Introduction
The phospholipase C-initiated synthesis of the diphosphoinositol polyphosphates (e.g. PP-InsP4, PP-InsP5 and (PP)2-InsP4; Fig. 1) is proposed to be a primordial regulatory pathway that predates the evolution of the much better known Ins(1,4,5)P3-activated Ca2+-mobilization cascade (Seeds et al., 2007). The diphosphoinositol polyphosphates have been proposed to regulate a variety of cellular activities (for reviews see refs (Barker et al., 2009; Burton et al., 2009; Shears 2009)), including apoptosis, vesicle trafficking, cytoskeletal dynamics, exocytosis, telomere maintenance, and adaptations to environmental stress. This work is well-covered by several other recent reviews (Barker et al., 2009; Burton et al., 2009; Shears 2009). The main purpose of this current review is to focus on possible molecular mechanisms by which the diphosphoinositol polyphosphates might act. Traditionally, small diffusible intracellular messengers bind to specific receptors in order to elicit their biological effects. In this review we will note that, with one notable exception, there is little solid information concerning the nature of “receptors” for the diphosphoinositol polyphosphates. Into this void has stepped an unconventional molecular mechanism of action, namely, protein-kinase independent transfer of a “high-energy” phosphate from the diphosphoinositol polyphosphate to a target protein (Saiardi et al., 2004). In this review we shall also discuss the latest information concerning this “transphosphorylation” hypothesis.
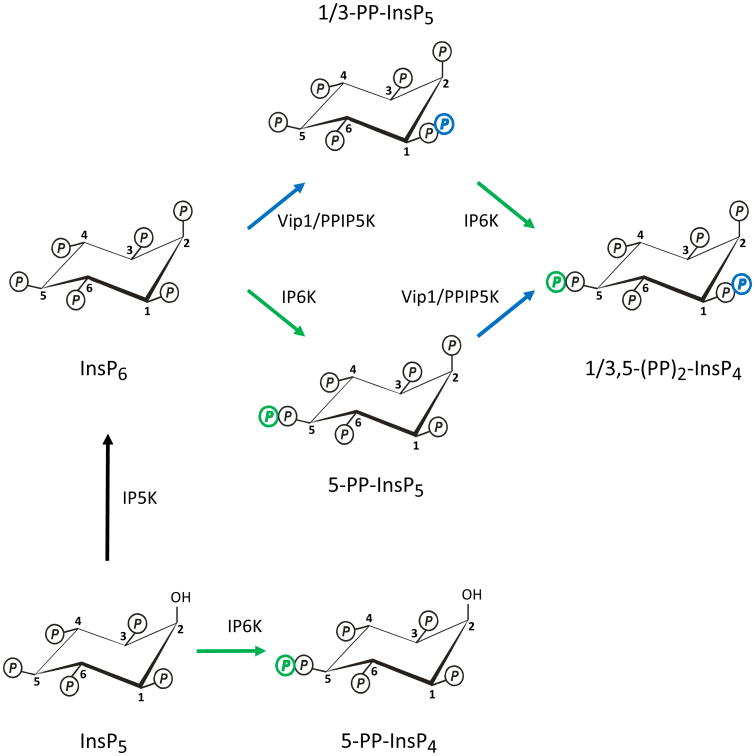
Fig. 1. Synthesis of diphosphoinositol polyphosphates.
These metabolic reactions account for the synthesis of diphosphoinositol polyphosphates in both yeasts and mammalian cells. In the abbreviations of the chemical structures, “Ins” indicates the myo-inositol skeleton. The number of monophosphates around the inositol ring is denoted as a suffix after the ‘P’. The prefixes denote the number of diphosphates (PP). The schematic shows reactions catalyzed by the IP5K (also known as IPK1; black arrow), the IP6Ks (green arrows), and the Vip1/PPIP5Ks (blue arrows). The position of the diphosphate at the 1-position is an arbitrary choice between the two available options, namely, the 1- and 3-positions (see text for details). So as to simplify the figure, the reactions catalyzed by the DIPP phosphatases are not shown.
Metabolism of the diphosphoinositol polyphosphates
A brief overview of the metabolism of this group of molecules should, hopefully, provide a helpful introduction to the newcomer to this field. With the sole exception - so far - of certain Dictyostelids (Laussmann et al., 1998), eukaryotic cells appear to contain just one isomer of (PP)2-InsP4, and it possesses diphosphates at the 1/3 and 5 positions, see (Lin et al., 2009). This 1/3,5-(PP)2-InsP4 is synthesized from either of two isomers of PP-InsP5 (the 1/3- and 5-isomers). PP-InsP5 and (PP)2-InsP4 will frequently be encountered in the literature by their “street-names”: InsP7 and InsP8, respectively. Such fraternization is avoided here in the interests of technical accuracy; eight phosphates can be added to an inositol ring by including a triphosphate group (Draskovic et al., 2008), but that beast is neither a diphosphoinositol (as denoted by the abbreviation “(PP)”), nor an “inositol pyrophosphate”.
Two families of enzymes collaborate to synthesize this group of molecules (Fig. 1): the IP6Ks (E.C. 2.7.4.21, Kcs1 in yeasts; (Saiardi et al., 1999)) and the PPIP5Ks (E.C. 2.7.4.24, Vip1 in yeasts; (Mulugu et al., 2007)). The Kcs1/IP6K family (Saiardi et al., 2001) are responsible for adding an extra phosphate to the 5-position of three different substrates: Ins(1,3,4,5,6)P5, InsP6 and 1/3-PP-InsP5 (Draskovic et al., 2008; Lin et al., 2009). These particular kinases possess a PxxxDxKxG catalytic domain, and a remote, catalytically-essential SSLL (or similar) tetrapeptide. Vip1 (Mulugu et al., 2007) and the two mammalian PPIP5Ks (Choi et al., 2007; Fridy et al., 2007) add a 1/3-diphosphate to both InsP6 and 5-InsP6 (Lin et al., 2009). The uncertainty concerning the precise placement of the 1/3-diphosphate group reflects an analytical impediment created by the axis of symmetry in the inositol ring. That is, 1-PP-InsP5 and 3-PP-InsP5 are an enantiomeric pair, and a stereoselective technique is needed in order to distinguish between them. A recent molecular modeling study tentatively proposed that the Vip1/PPIP5Ks are 3-kinases (Thorsell et al., 2009); on the contrary, some unpublished data that were cited in a recent review (Best et al., 2010) suggest a 1-kinase specificity. The placement of this diphosphate at the 1-position in Fig. 1 is completely arbitrary and should not be taken as indicative of any preference that we might have about which one of the two alternative proposed structures is correct. Unlike the IP6Ks, the PPIP5Ks do not phosphorylate Ins(1,3,4,5,6)P5 (Choi et al., 2007).
The mammalian PPIP5Ks are veritable behemoths of around 120-160 kDa (Choi et al., 2007; Fridy et al., 2007). The PPIP5Ks utilize an ATP-grasp catalytic domain which has been mapped to just the N-terminal one third of these proteins (Mulugu et al., 2007). What, then, is the significance of the other two-thirds of these proteins, including an intriguing, catalytically-inactive phosphatase-like domain? It's a fair bet that the answer(s) to that question will illuminate some important cell-signaling functions of the PPIP5Ks.
