Abstract
The post translational modification of histones has significant effects on overall chromatin function. One such modification is citrullination, which is catalyzed by the protein arginine deiminases (PADs), a unique family of enzymes that catalyzes the hydrolysis of peptidyl-arginine to form peptidyl-citrulline on histones, fibrinogen, and other biologically relevant proteins. Overexpression and/or increased PAD activity is observed in several diseases, including rheumatoid arthritis, Alzheimer’s disease, multiple sclerosis, lupus, Parkinson’s disease, and cancer. This review discusses the important structural and mechanistic characteristics of the PADs, as well as recent investigations into the role of the PADs in increasing disease severity in RA and colitis and the importance of PAD activity in mediating neutrophil extracellular trap (NET) formation through chromatin decondensation. Lastly, efforts to develop PAD inhibitors with excellent potency, selectivity and in vivo efficacy are discussed, highlighting the most promising inhibitors.
Introduction
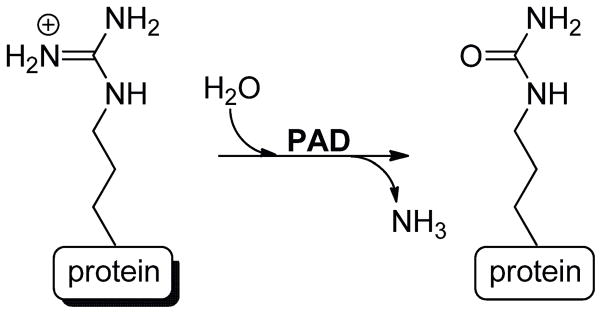
Serving as the scaffold for DNA in eukaryotes, histones are the essential building blocks of chromatin. As such, alterations in histone structure subsequently affects their interaction with DNA (and other nuclear proteins), with consequent effects on overall chromatin function, e.g. gene transcription. Histones undergo a host of post-translational modifications (PTMs), including phosphorylation, methylation, acetylation, ubiquitination, and citrullination.1,2 Citrullination, often referred to as deimination, is the hydrolytic conversion of peptidyl-arginine to peptidyl-citrulline (Figure 1). This modification, which is catalyzed by the protein arginine deiminase (PAD) family of enzymes,3–5 is particularly interesting because PAD overexpression and upregulated enzyme activity has been observed in several diseases, including rheumatoid arthritis (RA), Alzheimer’s disease (AD), multiple sclerosis (MS), lupus, Parkinson’s disease, and cancer.4,6–10
Figure 1.
PADs catalyze the hydrolysis of peptidyl-arginine to peptidyl-citrulline.
In humans, the PAD family is composed of five, calcium dependent isozymes (PADs 1–4 and 6),4 which share roughly 50% sequence similarity.11 PADs are found in a myriad of cell and tissue types, including the epidermis and uterus (PAD1), skeletal muscle, brain, inflammatory cells, several cancer cell lines, and secretory glands (PAD2), hair follicles and keratinocytes (PAD3), granulocytes and several types of cancer (PAD4), and oocytes and embryos (PAD6).3,4 Important to note is that while all of the PADs are found in the cytoplasm of a cell, PAD4 is the only isozyme that has been confirmed to play a role in histone deimination, 3,4 although recent reports suggest that PAD2 may also deiminate histones.12,13 In addition to the cytoplasm and nucleus, evidence is emerging to suggest that PAD isozymes are also present in granules, likely PAD4,14 and mitochondria, likely PAD2.13 Although histone deimination is well characterized, evidence suggests that fibrinogen, fillagrin, and actin are also PAD substrates, and the citrullination of these proteins is known to occur in rheumatoid arthritis.15,16 PAD4, the best characterized isozyme, has also been shown to citrullinate a number of other proteins, including p300,17 ING4,18 RPS2,19 lamin C,20 nucleophosmin,21 and a host of other less well characterized substrates that were recently identified by screening protein arrays.19
Given the focus of previous reviews on the PADs,5,22–24 the scope of this review will be three part, focusing on: (1) the structure and mechanism of the PADs; (2) recent investigations into the role of the PADs in several diseases; and (3) an update on efforts to develop isozyme-specific PAD inhibitors.
