, 1992; Mueller et al., 1993). These substances can activate the expression of several genes, leading to the accumulation of their products, which are referred to as jasmonate-induced proteins. The best-studied jasmonate-induced proteins include proteinase inhibitors, thionins, vegetative storage proteins, lipoxygenases, ribosome-inactivating proteins, enzymes of phenylpropanoid metabolism, and others (Koda, 1992; for review, see Reinbothe et al., 1994). The jasmonates can also repress the expression of genes related to photosynthesis at the transcriptional and translational levels (Reinbothe et al., 1994). It has been demonstrated that MeJA induces a shift in the length of the plastid rbcL transcript in barley, thus impairing translation initiation (Reinbothe et al., 1993). However, little is known about the mechanisms by which jasmonates control gene expression and the signal cascade that mediates this response. New evidence suggests that protein phosphorylation is required for the activation of certain wound-inducible genes that respond to JA (Damman et al., 1997).
Coronatine is a phytotoxin produced by several pathovars of Pseudomonas syringae (Ichihara et al., 1977; Mitchell and Young, 1978; Mitchell, 1982). Its biological effects include induction of leaf chlorosis and inhibition of root growth (Nishiyama et al., 1976; Ferguson and Mitchell, 1985; Kenyon and Turner, 1990), and it has been suggested to play a role in disease development as a virulence factor produced by the bacteria during infection. Mutations that abolished coronatine production in P. syringae pv tomato reduced the capacity of this pathogen to produce necrotic lesions on tomato (Lycopersicon esculentum) leaves (Bender et al., 1987). Moreover, coronatine production was required for the successful infection of Arabidopsis leaves by P. syringae pv tomato, and this was attributed to the suppression of defense-related genes by the toxin (Mittal and Davis, 1995).
Coronatine acts as a mimic of MeJA in plants (Weiler et al., 1994), and the Arabidopsis thaliana mutant insensitive to coronatine (coi1) is also insensitive to MeJA and is male sterile (Feys et al., 1994; Benedetti et al., 1995). In addition, coi1 plants are more sensitive to wounding than are the wild type, but they are resistant to P. syringae infection (Feys et al., 1994). These findings suggest that both coronatine and MeJA may interact with a common receptor and that MeJA is required for pollen development and mediates at least part of the wound response. However, there is evidence that one of the MeJA responses in Arabidopsis is needed to induce the symptoms caused by a coronatine-producing strain of P. syringae (Feys et al., 1994). This apparent paradox remains to be clarified, since MeJA is suggested to play a role in plant-defense responses (Coehn et al., 1993; Mueller et al., 1993; Reinbothe et al., 1994). Nevertheless, no correlation could be found between jasmonates and defense responses in plant-pathogen interactions (Schweizer et al., 1993; Kogel et al., 1995; Schweizer et al., 1997). How coronatine could function as a virulence factor by mimicking MeJA in a bacteria-plant interaction is an open question.
In an attempt to clarify this issue we are studying coronatine and MeJA responses in Arabidopsis by identifying genes that are rapidly activated by coronatine, MeJA, or wounding. To detect and clone such genes we are using the mRNA differential display technique (Liang and Pardee, 1992). We present here the initial characterization of a novel Arabidopsis gene that is induced by coronatine, MeJA, and wounding, the expression of which is affected by the COI1 gene.
MATERIALS AND METHODS
Biological Material and Chemicals
Wild-type Arabidopsis thaliana ecotype Columbia (Col-0) was used. The coi1 mutant was described previously (Feys et al., 1994) and was donated by Dr. John G. Turner (University of East Anglia, UK). Coronatine and MeJA were obtained as previously described (Feys et al., 1994).
Plant Growth
Seeds of wild-type Arabidopsis were germinated in MS medium (Murashige and Skoog, 1962), whereas coi1 seeds from an F2 population segregating for the Coi phenotype were first germinated in MS containing 1 μm coronatine to select for homozygous coi1 plants. Seedlings were grown in white light (70 μE m−2 s−1) for 1 week in a growth cabinet with a 16-h day/8-h night photoperiod at 22°C, after which they were transferred to fresh MS and grown for another 1 week. Seedlings were then either transferred to fresh MS for coronatine and MeJA treatments or moved to soil to grow to maturity.