A variety of tri- and putative tetra-phosphate products have been shown to be produced in vitro upon the prolonged incubation of either the Kcs1/IP6Ks or the Vip1/PPIP5Ks with their respective substrates (Draskovic et al., 2008; Losito et al., 2009; Saiardi et al., 2000). Some of these products have also been observed when the relevant kinases are over-expressed in yeasts (Draskovic et al., 2008). We have not included these molecules in Fig. 1, out of concern that their detection in intact cells relies upon kinase over-expression, and so we consider that their physiological significance remains to be proven. It has been proposed (Draskovic et al., 2008) that the cellular levels of these particular polyphosphates have been underestimated due to their instability, both in the acidic conditions used to quench cells and also at the low pH typically employed for HPLC analysis. However, we have not found that to be the case with the techniques in routine use in our laboratory (unpublished data).
When the diphosphoinositol polyphosphates were first discovered, attention was drawn to their rapid rates of turnover through kinase/phosphatase substrate cycles (Menniti et al., 1993; Stephens et al., 1993b). Indeed, the family of phosphatases (DIPPs in mammals, E.C. 3.6.1.52, Ddp1 in yeast) that degrade the polyphosphates exhibit values for their specificity constants (kcat/Km) that are sufficiently large as to approach the theoretical limit, as defined by diffusion-controlled encounter between enzyme and substrate (Caffrey et al., 2000; Safrany et al., 1998a). While the rapid turnover of the diphosphoinositol polyphosphates is surely functionally important - because otherwise it would be a wasteful ongoing expenditure of cellular energy - its significance has yet to be explained. Each of the DIPPS can remove a diphosphate group from all of the diphosphoinositol polyphosphates (Caffrey et al., 2000; Hua et al., 2003; Leslie et al., 2002; Safrany et al., 1998a). The Human Genome Organization Gene Nomenclature Committee (HGNC) has allocated the DIPPs to the NUDT gene family (NUDT = “Nudix [Nucleoside Diphosphate attached moiety ‘x’]-Type motif), in honor of their eponymous catalytic site (Gx5Ex5[UA]×REx2EExGU, or similar, in which U is an aliphatic, hydrophobic residue (McLennan 2006)).
It is unclear what are the relative rates of flux in vivo through the two complementary pathways of (PP)2-InsP4 synthesis (see Fig. 1). It is known that steady-state levels of 5 PP-InsP5 greatly exceed those of 1/3-PP-InsP5 (Albert et al., 1997), but that observation by itself is not indicative of relative metabolic fluxes. Some kinetic data obtained from assays with the recombinant kinases (Choi et al., 2007), and some pharmacological data obtained from intact cells (Padmanabhan et al., 2009), together are consistent with 5-PP-InsP5 being the most important intermediate. However, the kinetic data do not take into account the possibility that subcellular compartmentalization of substrates and/or enzymes might occur in vivo. Also, the DIPPs may have hindered earlier attempts to determine which of the synthetic pathways is preferred in vivo (Padmanabhan et al., 2009), especially if the phosphatases show asymmetric activities against the two diphosphate groups in (PP)2-InsP4, as has been proposed (Shears et al., 1995).
Yeast and mammalian cells contain quite low concentrations of PP-InsP5, namely, 0.5 to 5 μM (Barker et al., 2004; Bennett et al., 2006; Fisher et al., 2002; Illies et al., 2007; Ingram et al., 2003). These concentrations for the diphosphoinositol polyphosphates may not sound like much, but they are similar to those seen for other bio-active inositol phosphates, such as Ins(1,4,5)P3 (Streb et al., 1983) and Ins(1,3,4,5)P4 (Huang et al., 2007). The levels of PP-InsP4 and (PP)2-InsP4 are rather lower, each about 10-20% of those of PP-InsP5 (Choi et al., 2005; Choi et al., 2008; Glennon et al., 1993). Thus, it is of some functional significance - as we shall see below - that the metabolic precursors for the diphosphoinositol polyphosphates, namely, InsP5 and InsP6 (Fig. 1), are normally present in cells at concentrations of 15-60 μM (Barker et al., 2004; Ingram et al., 2003; Irvine et al., 2001; Letcher et al., 2008; Pittet et al., 1989; Szwergold et al., 1987). In particular, InsP6 is typically 20-times (or more) abundant than any of the diphosphoinositol polyphosphates.
Signaling by Diphosphoinositol Polyphosphates?
Considering that diphosphoinositol polyphosphates have been implicated in regulating so many diverse biological activities (see the Introduction), it may seem surprising that the title of a recent review of the field asked if these molecules were really signaling entities (Burton et al., 2009). What the authors of that review were acknowledging is that a diffusible, intracellular signal typically changes its intracellular concentrations in response to the activation of a defined signal transduction pathway. However, it is not yet known how the turnover of the diphosphoinositol polyphosphates is regulated by any specific signaling pathway. That lack of information makes it difficult to place into a signaling context any of the reported actions of this group of polyphosphates. Nevertheless, we now know where we should look to find this information: by studying molecular responses to cellular stress.
For example, the levels of diphosphoinositol polyphosphates increase dramatically in Dictyostelium in response to starvation (Luo et al., 2003), although the significance of this adaptation is unknown. Slime molds are generally considered not to be a representative model organism for this field of research: this organism accumulates >100-fold higher levels of the diphosphoinositol polyphosphates than do yeast and mammalian cells, and the isomers are different; the 4/6,5-isomer of (PP)2-InsP4 is unique to Dictysotelium (Laussmann et al., 1996). Hence the current consensus (Burton et al., 2009) that whatever actions these polyphosphates are accomplishing in slime molds, they probably are not applicable to other eukaryotes.