PAD Structure and Mechanism
Although high resolution structures of PADs 1–3 and 6 have yet to be reported, significant work has been done to elucidate the structure of PAD4 both with and without calcium bound,11 as well as with PAD4 substrates (i.e., benzoyl-l-arginine amide (BAA),25 histone H3, and histone H4),11,25 and the PAD4 inhibitors F-amidine, Cl-amidine, o-F-amidine, o-Cl-amidine and TDFA.26–28 This work has indicated that PAD4 is divided into distinct N- and C-terminal domains and contains a total of five calcium binding sites. Two of the sites help bridge the N- and C-terminal domains, and the remaining three calcium binding sites are in the N-terminal domain, which is further divided into two immunoglobulin-like subdomains, one of which (i.e., subdomain 1) contains a nuclear localization signal (NLS). Important to note is that residues 158–171 of the N-terminal domain are highly disordered in the calcium free form, but form a highly ordered α-helix in the presence of calcium. It is hypothesized that this change in conformation in response to calcium regulates protein-protein interactions with the enzyme.11 In the C-terminal domain, calcium binding induces large conformational shifts, generating the active site cleft. In fact, Cys645, which acts as the active site nucleophile, is well removed from the active site in the calcium-free form of the enzyme, but moves roughly 5 Å in the calcium bound form, placing it in the proper position for nucleophilic attack on the bound substrate.11 Subsequent crystallographic studies with the aforementioned inhibitors, designed to mimic BAA, confirm that they bind to the active site of PAD4 in a similar manner as BAA, however the haloacetamidine warhead of these inhibitors forms a covalent adduct with Cys645, rendering the enzyme inactive.26,27
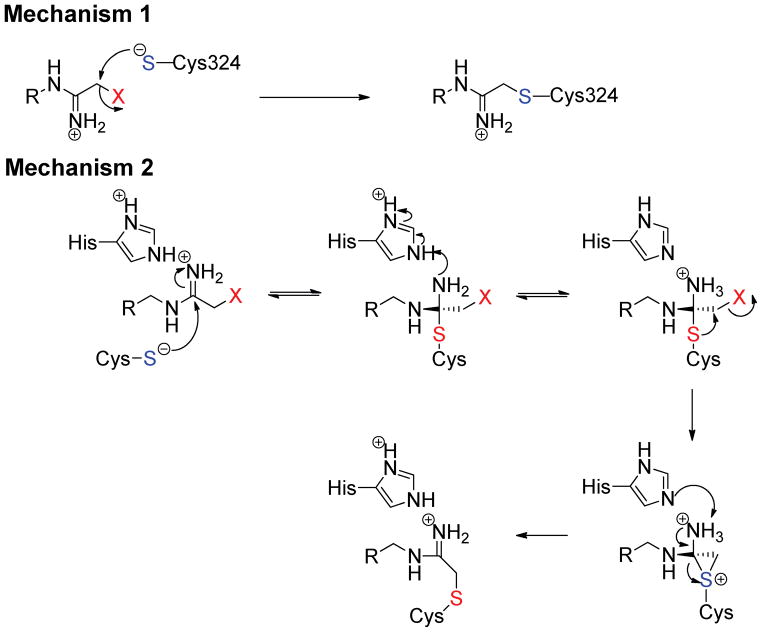
Significant work has been done to elucidate the catalytic mechanism of the PADs.8,27,29–32 This work has revealed that the PADs use a reverse protonation mechanism wherein the active site nucleophile, Cys645, in PAD4 exists as a thiolate in the active form of the enzyme. A second important active site residue, His471, acts as a general acid/base, facilitating the initial release of ammonia from the substrate guanidinium group and subsequently activating a water molecule to complete substrate hydrolysis.8,29,31 While the initial body of work focused on PAD4, mechanistic studies on PADs 1 and 3 have confirmed that they too proceed through a similar reverse protonation mechanism.30 Although modeling QM/MM studies33,34 have suggested that the nucleophilic species is the thiol form of Cys645, it is hard to reconcile this data with the inverse solvent isotope effect observed on kcat/Km with PAD4, as well as PADs 1 and 3.29,31 Based on these mechanistic studies, as well as mechanistic studies with F-amidine, Cl-amidine, 2-fluoracetamidine, and 2-chloroacetamidine, which show bell shaped pH inactivation rate profiles, we recently proposed that inactivation by F-amidine, Cl-amidine, and 2-fluoroacetamidine occurs via the initial attack of the Cys645 thiolate on the amidinium carbon which results in the formation of a stable protonated tetrahedral intermediate that mimics the initial tetrahedral intermediate formed during substrate hydrolysis (Figure 2); His471 is the likely proton donor. The intramolecular halide displacement reaction then proceeds to generate a three-membered sulfonium ring. Deprotonation and collapse of the tetrahedral intermediate leads to a 1,2-shift that generates a thioether adduct,31 the existence of which has been verified crystallographically.26–28 2-chloroacetamidine likely inactivates PAD4 via the direct displacement of the halide (Figure 2).31
Figure 2.