Coronatine and MeJA Treatments
Two-week-old seedlings were transferred to MS (control seedlings) or MS containing either 1 μm coronatine or 10 μm MeJA. After different periods of incubation, they were frozen in liquid nitrogen and total RNA was extracted.
Wounding
For the wounding experiment, seeds were germinated in MS and grown for 2 weeks under short-day conditions (9-h day/15-h night at 22°C). Seedlings were then transferred to soil and grown for 2 weeks. Leaves of individual plants were wounded once with scissors. After the treatment, plants were returned to the growth cabinet, and wounded leaves were collected at different times, frozen in liquid nitrogen, and stored at −70°C for RNA extraction.
RNA Extraction and Differential Display
Total RNA from roots, seedlings, leaves, flowers, siliques, and wounded leaves was extracted according to the method of Verwoerd et al. (1989).
Differential display of mRNA was performed according to the method of Liang and Pardee (1992), with minor modifications. Total RNA (1 μg) from control and coronatine-treated seedlings of wild-type and coi1 plants were reverse transcribed with 100 units of reverse transcriptase in the presence of 2.5 μm T12VN as anchored primers and 20 μm dNTPs for 10 min at 25°C, followed by a 50-min incubation at 37°C. Two microliters of the reaction was then added to 18 μL of the PCR mixture consisting of 1× PCR buffer, 1.25 mm MgCl2, 1 μm anchored primer (T12VN), 1 μm arbitrary primer (10-mer from Operon, Alameda, CA), 2.0 μm dNTPs, 10 μCi [α-33P]ATP, and 1.5 units of Taq polymerase. PCR conditions were 40 cycles of 94°C for 30 s, 40°C for 2 min, and 72°C for 30 s, followed by 5 min of elongation at 72°C. PCR products were analyzed on a 6% acrylamide denaturing DNA-sequencing gel. Gels were dried at 80°C for 2 h, and radiographic films were aligned to them and exposed overnight at −70°C. Differentially displayed bands were cut off and eluted in 200 μL of water at 95°C for 15 min. DNA was ethanol precipitated to remove urea and reamplified by PCR using the same conditions as above, except that the final concentration of dNTPs was 20 μm. Reamplified DNA was analyzed on agarose gels, and bands of the expected size were purified and cloned into pMOSBlue vector (Amersham). Cloned fragments were sequenced and used to probe RNA blots and to screen a cDNA library of Arabidopsis flowers.
Northern Analysis
Total RNA (20 μg of each sample) was electrophoresed on formaldehyde-agarose gels (Sambrook et al., 1989), transferred onto nylon membranes (Hybond N+, Amersham) by capillary blot, and fixed by UV cross-linking according to the manufacturer's instructions. Blots were hybridized using the cloned fragments obtained from the display gels or fragments of the full-length cDNA clones as probes. Membranes were washed twice with 2.0× SSC containing 0.1% SDS for 10 min at 42°C and twice with 0.2× SSC containing 0.1% SDS for 10 min at 42°C.
cDNA Library Screening
A cDNA library of Arabidopsis (ecotype Landsberg erecta) flowers constructed in Lambda Zap II cloning vector (Stratagene) was kindly donated by Dr. Elliot M. Meyerowitz (California Institute of Technology, Pasadena). The library was screened following the manufacturer's protocols using the cloned fragments that showed differential expression as probes on northern analysis.
RACE
The 5′ end of the isolated cDNA was cloned by RACE according to the method of Frohman et al. (1988). Total RNA from wild-type flowers and from MeJA- and coronatine-treated seedlings was reverse transcribed using the P1 primer (5′-CCATTCTTACACATACAACC-3′). First-strand cDNA was purified and amplified by PCR using P1 and the (dT17)-adaptor primer (Frohman et al., 1988). After 20 PCR cycles (94°C for 30 s, 45°C for 1 min, and 72°C for 1 min), an aliquot was reamplified using the internal P2 primer (5′-CGTGATGGATGGGTCTAATG-3′) and the adapter primer in 20 PCR cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min (Frohman et al., 1988). PCR fragments were gel purified, cloned into pMOSBlue vector (Amersham), and sequenced.