Nevertheless, cellular stress is a recurring theme in studies into changes in the cellular levels of diphosphoinositol polyphosphates. In mammalian cells, (PP)2-InsP4 synthesis is up-regulated in cells subjected to hyperosmotic pressure (Pesesse et al., 2004). The general biological relevance in mammals of cells being exposed to osmotic stress has not always been appreciated. Instead it has been a popular opinion for much of the previous 70 years (Bourque et al., 1997; Darrow et al., 1935) that metazoan cells, with the obvious exception of those of the renal medulla, are largely protected from the potentially deleterious effects of anisosmotic gradients, by virtue of their being bathed in an osmotically stable extracellular fluid. However, it takes as little as 25-50 mOsM hyperosmotic stress to activate (PP)2-InsP4 synthesis in mammalian cells (Pesesse et al., 2004). Recent research has uncovered increases in extracellular osmolarity that can exceed 100 mOsM in the extracellular environment bathing airway epithelial cells, lymphocytes and the various cell-types in bones and cartilage (Alfieri et al., 2007; Go et al., 2004; Knothe Tate 2003). For example, lymphocyte development depends upon their ability to adapt to the hyperosmotic environment of the thymus (Go et al., 2004). Bones also provide an osmotically-stressful environment for cells: there are fluctuating osmotic pressure gradients between osteocytes and their surrounding proteoglycan filled lacunocanalicular system which are important for mechanochemical coupling and for driving bulk fluid flow through the bone tissue (Knothe Tate 2003). Hyperosmotic stress can also occur in airway epithelial cells when the composition of the airway surface liquid layer is compromised by inadequate airway humidification, such as during rapid breathing (e.g. during exercise), breathing of dry/cold air, and in some airway diseases (Song et al., 2001). More generally, even in cell types that do not routinely experience any of these significant fluctuations in extracellular osmolarity, it is now accepted that mechanisms must be in place to adapt to the alterations in intracellular osmolarity (50-100 mOsM) that inevitably accompany normal cellular activities: ion-transport across the plasma membrane, uptake and release of sugars and amino-acids, and polymerization/depolymerization of macromolecules such as glycogen and proteins (Schliess et al., 2002). It is our hypothesis that (PP)2-InsP4 plays some as yet undefined role in the mechanisms by which cells adapt to the inherent dangers of osmotic stress, which include strain upon the cytoskeleton, perturbation of chromatin structure, DNA damage, and inhibition of DNA repair (Chiasson et al., 2003; Kültz et al., 2001).
Thermal stress - both heat and cold - can elevate cellular levels of (PP)2-InsP4 (Choi et al., 2005). External organs, such as skin and testis, must be especially adaptable to substantial variations in environmental temperature. Potentially deleterious or even lethal elevations in core temperature can occur even in euthermic organisms, due to fever, environmental stress and exertional heat illness (Sonna et al., 2002; Sonna et al., 2004). Such conditions can increase the rate of protein degradation, there can be cytoskeletal disruption, and damaging alterations in membrane permeability and ion homeostasis (Dorion et al., 2002; Seno et al., 2004; Sonna et al., 2002). We (Choi et al., 2005) have suggested that diphosphoinositol polyphosphates might mediate adaptive responses to these consequences of thermal stresses.
The molecular mechanisms that maintain energy homeostasis are a fundamental necessity for cell survival (Hardie 2004). Some recent developments ((Bennett et al., 2006; Choi et al., 2008; Lee et al., 2007), for example) have led to the suggestion (Shears 2009) that diphosphoinositol polyphosphates might couple signaling pathways to the energetic status of the cell. This hypothesis envisages that the levels of PP-InsP5and (PP)2-InsP4 in a cell are directly tied to its metabolic health. For example, we have found that (PP)2-InsP4 synthesis is inhibited by an elevation in cellular [AMP] (Choi et al., 2008), which is a sentinel for bioenergetic stress (Hardie 2004). (PP)2-InsP4 suffers the same fate when cells are incubated with 5-aminoimidazole-4-carboxamide ribonucleoside (Choi et al., 2008), which is a pharmacological mimic of bioenergetic stress (Hardie 2004). The molecular mechanisms that are involved have not been established, but it has been determined that AMPK is not involved (Choi et al., 2008).
If the cellular levels of the diphosphoinositol polyphosphates truly reflect - or even regulate - the metabolic health of the cell, this may eventually help us understand how and why an elevated [cAMP] can reduce (PP)2-InsP4 levels (Safrany et al., 1998b). By using several commercially available cAMP analogues as pharmacological tools, we have excluded both PKA (Safrany et al., 1998b) and EPAC (unpublished data) from mediating this effect of the cyclic nucleotide upon (PP)2-InsP4 turnover. So perhaps decreased (PP)2-InsP4 synthesis is a downstream consequence of high levels of cAMP sometimes being registered as a metabolic crisis. That proposal is made because others (Daval et al., 2005; Yin et al., 2003) have shown that, in adipocytes, elevated [cAMP] can activate AMPK, which is normally indicative of a cell undergoing metabolic stress (Hardie 2004). We have found a similar effect of cAMP upon AMPK in DDT1-MF2 cells (unpublished data). We are currently using proteomic techniques to identify sites on the IP6Ks and PPIP5Ks that might undergo covalent modification in response to cellular stress.
Yeasts may use diphosphoinositol polyphosphates to respond to a shortage of certain nutrients. This is a conclusion to emerge from the work of O'Shea and colleagues (Lee et al., 2007), who have shown that the synthesis of total PP-InsP5 levels are up-regulated in yeast cells grown for 1 to 2 hr in low-phosphate media. Genetic evidence indicates that it is the 1/3-isomer of PP-InsP5 that must have been elevated (Lee et al., 2007), although this was not directly confirmed. Again, the mechanism that regulates PP-InsP5 synthesis is unknown.
The changes in levels of diphosphoinositol polyphosphates that are described above are clearly of a global nature, in the sense that they can be observed within a cell population. Additionally, we should consider the possibility that there may be more subtle alterations in metabolic turnover in specific regions of single cells; such effects are difficult to record in the absence of the appropriate molecular probes.
The above discussion presents the case that the functions of diphosphoinositol polyphosphates are relevant to the cell's ability to survive cellular stress. And maybe also its decision not too: consider the response of an ovarian carcinoma cell line to its exposure to cisplatin (Nagata et al., 2005), a platinum-based chemotherapeutic agent that not only cross-links DNA, but also impairs cellular bioenergetic health (Rodriguez-Enriquez et al., 2009). Cisplatin causes the rate of synthesis of 5-PP-InsP5 to initially increase, apparently due to persistent activation of IP6K2 (Nagata et al., 2005). Another study (Morrison et al., 2009) indicated that apoptosis could even be induced by microinjection into a cell of 25 μM 5-PP-InsP5. If an increased concentration of PP-InsP5 is normally a signal of bioenergetic health (as proposed above), is it possible that a stress-associated activation of IP6K2 “misleads” the cell, preventing it from correcting metabolic imbalance? Apoptosis is frequently the ultimate outcome of a sustained failure to adapt to metabolic stress (Jin et al., 2007). Perhaps if a cell experiences a metabolic crisis – and at the same time an IP6K is activated – this “double-whammy” for cellular energy-imbalance is a means by which an apoptotic program is activated.
The search for receptors for diphosphoinositol polyphosphates
How are changes in the levels of a particular diphosphoinositol polyphosphate transduced into a biological response? In early efforts to answer that question, we worked with colleagues to screen for “receptors”, that is, proteins that might bind the polyphosphates with high affinity. We uncovered several candidates that all have in common a role in regulating vesicular traffic: Coatomer, AP-2, and AP180 (previously sometimes called “AP-3”) each bind PP-InsP5 with high affinity (Ali et al., 1995; Fleischer et al., 1994; Shears et al., 1995; Ye et al., 1995). This work is the origin of the proposal that diphosphoinositol polyphosphates might regulate vesicle traffic, and it has prompted subsequent studies in which kcs1Δ yeast was found to exhibit endosomal mis-sorting (Saiardi et al., 2002) and disrupted vacuole biogenesis (Dubois et al., 2002; Saiardi et al., 2000). It was proposed (Saiardi et al., 2002) that this phenotype might reflect disruption of a putative regulatory process that arises from diphosphoinositol polyphosphates competing with inositol lipids for binding to AP-180; such ligand competition was indeed observed in vitro (Hao et al., 1997). However, AP-180 has only a 5-fold lower affinity for InsP6 than PP-InsP5 (Ye et al., 1995); considering the much higher cellular levels of InsP6 (see above), it seems that molecule would overwhelm any potential regulatory ligand-binding role for the diphosphoinositol polyphosphates.