The proposed mechanisms of PAD inactivation by halo-acetamidine based inhibitors.
Important to note is that recent studies have indicated that PAD4 is capable of autodeimination.35,36 Although this modification has been reported to abolish enzyme activity,35 confirmatory studies from our lab have shown that this is not the case.36 However, autodeimination does regulate protein-protein interactions with the enzyme.36 For example, autodeimination of PAD4 diminishes the affinity of the enzyme for histone deacetylase 1 (HDAC1), protein arginine methyltransferase 1 (PRMT1) and citrullinated histone H3 but not p53 or unmodified H3.36
Medical Relevance
A recent review has highlighted in detail the role of the PADs in RA, MS, cancer and other diseases,4 and as such, this review will focus on the most recent insights and advances in understanding the role of the PADs in diseases and how inhibition of these enzymes affects disease severity. Work completed within the past few years has indicated that PADs play crucial roles in the generation of neutrophil extracellular traps (NETs) and the loss of neural regenerative abilities.37–41 Furthermore, the pan-PAD inhibitor Cl-amidine has been shown to reduce disease severity in mouse models of ulcerative colitis and RA.42,43
PAD4 Mediates Bacterially Induced Neutrophil Extracellular Trap (NET) Formation
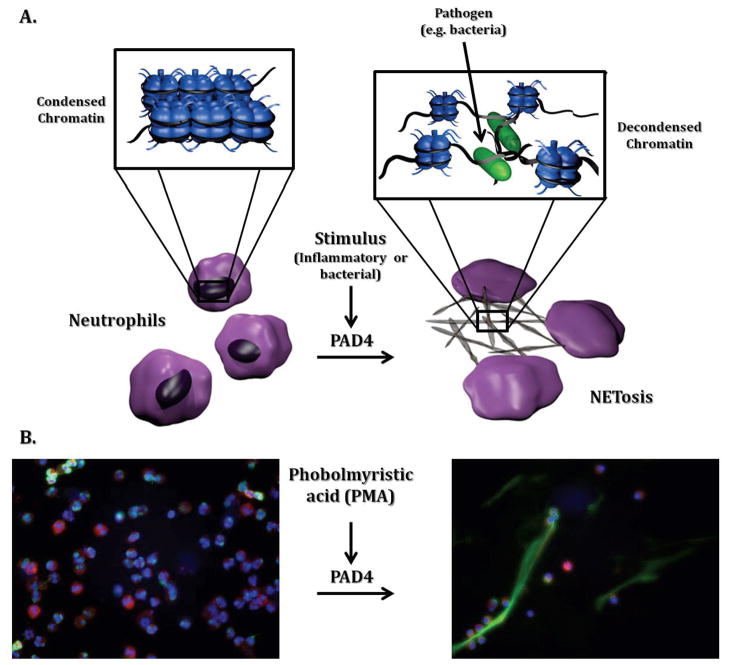
Neutrophils are among the first responders to a bacterial infection and perform several functions in the defense against pathogens, including bacteria phagocytosis, secretion of microbicidal agents, and recruitment of other immune cells.44 In addition, neutrophils can generate large extracellular structures of decondensed chromatin termed neutrophil extracellular traps (NETs) (Figure 3).45 NETs work by trapping bacteria, disarming them with specific proteases, and subsequently killing them with the help of NET immobilized histones, which have inherent antimicrobial properties.44,46 Although NETs were discovered in 2004,44 it was only more recently shown that PAD4 has a regulatory role in NET formation by mediating chromatin decondensation through histone citrullination.38,40,41
Figure 3.