RESULTS
A DNA fragment of about 280 bp was detected by differential display in wild-type but not coi1 seedlings upon induction with coronatine (not shown). DNA from the corresponding region of the display gel was reamplified, cloned, and sequenced.
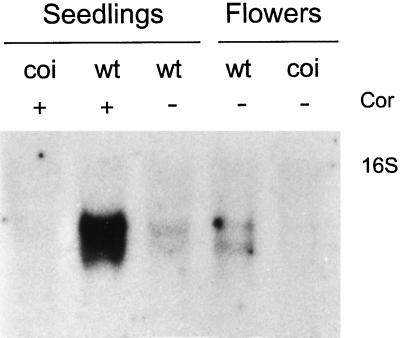
This clone, TGCOPP9–280, was used to probe a northern blot of total RNA extracted from seedlings and flowers of wild-type and coi1 mutant plants (Fig. 1). The probe confirmed the differential expression of a major transcript of approximately 1.3 kb that was induced by coronatine in wild-type but not in coi1 seedlings. Apparently, two transcripts of similar molecular weight were also detected in lower levels in untreated wild-type seedlings and flowers but not in male sterile flowers of coi1 (Fig. 1).
Figure 1.
Northern analysis of total RNA extracted from flowers and seedlings of wild-type (wt) and coi1 mutant seedlings (coi) growing in MS or in MS containing 1 μm coronatine (Cor) for 4 h, hybridized with TGCOPP9–280. The display probe detected a major transcript of about 1.3 kb that is induced by coronatine in wild-type but not coi1 seedlings. Transcripts of similar molecular weight are observed in lower levels in seedlings and flowers of the wild type but not in male-sterile flowers of coi1. The position of the 16S ribosomal band is indicated.
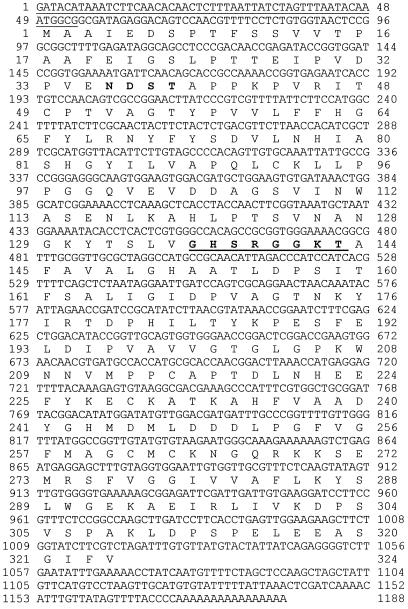
The clone TGCOPP9–280 was used to screen a cDNA library from Arabidopsis flowers. Five independent cDNA clones were isolated and sequenced. The sequences of all clones were identical and contained the entire sequence of TGCOPP9–280. However, unexpectedly, we found that the anchored primer T12GC did not prime at the poly(A+) tail of the mRNA but within the gene in an A-rich region. In addition, all of the isolated cDNAs were truncated at their 5′ ends. To obtain a full-length clone, the 5′ end of the gene was generated by RACE from mRNAs extracted from flowers or seedlings treated with either coronatine or MeJA. The sequences of these different RACE products were identical, indicating that the transcripts induced by coronatine or MeJA and those expressed in flowers were the same. The RACE products were also identical to the corresponding 5′ end of the cDNAs isolated from the library, except they contained an extra 50 bp in their 5′ ends (Fig. 2). A full-length cDNA clone was then obtained by ligating the 5′ end of the RACE product to the original cDNA isolated from the library using a SacI site upstream of the P2 primer (not shown). The resulting clone, ATHCOR1 (Arabidopsis thaliana coronatine-induced gene 1, accession no. AF021244), was chosen for further analysis.
Figure 2.
Sequence of ATHCOR1 cDNA (AF021244) and its predicted protein. The 5′ end of the untranslated sequence obtained from mRNAs by RACE is underlined. The first guanosine residue was not found in the genomic clone (not shown) and was interpreted as the mRNA cap. Amino acids in bold represent a possible N-glycosylation site and the bold, underlined sequence represents a potential ATP-/GTP-binding site motif A (P-loop).