We also have to consider that we may have been somewhat misled by our initial ligand-binding studies (Fleischer et al., 1994; Shears et al., 1995; Ye et al., 1995) being performed in the absence of Mg2+. Since that cation chelates some of the phosphate group's negative charge, the lack of Mg2+ may have led to an over-estimation of the affinity with which PP-InsP5 binds to a protein (Shears 2009). Moreover, a highly-phosphorylated molecule (such as PP-InsP5) can interact with a protein through delocalized and non-specific electrostatic interactions (Lemmon et al., 2002). These can substitute for the more physiologically-relevant and specific ligand interactions that are normally driven by a geometrically-precise arrangement of fewer phosphate groups (Lemmon et al., 2002). Perhaps in this particular series of experiments (Fleischer et al., 1994; Shears et al., 1995; Ye et al., 1995) the detection of PP-InsP5 as a ligand in vitro merely reflected the occurrence of inositol lipid binding in vivo. Indeed, most of the current literature on AP-180 is consistent with the inositol lipids being the biologically-significant ligands, and the binding of the diphosphoinositol polyphosphates is now largely ignored (Legendre-Guillemin et al., 2004).
Snyder and colleagues (Luo et al., 2003) have reported that PP-InsP5 competes with PtdIns(3,4,5)P3 for binding to certain PH domains (Luo et al., 2003), but Downes and colleagues (Downes et al., 2005; Komander et al., 2004) have been unable to reproduce that observation. In any case, in mammalian cells the physiological levels of PP-InsP5 (<5 μM, see above) make it an unlikely competitor for PtdIns(3,4,5)P3, the concentration of which may increase to approx. 200 μM after activation of the PtdIns 3-kinase pathway (Stephens et al., 1993a). Furthermore, the plasma membrane residence of proteins endowed with a PH-domain does not entirely depend upon electrostatic interactions, but also involves hydrophobic interactions between protein and membrane (Manna et al., 2007). The latter phenomenon will reduce the significance of any electrostatic competition between PP-InsP5 and PtdIns(3,4,5)P3 that might occur.
To date, the most promising intracellular “receptor” for any diphosphoinositol polyphosphate is the Pho80/Pho85/Pho81 cyclin-dependent kinase/cyclin kinase inhibitor complex in S. cerevisiae (Lee et al., 2007; Lee et al., 2008). (We should note here that the ligand in question is 1/3-PP-InsP5 (Lin et al., 2009) and not the 4/6-isomer of PP-InsP5 that, initially, it was tentatively suggested to be (Mulugu et al., 2007)). The binding of 1/3-PP-InsP5 to the multimeric cyclin kinase complex is unaffected by a 50-fold excess of InsP6 (Lee et al., 2008), making it an especially specific interaction. It appears that 1/3-PP-InsP5 either binds to a site that is constructed from both Pho81 and Pho80-Pho85, or the polyphosphate may instead induce structural changes that stabilize interactions between Pho81 and Pho80-Pho85 (Lee et al., 2008). The significance of these interactions may lie in the yeast's adaptive responses to phosphate starvation. When inorganic phosphate is limiting, Pho81 inhibits cyclin kinase activity, so that it no longer hyperphosphorylates the transcription factor Pho4 (Kaffman et al., 1994). The latter then becomes competent to enter the nucleus to drive the transcription of genes important for phosphate generation and assimilation, such as a phosphate transporter and a secreted acid phosphatase (Springer et al., 2003). O'Shea and colleagues demonstrated that 1/3-PP-InsP5 augments the inhibitory activity of Pho81 (Lee et al., 2007; Lee et al., 2008). Moreover, the levels of this PP-InsP5 were reported to rise dramatically in response to phosphate starvation (Lee et al., 2007). However, it was not directly confirmed that it was the synthesis of the 1/3-PP-InsP5 that was elevated, rather than 5-PP-InsP5 isomer; that point is significant because 5-PP-InsP5 does not significantly inhibit the Pho complex (Lee et al., 2007). It has further been argued (Shears 2009) that the levels of 1/3-PP-InsP5 in phosphate-starved yeast were over-estimated by O'Shea and colleagues (Lee et al., 2007), which raises the possibility that the levels of the polyphosphate may be insufficient to regulate the Pho complex in vivo, although this concern could be alleviated if there were to be compartmentalization of 1/3-PP-InsP5 synthesis. Perhaps more worryingly, one group (Burton et al., 2009) has indicated that they are unable to reproduce the observation that levels of 1/3-PP-InsP5 actually increase in phosphate-starved S. cerevisiae. Thus, further studies would be useful to validate the biological significance of 1/3-PP-InsP5 as a signal in this context.
Protein Phosphorylation by Diphosphoinositol Polyphosphates?
The diphosphoinositol polyphosphates are clearly molecules that endure severe electrostatic and steric congestion. The relief of these molecular constraints following hydrolysis of their diphosphate groups has long-been viewed as a “high-energy” reaction that ought to have biological significance. Hence, for example, the origin of the idea that diphosphoinositol polyphosphates might phosphorylate proteins (Hand et al., 2007; Laussmann et al., 1996; Stephens et al., 1993b; Voglmaier et al., 1996). Snyder and colleagues have actively pursued this idea. This group (Bhandari et al., 2007; Saiardi et al., 2004) has shown, at least in vitro, that diphosphoinositol polyphosphates can phosphorylate certain proteins. The consensus phosphorylation site is a serine that is surrounded by acidic residues (Saiardi et al., 2004). The appropriate target sequence is especially well-represented in several nucleolar proteins, including Nsr1 (yeast nucleolin), NOPP140 and TCOF1 (Saiardi et al., 2004), although there is as yet no suggestion that diphosphoinositol polyphosphates regulate nucleolar function. One somewhat puzzling aspect of this work is that each of the individual diphosphoinositol polyphosphates have similar abilities to phosphorylate proteins in vitro (Bhandari et al., 2007). Why should the cell invest resources in synthesizing several highly-phosphorylated molecules that all have an identical mechanism of action? Perhaps 5-PP-InsP5 is the only one from this group of molecules that is competent to perform this action in vivo, because it is the most abundant (see above).