In neutrophils, inflammatory stimuli (e.g., LPS, PMA, and/or chemokines) result in increased PAD activity, histone citrullination, chromatin decondensation, and NET formation. (A.) Schematic depiction of a neutrophil undergoing NET formation. (B.) Representative images of control neutrophils isolated from peripheral blood and analyzed at baseline or after stimulation for 2 h with PMA. Cells were stained with Myeloperoxidase (red), elastase (green) or Hoechst 33342 to detect DNA (blue). Upon PMA stimulation, clear NET formation is detected. Original magnification ×40. We thank Mariana Kaplan for kindly providing the images of neutrophils undergoing NET formation and Heather L. Rust for the preparation of the schematic depiction of neutrophils undergoing NET formation.
Citrullinated histone H3 was first detected within NET structures by confocal microscopy using citrullinated H3 specific antibodies; neutrophils were stimulated with lipopolysaccharide (LPS) to produce NETs.40 Subsequently, H3 citrullination was shown to occur in response to a suite of bacterial and inflammatory signaling molecules, such as LPS, tumor necrosis factor (TNF), N-formyl-methionine-leucine-phenylalanine (f-MLP), lipoteichoic acid (LTA), and hydrogen peroxide.40 Although unknown at the time, it was believed that histone citrullination in stimulated neutrophils may have several roles, including increased histone bactericidal properties, immune system signaling, and chromatin decondensation to aid NET formation.40 Further studies confirmed that PAD4 catalyzed histone citrullination is crucial for chromatin decondensation and subsequent NET formation.41 Stimulation of HL-60 granulocytes with a calcium ionophore (A23187), or other agents that are more relevant to the situation in vivo, i.e. IL-8 and Shigella felxneri, results in increased histone citrullination and NET formation. However, incubation of the granulocytes with Cl-amidine prior to either stimulus leads to a significant reduction in histone citrullination and eliminated the generation of NETs, indicating that PAD4 activity is important for NET formation; PAD4 is the primary PAD isozyme expressed in neutrophils. Further supporting a role for PAD4 in this process is the fact that administration of wild type PAD4 protein, but not an inactive PAD4C645S mutant, to permeablilized granulocytes resulted in chromatin decondensation.41 Other studies confirm that the pro-inflammatory signaling molecules LPS, PMA, and hydrogen peroxide stimulated NET formation in PAD4+/+ neutrophils versus PAD4−/− neutrophils and that administration of Cl-amidine decreases NET formation to near control levels.38 In necrotizing faciitis caused by Streptococcus pyogenes M1 GAS, the DNase Sda1 provides a means for degrading NETs and promoting bacterial growth. In a mouse model of necrotizing faciitis, knockout of this DNase (M1 ΔSda1 GAS) significantly reduced lesion size in PAD4+/+ mice but not in PAD4−/− mice.38 These data taken together confirm the important role of PAD4 in mediating NET formation in response to inflammatory stimuli through histone citrullination. Given that increased NET formation is a hallmark of several diseases, e.g. lupus and ulcerative colitis,47,48 the fact that PAD4 activity is required for NET formation indicates that the therapeutic potential of a PAD4 inhibitor is likely to be quite broad.