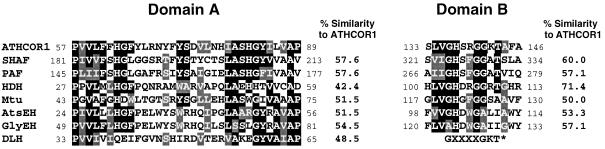
The sequence of ATHCOR1 cDNA has 1172 bp and may encode a novel protein of 324 amino acids with a molecular mass of 34.8 kD (Fig. 2). The predicted polypeptide has no significant homology to other proteins in the database, except in two small and conserved domains (A and B) found in microbial, plant, and mammalian hydrolases. These include the Synechocystis sp. dienelactone hydrolase, Moraxela sp. haloacetate dehalogenase, guinea pig platelet-activating factor acetylhydrolase, and epoxide hydrolases from soybean, potato, and Arabidopsis (Fig. 3). In the literature, no biochemical function has been attributed to domain A in these different hydrolases; however, domain B is a potential ATP-/GTP-binding site known as the P-loop, which is common to many ATP-/GTP-binding proteins (Saraste et al., 1990).
Figure 3.
Protein alignments between regions of the translated peptide from ATHCOR1 with two conserved domains (A and B) found in different hydrolases. SHAF, Similar to human-activating factor acetylhydrolase from C. elegans, U64598; PAF, platelet-activating factor acetylhydrolase from Cavia porcellus, JC5021; HDH, haloacetate dehalogenase from Moraxella sp., A44856; Mtu, unknown Mycobacterium tuberculosis Z95389; AtsEH, Arabidopsis epoxide hydrolase, D16628; GlyEH, soybean epoxide hydrolase, D63781; and DLH, dienelactone hydrolase from Synechocystis sp. dienelactone hydrolase, D90904. Black boxes indicate conserved amino acids at the minimum of 50%. Gray boxes indicate changes by similar residues. The percentage of similarity between ATHCOR1 and each of the sequences is presented. The consensus for the putative ATP-/GTP-binding site described in the literature (Saraste et al., 1990) is indicated by an asterisk. Sequences were aligned by the CLUSTAL W program (Thompson et al., 1994) and shaded by the BOXSHADE program (used at the Bioinformatics Group WWW site at the Swiss Institute for Experimental Cancer Research, Lousanne, Switzerland).
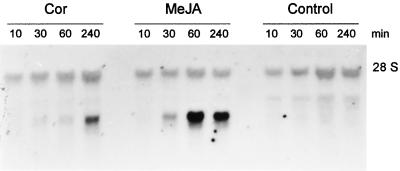
Northern blots using ATHCOR1 as a probe confirmed the same pattern of coronatine induction observed with the TGCOPP9–280 probe shown in Figure 1, except that the cDNA detected a single, approximately 1.3-kb band on northern analysis. To determine whether this gene was also induced by MeJA, wild-type and coi1 seedlings were grown in the presence of 10 μm MeJA, which produces the same phenotype as 1 μm coronatine (e.g. inhibition of root growth and anthocyanin accumulation; Feys et al., 1994). Figure 4 shows that the gene is rapidly induced by MeJA and that high levels of the transcript accumulate in the first 4 h of treatment. A similar pattern of induction was observed with 1 μm coronatine; however, the toxin was apparently a less efficient inducer of the gene in comparison with MeJA (Fig. 4).
Figure 4.
Time-course induction of total RNA extracted from seedlings of wild-type Arabidopsis growing in MS medium (Control) or MS containing 1 μm coronatine (Cor) or 10 μm MeJA. ATHCOR1 was rapidly induced by MeJA after 30 min of induction. A weaker induction of ATHCOR1 was observed with coronatine treatment. A background hybridization with the 28S ribosomal band is shown.