The transfer of the phosphate group from the diphosphoinositol polyphosphate to the protein substrate is an especially remarkable phenomenon because it occurs independently of protein kinase activity (Saiardi et al., 2004). It seems that the co ordination of the phosphate's negative charge by Mg2+ is key to enabling the transphosphorylation to occur (Bhandari et al., 2007; Saiardi et al., 2004). There is also a requirement that the target proteins must first be “primed” by an initial casein kinase 2 (CK2)-dependent phosphorylation event (Bennett et al., 2006; Bhandari et al., 2007). In fact, experimental evidence now points to the diphosphoinositol polyphosphates actually further phosphorylating the same serine residue that is initially phosphorylated by CK2 (Bhandari et al., 2007). That is, the protein target becomes diphosphorylated, which is a novel means of covalent modification.
These in vitro data are very impressive and, potentially, they fill a significant gap in our understanding of how diphosphoinositol polyphosphates can regulate cellular function. Furthermore, because all of the diphosphoinositol polyphosphates can transphosphorylate proteins (Bhandari et al., 2007), there is less concern when an apparent biological function of these molecules shows no specificity for one specific diphosphate isomer. Such as was the case when all isomers of PP-InsP5 were found to be equally effective at enhancing insulin secretion from pancreatic beta-cells (Illies et al., 2007). In such an event, specificity may not matter because 5-PP-InsP5 is the only isomer that is present at a high enough concentration to elicit the biological effect. Incidentally, 5-PP-InsP5 appears to act by increasing the size of the readily-releasable pool of insulin granules (Illies et al., 2007), which offers some possible directions for further homing in on the molecular mechanisms involved. Another notable feature of this study is that InsP6 did not imitate the effects of the 5-PP-InsP5 (Illies et al., 2007).
However, it has not yet proved possible to obtain direct and convincing evidence that this transphosphorylation process actually occurs in vivo. Despite recent improvements in the ability of mass spectrometry to measure changes in protein phosphorylation (Steen et al., 2006), it remains challenging to unequivocally identify a serine diphosphate in a peptide fragment obtained from a cell extract. Perhaps in the future it might be possible to develop antibodies against diphospho-serine. In the meantime, Snyder, Saiardi and colleagues (Azevedo et al., 2009; Bhandari et al., 2007; Saiardi et al., 2004) have tried indirect approaches to investigate if protein phosphorylation by the diphosphoinositol polyphosphates is physiologically relevant. For example, in some experiments they used yeast cells in which the InsP6 kinase (Kcs1) that synthesizes 5-PP-InsP5 was genetically eliminated (Saiardi et al., 2004). It was the absence of the phosphate donor activity of the PP-InsP5 that was proposed to account for the lowered degree of phosphorylation of endogenous Nsr1. This is certainly an intriguing observation, but a change in the phosphorylation status of Nsr1 could instead arise independently of PP-InsP5 synthesis per se, and might instead reflect one of the cell's many and complex adjustments that compensate for the kcs1Δ genotype.
It has also been observed that the deletion of the Nsr1 gene in S. cerevisiae caused a doubling of intracellular levels of PP-InsP5 and (PP)2-InsP4 (Saiardi et al., 2004). This increase was proposed to reflect a reduced demand for diphosphoinositol polyphosphate turnover, since one of the proposed targets of phosphorylation was now eliminated (Saiardi et al., 2004). However, this proposal might now be questioned by the expansion of the number of proteins now put forward as substrates for transphosphorylation (Bhandari et al., 2007). If there really are such a large number of protein substrates, removing just one of them would not be expected to significantly impact the cellular levels of diphosphoinositol polyphosphates. Especially if the putative serine-diphosphate is long-lived, as has been proposed (Burton et al., 2009), since this also limits the impact of the phosphorylation process upon the turnover of the phosphate donors.
In the absence of a reliable method for directly assaying diphosphorylation of proteins, we, too, have looked for indirect evidence of its occurrence in vivo. Using the human homologue of Nsr1 - nucleolin - as a model, we (Yang et al., 2008) searched for evidence that its phosphorylation by diphosphoinositol polyphosphates might be physiologically relevant. We made the assumption that, if Snyder and colleagues (Bhandari et al., 2007; Saiardi et al., 2004) are correct, the degree of nucleolin phosphorylation should increase as the cellular levels of (PP)2-InsP4 and/or PP-InsP5 are elevated. We also noted previous experiments demonstrating that the phosphorylation of nucleolin is associated with its transfer from the nucleolus into the nucleoplasm (Kim et al., 2005). Thus, the extent to which nucleolin accumulates in the nucleoplasm can be anticipated to provide a readout of its degree of phosphorylation. We therefore manipulated cellular levels of diphosphoinositol polyphosphates in an osteosarcoma cell line using a combination of hyperosmotic stress, and some pharmacological tricks (Yang et al., 2008). We found that a hyperosmotic challenge caused nucleolin to accumulate in the nucleoplasm -- suggesting its degree of phosphorylation was increased -- but this response occurred independently of changes in levels of diphosphoinositol polyphosphates (Yang et al., 2008).
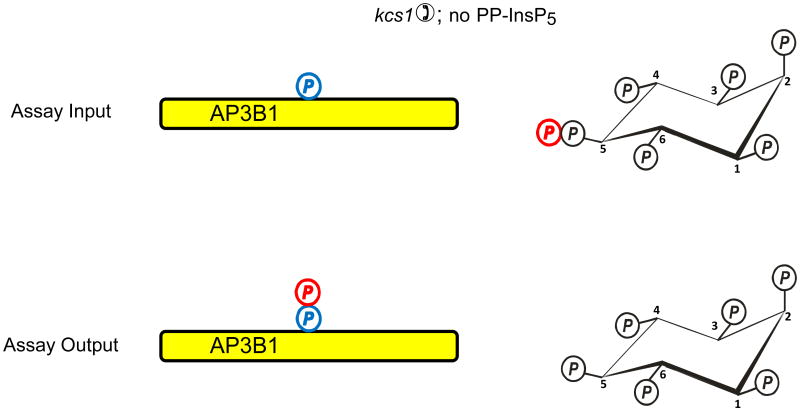
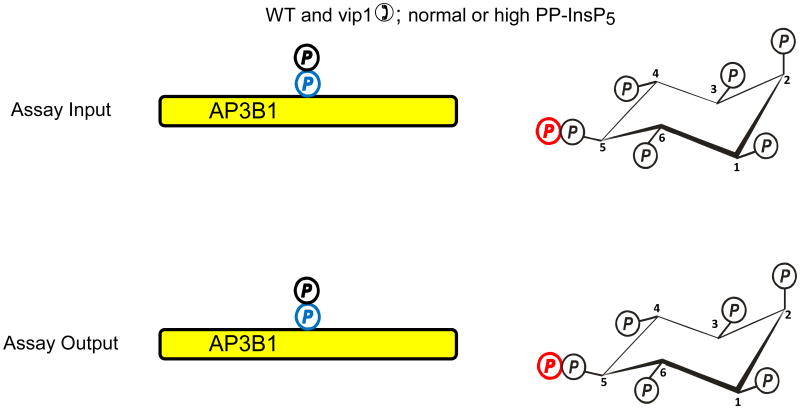
Azevedo et al. (Azevedo et al., 2009) recently used a “back-phosphorylation” assay to study if protein diphosphorylation might occur in vivo. The protein target that was studied was AP3B1, the β-subunit of the AP3 adaptor complex. The basis of this assay is that following the isolation of AP3B1 from intact cells, it would only be diphosphorylated by [32P]PP-InsP5 in vitro if it had not already been diphosphorylated by PP-InsP5 in vivo (Fig. 2a). So, AP3B1 was exogenously expressed in a kcs1Δ strain of S. cerevisiae, in which diphosphoinositol polyphosphates and hence the capacity for transphosphorylation were both virtually eliminated. When AP3B1 was extracted from this strain of yeast and incubated in vitro with [32P]PP-InsP5, there was considerable transphosphorylation of the adaptor (Fig. 2a). AP3B1 was also expressed in either wild-type S. cerevisiae, or in a strain (vip1Δ) that has elevated PP-InsP5 levels. The AP3B1 obtained from these strains exhibited little or no transphosphorylation in vitro. Thus, the authors argued that the adaptor protein must already have been diphosphorylated by cellular PP-InsP5 in vivo (Fig. 2b). However, there is another interpretation of these results that becomes clear once it is recalled that in vitro, diphosphoinositol polyphosphates can only phosphorylate an appropriate Ser residue that is first primed by phosphorylation by casein-kinase II (CK2) (Bhandari et al., 2007). It is certainly the case that the AP3B1 that was isolated from kcs1Δ yeast must have already been mono-phosphorylated by CK2 in vivo, or the transphosphorylation by PP-InsP5 would not have occurred in vitro (Fig. 2a). So, let us suppose that for some reason the expression of kcs1 in intact cells causes AP3B1 not to be monophosphorylated by CK2 in vivo (Fig. 2c). In such a situation, AP3B1 cannot then be transphosphorylated by PP-InsP5 in vitro (Fig. 2c). In fact, genetic interaction studies (Fiedler et al., 2009), which measure the extent to which the function of one gene depends on the presence of a second gene, have found an association between Kcs1 and casein kinase (Cka2) in S cerevisiae. Therefore, it is possible that the back-phosphorylation assay actually could be recording the degree of CK2-mediated monophosphorylation of the appropriate Ser in AP3B1 in vivo (Fig. 2).
Fig. 2. Evidence that diphosphoinositol polyphosphates transphosphorylate proteins in intact cells?
The graphic depicts one particular site on AP3B1 (colored blue) that, in vivo, casein kinase II can mono-phosphorylate, thereby priming it to be transphosphorylated by PP-InsP5 (AP3B1 has other potential phosphorylation sites (Azevedo et al., 2009) that are not illustrated here). Panel a illustrates that Azevedo et al., (Azevedo et al., 2009) heterologously expressed AP3B1 in a kcs1Δ strain of S. cerevisiae. The protein was then extracted and incubated with [32P]PP-InsP5 in vitro (“assay input”). The [32P] (colored red) was transferred from PP-InsP5 to AP3B1 (“assay output”) which can only have occurred if AP3B1 were to have already been mono-phosphorylated by casein kinase II in vivo. Panels b, c depict two alternative explanations for the outcome of separate experiments in which AP3B1 was expressed in either wild-type or vip1Δ S. cerevisiae, which respectively contain either normal or elevated levels of PP-InsP5. AP3B1 was then extracted and incubated with [32P]PP-InsP5 in vitro (“assay input”). Little or no [32P] was transferred from [32P]PP-InsP5 to AP3B1 (“assay output”). There are two explanations for that result. Either (panel b) the AP3B1 was already transphosphorylated (colored black) in vivo, as argued by Azevedo et al., (Azevedo et al., 2009), or (panel c) as we alternately propose, the AP3B1 may not have been monophosphorylated by casein kinase II in vivo. The data do not distinguish between these two possibilities.
InsP6 is quite an effective inhibitor of protein phosphorylation by diphosphoinositol polyphosphates (Saiardi et al., 2004). In eukaryotic cells the cellular levels of InsP6 are typically at least 20-fold higher than the diphosphoinositol polyphosphates, so the latter will likely only be capable of phosphorylating proteins in an a microenvironment from which InsP6 is relatively excluded. This scenario is plausible. There is certainly evidence that some InsP6 is divided into metabolically-separated “pools” (Otto et al., 2007). Other data showing a punctate distribution of the InsP5 2-kinase within certain cellular structures such as nucleoli and stress-granules also indicates that intracellular InsP6 synthesis is compartmentalized (Brehm et al., 2007). Thus, future studies into the possibility that there is compartmentalization of diphosphoinositol polyphosphate synthesis could have a significant impact on the future of the transphosphorylation hypothesis.
It can be anticipated that if diphosphoinositol polyphosphates were indeed to phosphorylate proteins in vivo, then the reverse reaction - dephosphorylation of the protein - might also be a regulated event. Yet, so far, no such phosphatase activity has been observed, and in fact, the diphosphorylated proteins are notably resistant to dephosphorylation when added to cell lysates (Bhandari et al., 2007). This metabolic stability has been argued to be biologically significant by ensuring that signaling through this process is long-lived (Burton et al., 2009). Nevertheless, the identification of the requisite phosphatase, even if it is not very active, is key to bolstering the credentials of this hypothesis.
Summary
In countries where being 18 years old grants you all the benefits of adulthood, 2011 can be the point at which the diphosphoinositol polyphosphates might formally be described as “coming of age”, since these molecules were first fully defined in 1993 (Menniti et al., 1993; Stephens et al., 1993b). But from a biological perspective, these polyphosphates cannot quite be considered to have matured into the status of being independently-acting intracellular signals. This review has discussed several of the published proposals for mechanisms by which the diphosphoinositol polyphosphates might act. We have argued that all of these hypotheses need further development. We also still do not know a single molecular mechanism by which a change in the levels of a particular diphosphoinositol polyphosphate can be controlled. Yet, despite all these gaps in our understanding, there is an enduring anticipation that these molecules have great potential in the signaling field. Reflecting our expectations of all teenagers, it should be our earnest hope that in the near future the diphosphoinositol polyphosphates will finally grow up.