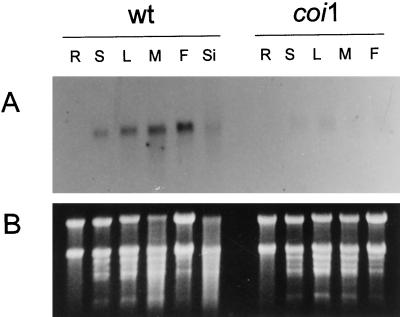
ATHCOR1 was used to probe northern blots of total RNA extracted from different plant organs (Fig. 5). The coronatine-/MeJA-induced gene was normally expressed in low levels in young and mature leaves and was apparently expressed in higher levels in flowers, but not detected at all in roots. Its expression was dramatically reduced in all tissues of coi1 plants (Fig. 5).
Figure 5.
A, Northern analysis of total RNA extracted from different organs of wild-type (wt) and coi1 (coi1) mutant plants. ATHCOR1 is normally expressed in seedlings (S), young (L) and mature (M) leaves, flowers (F), and siliques (Si), but it could not be detected in roots (R). Very low levels of the corresponding transcript were detected in coi1 tissues. B, Total RNA stained with ethidium bromide before being transferred onto the membrane.
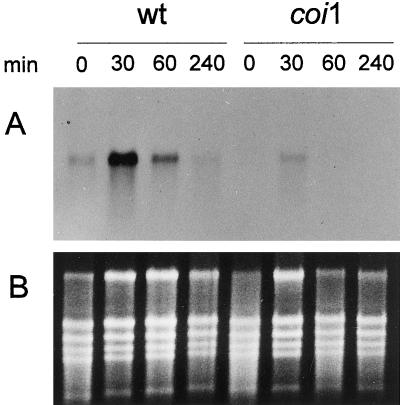
Wounding of wild-type leaves produced a rapid induction of ATHCOR1 (Fig. 6). Its expression peaked about 30 min after wounding and returned to basal levels in the following 4 h. Wounded leaves of coi1 showed a similar pattern of induction, but transcripts accumulated at much lower levels (Fig. 6).
Figure 6.
A, Northern analysis of total RNA extracted from leaves of wild-type (wt) and coi1 (coi1) mutant plants after wounding. ATHCOR1 was highly induced in wild-type leaves by wounding. Its expression peaked about 30 min after leaves were injured and decreased to normal levels after the first 4 h. The same pattern of induction was observed in the leaves of coi1, although at much lower levels. B, Total RNA stained with ethidium bromide before being transferred onto the membrane.
DISCUSSION
The coi1 mutant of Arabidopsis has proved to be an excellent model for the identification and analysis of coronatine- and MeJA-responsive genes. By means of differential display we have successfully identified ATHCOR1, a novel Arabidopsis gene that is induced by the phytotoxin coronatine in wild-type plants but not in the coi1 mutant. ATHCOR1 is expressed in leaves, flowers, and siliques, but it could not be detected in roots. Treatment with MeJA or wounding also increased its expression in wild-type but not in coi1 plants. However, the time-course induction of ATHCOR1 by exogenously applied MeJA was different from that of wounding, which seems to follow a transient stimulus.
ATHCOR1 has no strong similarities to other proteins, except in two conserved domains present in several hydrolases, including the dienelactone hydrolase, haloacetate dehalogenase, epoxide hydrolase, and acetylhydrolase. Epoxide hydrolase is the only one that has been described in plants. The microbial dienelactone hydrolase hydrolyzes dienelactone to maleylacetate and has esterase activity toward other substrates (Pathak et al., 1991; Beveridge and Ollis, 1995). Haloacetate dehalogenase acts specifically on halogenated acetates to yield glycolate as a carbon source in microorganisms (Kawasaki et al., 1992). Epoxide hydrolase catalyzes the conversion of epoxides to diols. In plants epoxide hydrolase has been suggested to play a role in the biosynthesis of monomers of cutin, a polymer that accumulates in the cell wall of wounded tissues (Kolattukudy, 1984; Blée and Schuber, 1993).