Acknowledgments
Work in the authors' laboratory was supported by the Intramural Research Program of the NIH/National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy KK, Falck JR, et al. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri RR, Petronini PG. Hyperosmotic stress response: comparison with other cellular stresses. Pflugers Arch. 2007;454:173–185. doi: 10.1007/s00424-006-0195-x. [DOI] [PubMed] [Google Scholar]
- Ali N, Duden R, Bembenek ME, Shears SB. The interaction of coatomer with inositol polyphosphates is conserved in Saccharomyces cerevisiae. Biochem J. 1995;310:279–284. doi: 10.1042/bj3100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Natl Acad Sci U S A. 2009;106:21161–21166. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CJ, Illies C, Gaboardi GC, Berggren PO. Inositol pyrophosphates: structure, enzymology and function. Cell Mol Life Sci. 2009;66:3851–3871. doi: 10.1007/s00018-009-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CJ, Wright J, Hughes PJ, Kirk CJ, Michell RH. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell Mol Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best MD, Zhang H, Prestwich GD. Inositol polyphosphates, diphosphoinositol polyphosphates and phosphatidylinositol polyphosphate lipids: Structure, synthesis, and development of probes for studying biological activity. Nat Prod Rep. 2010 doi: 10.1039/b923844c. [DOI] [PubMed] [Google Scholar]
- Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci U S A. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque CW, Oliet SH. Osmoreceptors in the central nervous system. Annu Rev Physiol. 1997;59:601–619. doi: 10.1146/annurev.physiol.59.1.601. [DOI] [PubMed] [Google Scholar]
- Brehm MA, Schenk TM, Zhou X, Fanick W, Lin H, Windhorst S, et al. Intracellular localization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. Biochem J. 2007;408:335–345. doi: 10.1042/BJ20070382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Hu X, Saiardi A. Are Inositol Pyrophosphates Signalling Molecules? J Cell Physiol. 2009;220:8–15. doi: 10.1002/jcp.21763. [DOI] [PubMed] [Google Scholar]
- Caffrey JJ, Safrany ST, Yang X, Shears SB. Discovery of Molecular and Catalytic Diversity Among Human Diphosphoinositol Polyphosphate Phosphohydrolases: An Expanding NUDT Family. J Biol Chem. 2000;275:12730–12736. doi: 10.1074/jbc.275.17.12730. [DOI] [PubMed] [Google Scholar]
- Chiasson JL, Aris-Jilwan N, Belanger R, Bertrand S, Beauregard H, Ekoe JM, et al. Diagnosis and treatment of diabetic ketoacidosis and the hyperglycemic hyperosmolar state. CMAJ. 2003;168:859–866. [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Choi JH, Shears SB. Cellular Energetic Status Supervises the Synthesis of Bis-Diphosphoinositol Tetrakisphosphate Independently of AMP-Activated Protein Kinase. Mol Pharmacol. 2008;74:527–536. doi: 10.1124/mol.107.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Darrow DC, Yannet H. the Changes in the Distribution of Body Water Accompanying Increase and Decrease in Extracellular Electrolyte. J Clin Invest. 1935;14:266–275. doi: 10.1172/JCI100674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daval M, Diot-Dupuy F, Bazin R, Hainault I, Viollet B, Vaulont S, et al. Anti-lipolytic action of AMP-activated protein kinase in rodent adipocytes. J Biol Chem. 2005;280:25250–25257. doi: 10.1074/jbc.M414222200. [DOI] [PubMed] [Google Scholar]
- Dorion S, Landry J. Activation of the mitogen-activated protein kinase pathways by heat shock. Cell Stress Chaperones. 2002;7:200–206. doi: 10.1379/1466-1268(2002)007<0200:aotmap>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes CP, Gray A, Fairservice A, Safrany ST, Batty IH, Fleming I. The regulation of membrane to cytosol partitioning of signalling proteins by phosphoinositides and their soluble headgroups. Biochem Soc Trans. 2005;33:1303–1307. doi: 10.1042/BST0331303. [DOI] [PubMed] [Google Scholar]
- Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, et al. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Dubois E, Scherens B, Vierendeels F, Ho MWY, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, et al. Functional organization of the Scerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DI, Safrany ST, McLennan AG, Cartwright JL. Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J Biol Chem. 2002;277:47313–47317. doi: 10.1074/jbc.M209795200. [DOI] [PubMed] [Google Scholar]
- Fleischer B, Xie J, Mayrleitner M, Shears SB, Palmer DJ, Fleischer S. Golgi coatomer binds, and forms K+-selective channels gated by, inositol polyphosphates. J Biol Chem. 1994;269:17826–17832. [PubMed] [Google Scholar]
- Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J. 1993;293:583–590. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go WY, Liu X, Roti MA, Liu F, Ho SN. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc Natl Acad Sci U S A. 2004;101:10673–10678. doi: 10.1073/pnas.0403139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand CE, Honek JF. Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: a semi-empirical investigation. Bioorg Med Chem Lett. 2007;17:183–188. doi: 10.1016/j.bmcl.2006.09.066. [DOI] [PubMed] [Google Scholar]
- Hao W, Tan Z, Prasad K, Reddy KK, Chen J, Prestwich GD, et al. Regulation of AP-3 function by inositides. Identification of phosphatidylinositol 3,4,5-trisphosphate as a potent ligand. J Biol Chem. 1997;272:6393–6398. doi: 10.1074/jbc.272.10.6393. [DOI] [PubMed] [Google Scholar]
- Hardie DG. The AMP-activated protein kinase pathway--new players upstream and downstream. J Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- Hua LV, Hidaka K, Pesesse X, Barnes LD, Shears SB. Paralogous murine Nudt10 and Nudt11 genes have differential expression patterns but encode identical proteins that are physiologically competent diphosphoinositol polyphosphate phosphohydrolases. Biochem J. 2003;373:81–89. doi: 10.1042/BJ20030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, et al. Positive regulation of Itk PH domain function by soluble IP4. Science. 2007;316:886–889. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, et al. Inositol pyrophosphates determine exocytic capacity. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Ingram SW, Safrany ST, Barnes LD. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene and the effects on growth rate, morphology, and intracellular diadenosine 5′, 5′-P1, P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem J. 2003;369:519–528. doi: 10.1042/BJ20020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF, Schell M. Back in the water: the return of the inositol phosphates. Nature Reviews Molecular Cell Biology. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Jin S, DiPaola RS, Mathew R, White E. Metabolic catastrophe as a means to cancer cell death. J Cell Sci. 2007;120:379–383. doi: 10.1242/jcs.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, Herskowitz I, Tjian R, O'Shea EK. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Kim K, Dimitrova DD, Carta KM, Saxena A, Daras M, Borowiec JA. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation. Mol Cell Biol. 2005;25:2463–2474. doi: 10.1128/MCB.25.6.2463-2474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knothe Tate ML. “Whither flows the fluid in bone?” An osteocyte's perspective. J Biomech. 2003;36:1409–1424. doi: 10.1016/s0021-9290(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter DC, et al. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kültz D, Chakravarty D. Maintenance of genomic integrity in mammalian kidney cells exposed to hyperosmotic stress. Comp Biochem Physiol A Mol Integr Physiol. 2001;130:421–428. doi: 10.1016/s1095-6433(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Laussmann T, Eujen R, Weisshuhn CM, Thiel U, Falck JR, Vogel G. Structures of diphospho-myo-inositol pentakisphosphate and bisdiphospho-myo-inositol tetrakisphosphate from Dictyostelium by NMR analysis. Biochem J. 1996;315:715–725. doi: 10.1042/bj3150715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laussmann T, Hansen A, Reddy KM, Reddy KK, Falck JR, Vogel G. Diphospho-myo-inositol phosphates in Dictyostelium and Polysphondylium: identification of a new bisdiphospho-myo-inositol tetrakisphosphate. FEBS Lett. 1998;426:145–150. doi: 10.1016/s0014-5793(98)00329-9. [DOI] [PubMed] [Google Scholar]
- Lee YS, Huang K, Quiocho FA, O'Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O'Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 2002;513:71–76. doi: 10.1016/s0014-5793(01)03243-4. [DOI] [PubMed] [Google Scholar]
- Leslie NR, McLennan AG, Safrany ST. Cloning and characterization of hAps1 and hAps2, human diadenosine polyphosphate-metabolizing Nudix hydrolases. BMC Biochemistry. 2002;3:20–33. doi: 10.1186/1471-2091-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letcher AJ, Schell MJ, Irvine RF. Do mammals make all their own inositol hexakisphosphate? Biochem J. 2008;416:263–270. doi: 10.1042/BJ20081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, et al. Structural analysis and detection of biological inositol pyrophosphates reveals that the VIP/PPIP5K family are 1/3-kinases. J Biol Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losito O, Szijgyarto Z, Resnick AC, Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE. 2009;4:e5580. doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, et al. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- Manna D, Albanese A, Park WS, Cho W. Mechanistic basis of differential cellular responses of phosphatidylinositol 3,4-bisphosphate- and phosphatidylinositol 3,4,5-trisphosphate-binding pleckstrin homology domains. J Biol Chem. 2007;282:32093–32105. doi: 10.1074/jbc.M703517200. [DOI] [PubMed] [Google Scholar]
- McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, Jr, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, et al. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- Otto JC, Kelly P, Chiou ST, York JD. Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc Natl Acad Sci U S A. 2007;104:15653–15658. doi: 10.1073/pnas.0705729104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesse X, Choi K, Zhang T, Shears SB. Signalling by higher inositolpolyphosphates: Synthesis of bis-diphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J Biol Chem. 2004;279:43378–43381. doi: 10.1074/jbc.C400286200. [DOI] [PubMed] [Google Scholar]
- Pittet D, Schlegel W, Lew DP, Monod A, Mayr GW. Mass changes in inositol tetrakis-and pentakisphosphate isomers induced by chemotactic peptide stimulation in HL-60 cells. J Biol Chem. 1989;264:18489–18493. [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Marin-Hernandez A, Gallardo-Perez JC, Carreno-Fuentes L, Moreno-Sanchez R. Targeting of cancer energy metabolism. Mol Nutr Food Res. 2009;53:29–48. doi: 10.1002/mnfr.200700470. [DOI] [PubMed] [Google Scholar]
- Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, et al. A novel context for the “MutT” module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998a;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany ST, Shears SB. Turnover of bis-diphosphoinositol tetrakisphosphate in a smooth muscle cell line is regulated by b2- adrenergic receptors through a cAMP-mediated, A-kinase-independent mechanism. EMBO J. 1998b;17:1710–1716. doi: 10.1093/emboj/17.6.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A, Bhandari A, Resnick R, Cain A, Snowman AM, Snyder SH. Inositol Pyrophosphate: Physiologic Phosphorylation of Proteins. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The Inositol Hexakisphosphate Kinase Family: Catalytic Flexibility, and Function in Yeast Vacuole Biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Erdjument-Bromage H, Snowman A, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Current Biology. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Nat Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliess F, Haussinger D. The cellular hydration state: a critical determinant for cell death and survival. Biol Chem. 2002;383:577–583. doi: 10.1515/BC.2002.059. [DOI] [PubMed] [Google Scholar]
- Seeds AM, York JD. Inositol polyphosphate kinases: regulators of nuclear function. Biochem Soc Symp. 2007:183–197. doi: 10.1042/BSS0740183. [DOI] [PubMed] [Google Scholar]
- Seno JD, Dynlacht JR. Intracellular redistribution and modification of proteins of the Mre11/Rad50/Nbs1 DNA repair complex following irradiation and heat-shock. J Cell Physiol. 2004;199:157–170. doi: 10.1002/jcp.10475. [DOI] [PubMed] [Google Scholar]
- Shears SB. Diphosphoinositol polyphosphates: metabolic messengers? Mol Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Ali N, Craxton A, Bembenek ME. Synthesis and metabolism of bis-diphosphoinositol tetrakisphosphate in vitro and in vivo. J Biol Chem. 1995;270:10489–10497. doi: 10.1074/jbc.270.18.10489. [DOI] [PubMed] [Google Scholar]
- Song Y, Jayaraman S, Yang B, Matthay MA, Verkman AS. Role of aquaporin water channels in airway fluid transport, humidification, and surface liquid hydration. J Gen Physiol. 2001;117:573–582. doi: 10.1085/jgp.117.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonna LA, Fujita J, Gaffin SL, Lilly CM. Invited review: Effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- Sonna LA, Wenger CB, Flinn S, Sheldon HK, Sawka MN, Lilly CM. Exertional heat injury and gene expression changes: a DNA microarray analysis study. J Appl Physiol. 2004;96:1943–1953. doi: 10.1152/japplphysiol.00886.2003. [DOI] [PubMed] [Google Scholar]
- Springer M, Wykoff DD, Miller N, O'Shea EK. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 2003;1:E28. doi: 10.1371/journal.pbio.0000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- Stephens LR, Jackson TR, Hawkins PT. Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim Biophys Acta. 1993a;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- Stephens LR, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, et al. The detection, purification, structural characterization and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993b;268:4009–4015. [PubMed] [Google Scholar]
- Streb H, Irvine RF, Berridge MJ, Schulz I. Release of Ca2+ from a nonmitochondrial store in pancreatic cells by inositol-1,4,5-trisphosphate. Nature. 1983;306:67–68. doi: 10.1038/306067a0. [DOI] [PubMed] [Google Scholar]
- Szwergold BS, Graham RA, Brown TR. Observation of inositol pentakis- and hexakis-phosphates in mammalian tissues by 31P NMR. Biochem Biophys Res Commun. 1987;149:874–881. doi: 10.1016/0006-291x(87)90489-x. [DOI] [PubMed] [Google Scholar]
- Thorsell AG, Persson C, Graslund S, Hammarstrom M, Busam RD, Hallberg BM. Crystal structure of human diphosphoinositol phosphatase 1. Proteins. 2009;77:242–246. doi: 10.1002/prot.22489. [DOI] [PubMed] [Google Scholar]
- Voglmaier SM, Bembenek ME, Kaplin AI, Dormán G, Olszewski JD, Prestwich GD, et al. Purified inositol hexakisphosphate kinase is an ATP synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc Nat Acad Sci USA. 1996;93:4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Reece JM, Cho J, Bortner CD, Shears SB. The nucleolus exhibits an osmotically regulated gatekeeping activity that controls the spatial dynamics and functions of nucleolin. J Biol Chem. 2008;283:11823–11831. doi: 10.1074/jbc.M800308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high-affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995;270:1564–1568. [PubMed] [Google Scholar]
- Yin W, Mu J, Birnbaum MJ. Role of AMP-activated protein kinase in cyclic AMP-dependent lipolysis In 3T3-L1 adipocytes. J Biol Chem. 2003;278:43074–43080. doi: 10.1074/jbc.M308484200. [DOI] [PubMed] [Google Scholar]