An epoxide hydrolase from A. thaliana (AtsEH) has been cloned (Kiyosue et al., 1994). AtsEH is only 20% similar to ATHCOR1 and is not expressed in flowers (Kiyosue et al., 1994). However, an epoxide hydrolase from potato, which is highly homologous to AtsEH, is induced by MeJA and wounding (Stapleton et al., 1994). Since ATHCOR1 is rapidly expressed in response to wounding, and because its expression is significantly reduced in coi1 plants, which are more sensitive to wounding, we speculate that this gene may be important in the process of healing injured tissue. It is possible that ATHCOR1 belongs to a family of related enzymes involved in the biosynthesis/hydrolysis of plant cell wall components. ATHCOR1 may also function during anther development, since male sterile-flowers of coi1 have much lower levels of its transcripts.
It is interesting to note that the predicted protein of ATHCOR1 has a potential ATP-/GTP-binding site. This would suggest that the protein could hydrolyze ATP or GTP to exert its function or it could simply be modulated by the binding of nucleotides in a regulatory fashion. We are currently investigating these possibilities. Further characterization of ATHCOR1 will be necessary to understand its function and regulation by MeJA, as well as its possible role in the wounding response and perhaps in disease development of Arabidopsis infected by coronatine-producing strains of P. syringae.
ACKNOWLEDGMENTS
We wish to thank Dr. John G. Turner for donating the coi1 mutant and Dr. Elliot M. Meyerowitz for providing the Arabidopsis cDNA library. We also thank Dr. Adilson Leite and Dr. Ivan Maia for their helpful discussions.
Abbreviations:
- dNTP
deoxyribonucleotide triphosphate
- JA
jasmonic acid
- MeJA
methyl jasmonate
- MS
Murashige-Skoog
- RACE
rapid amplification of cDNA ends
Footnotes
This work was supported by grant no. 95/6662–5 from Fundação de Amparo à Pesquisa do Estado de São Paulo. C.E.B. received a fellowship from Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (no. 300764/95–2) and Fundação de Amparo à Pesquisa do Estado de São Paulo (no. 97/0917–7). C.L.C. received a fellowship from Fundação de Amparo à Pesquisa do Estado de São Paulo (96/10274–3).
LITERATURE CITED
- Bender CL, Stone HE, Sims JJ, Cookey DA. Reduced pathogen fitness of Pseudomonas syringae pv tomato Tn5 mutants defective in coronatine production. Physiol Mol Plant Pathol. 1987;30:273–283. [Google Scholar]
- Benedetti CE, Xie D, Turner JG. COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine and methyl jasmonate. Plant Physiol. 1995;109:567–572. doi: 10.1104/pp.109.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge AJ, Ollis DL. A theoretical study of substrate-induced activation of dienelactone hydrolase. Protein Eng. 1995;8:135–142. doi: 10.1093/protein/8.2.135. [DOI] [PubMed] [Google Scholar]
- Blée E, Schuber F. Biosynthesis of cutin monomers: involvement of a lipoxygenase/peroxygenase pathway. Plant J. 1993;4:113–123. [Google Scholar]
- Coehn Y, Gisi U, Niderman T. Local and systemic protection against Phytophthora infestans induced in potato and tomato plants by jasmonic acid and jasmonic methyl ester. Phytopathology. 1993;83:1054–1062. [Google Scholar]
- Creelman RA, Tierney ML, Mullet JE. Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc Natl Acad Sci USA. 1992;89:4938–4941. doi: 10.1073/pnas.89.11.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damman C, Rojo E, Sánches-Serrano JJ. Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J. 1997;11:773–782. doi: 10.1046/j.1365-313x.1997.11040773.x. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson IB, Mitchell RE. Stimulation of ethylene production in bean leaf discs by the pseudomonad phytotoxin coronatine. Plant Physiol. 1985;77:969–973. doi: 10.1104/pp.77.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara A, Shiraishi K, Sato H, Sakamura K, Nishiyama K, Sakai R, Furusaki A, Matsumoto T. The structure of coronatine. J Am Chem Soc. 1977;99:636–637. [Google Scholar]
- Kawasaki H, Tsuda K, Matsushita I, Tonomura K. Lack of homology between two haloacetate dehalogenase genes encoded on a plasmid from Moraxella sp. strain B. J Gen Microbiol. 1992;138:1317–1323. doi: 10.1099/00221287-138-7-1317. [DOI] [PubMed] [Google Scholar]
- Kenyon JS, Turner JG. Physiological changes in Nicotiana tabacum leaves during development of chlorosis caused by coronatine. Physiol Mol Plant Pathol. 1990;37:463–477. [Google Scholar]
- Kiyosue T, Beetham JK, Pinot F, Hammock BD, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of an Arabidopsis cDNA for a soluble epoxide hydrolase gene that is induced by auxin and water stress. Plant J. 1994;6:259–269. doi: 10.1046/j.1365-313x.1994.6020259.x. [DOI] [PubMed] [Google Scholar]
- Koda Y. The role of jasmonic acid and related compounds in the regulation of plant development. Int Rev Cytol. 1992;135:155–199. doi: 10.1016/s0074-7696(08)62040-9. [DOI] [PubMed] [Google Scholar]
- Kogel KH, Ortel B, Jarosch B, Atzorn R, Schiffer R, Wasternack C. Resistance in barley against the powdery mildew fungus (Erysiphe graminis f. sp. hordei) is not associated with enhanced levels of endogenous jasmonates. Eur J Plant Pathol. 1995;101:319–332. [Google Scholar]
- Kolattukudy PE. Biochemistry and function of cutin and suberin. Can J Bot. 1984;62:2918–2933. [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Mitchell RE. Coronatine production by some phytopathogenic pseudomonads. Physiol Mol Plant Pathol. 1982;20:83–89. [Google Scholar]
- Mitchell RE, Young H. Identification of a chlorosis-inducing toxin of Pseudomonas glycinea as coronatine. Phytochemistry. 1978;17:2028–2029. [Google Scholar]
- Mittal S, Davis KR. Mol Plant-Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- Mueller MJ, Brodschelm W, Spannagl E, Zenk MH. Signaling in the elicitation process is mediated through the octadenoid pathway leading to jasmonic acid. Proc Natl Acad Sci USA. 1993;90:7490–7494. doi: 10.1073/pnas.90.16.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:493–497. [Google Scholar]
- Nishiyama K, Sakai R, Ezuka A, Ichihara A, Shiraishi K, Ogosawara M, Sato H, Mamura S. Phytotoxic effect of coronatine produced by Pseudomonas coronafaciens var. atropurpurea on leaves of Italian ryegrass. Ann Phytopathol Soc Jpn. 1976;42:613–614. [Google Scholar]
- Pathak D, Ashley G, Ollis D. Thiol protease-like active site found in the enzyme dienolactone hydrolase: localization using biochemical, genetic, and structural tools. Proteins. 1991;9:267–279. doi: 10.1002/prot.340090405. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Mollenhauer B, Reinbothe C. JIPs and RIPs: the regulation of plant gene expression by jasmonates in response to environmental cues and pathogens. Plant Cell. 1994;6:1197–1209. doi: 10.1105/tpc.6.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Heintzen C, Seidenbecher C, Parthier B. A methyl jasmonate-induced shift in the length of the 5′ untranslated region impairs translation of the plastid rbcL transcript in barley. EMBO J. 1993;12:1505–1512. doi: 10.1002/j.1460-2075.1993.tb05794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Schweizer P, Buchala A, Silverman P, Seskar M, Raskin I, Métraux J-P. Jasmonate-inducible genes are activated in rice by pathogen attack without a concomitant increase in endogenous jasmonic acid levels. Plant Physiol. 1997;114:79–88. doi: 10.1104/pp.114.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Gees R, Mösinger E. Effects of jasmonic acid on the interaction of barley (Hordeum vulgare L.) with the powdery mildew Erysiphe graminis f. sp. hordei. Plant Physiol. 1993;102:503–511. doi: 10.1104/pp.102.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton A, Beetham JK, Pinot F, Garbarino JE, Friedman M, Hammock BD, Belknap WR. Cloning and expression of soluble epoxide hydrolase from potato. Plant J. 1994;6:251–258. doi: 10.1046/j.1365-313x.1994.6020251.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F. The Pseudomonas phytotoxin coronatine mimics octadenoid signaling molecules of higher plants. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]