PADs Regulate the Response to Spinal Cord Injury
Spinal cord regeneration is a unique ability lost during embryonic development. The factors regulating the response to spinal cord injury are still poorly understood, however, the increased concentration of calcium present after injury, led Ferretti and colleagues to determine that PAD activity is activated during spinal cord injury in a chick embryo model.37 Importantly, they showed that the expression of PAD3 was elevated at the stage of embryonic development where regenerative abilities have been lost (embryonic day 15; (E15) versus E11). Furthermore, staining for citrullination and apoptosis (F95 antibody and TUNEL, respectively) indicated that citrullination and apoptosis/tissue loss coincided in spinal cord injury at E15 but not E11. The administration of Cl-amidine significantly reduced cavitation, apoptosis, and tissue loss at the site of injury when given at the time of injury or 2 h after. These data indicate that chicken PAD3 is an important regulator of the response to spinal cord injury. Whether these findings translate into humans remains an open question. However, it is tempting to speculate that PAD inhibitors may represent an avenue for reducing the severity of spinal cord injury.37
Cl-Amidine Reduces Disease Severity in Mouse Models of Rheumatoid Arthritis and Colitis
Rheumatoid Arthritis (RA), an autoimmune disorder, affects roughly 1% of the population, necessitating investigations into avenues for treating the causes and symptoms of RA.4 Increased levels of citrullinated proteins are common in RA patients, as are anti-citrullinated protein antibodies (ACPAs), and these markers are a hallmark of increased disease severity and joint damage.49,50 At the genetic level, a direct link between PAD4 mutations and RA development has only been confirmed in Asian populations,51 however, such a link is also hypothesized in other populations.4 More recently it was demonstrated that the administration of Cl-amidine, a potent pan-PAD inhibitor, to mice with collagen-induced arthritis (CIA) reduces disease severity, joint inflammation, and joint damage in a dose-dependent manner.43 Although Cl-amidine significantly reduced citrullination in synovial fluid and serum and reduced histological disease activity scores, it did not completely eliminate CIA. It is hypothesized that Cl-amidine does not work by preventing the onset of CIA, but rather reduces severity after disease onset by preventing protein citrullination and subsequent ACPA generation.43 Consistent with this possibility is the fact that Cl-amidine did not alter disease severity in the arthrogen model, which is thought to mimic the effector phase of the disease.
Cl-amidine has also proven useful in reducing disease severity in a mouse model of ulcerative colitis,42 another disease where expression of certain PADI4 haplotypes increases disease susceptibility and risk in specific populations.52 In these studies, 2% dextran sodium sulfate (DSS) was used to induce colitis in mice, resulting in increased citrullination of colon proteins. After one week on DSS, the administration of Cl-amidine (5, 25, and 75 mg/kg) by either intraperitoneal injection (IP) or oral gavage significantly increased colon length, increased mouse mobility and activity, and led to an overall reduction in disease severity compared to untreated mice.42 Furthermore, Cl-amidine increases p53 production in inflammatory cells in vitro and in vivo, increases inflammatory cell apoptosis in vitro and in vivo, and protects colonic epithelial cells from DNA damage in vivo, thereby supporting the hypothesis that Cl-amidine reduces ulcerative colitis disease severity by reducing inflammation.42 Important to note is that Cl-amidine showed no cytotoxic effects in either the RA or colitis studies mentioned above, and did not act as a general immunosuppressant, thereby indicating that PAD inhibitors have a unique mode of action.42,43
Development of New PAD Inhibitors
Although we and others have identified several reversible PAD inhibitors (e.g., taxol, minocycline, and streptomycin),31,53,54 these compounds are relatively weak PAD inhibitors, and the most potent inhibitors described to date irreversibly modify the enzymes.26–28,55,56 The lack of potency for the reversible inhibitors identified thus far likely relates to the small active site cavity that only accommodates the side chain of an arginine residue when the enzyme is bound to calcium. Given the lack of potency for reversible inhibitors, most of our efforts have focused on developing Cl-amidine analogs with improved potency, selectivity, and bioavailability.26,28,31 Structure-activity relationships between the active PAD isozymes (PADs 1, 2, 3, and 4) and F- and Cl-amidine identified the second generation PAD inhibitors o-F-amidine and o-Cl-amidine (Figure 4), which incorporate a carboxylate moiety at the ortho position on the benzoyl ring, and possess improved potencies and selectivities. For example, o-F-amidine is 65-fold more potent than F-amidine and preferentially inhibits PAD1 by at least 6-fold.28 Library approaches have also identified several novel PAD inhibitors.26,31 For example, a relatively small, 264 compound library was synthesized on the solid phase and then screened to identify PAD inhibitors. This approach identified a tripeptide (Thr-Asp-F-amidine; TDFA) as a highly selective (up to 65-fold) PAD4 inhibitor with excellent in vivo potency.26 Given the irreversible nature of the inhibition, TDFA was rapidly converted into a PAD4 specific Activity Based Proteomic Profiling (ABPP) tool by appending a biotin moiety onto the N-terminus of this tripeptide. Wang and colleagues recently reported a Cl-amidine analog, YW3-56. This compound shows improved bioavailability, likely due to the replacement of the Nα-benzoyl and Cα-amide groups with the more hydrophobic dimethylamide-naphthalene and benzylamide groups, respectively.56
Figure 4.
Structure, potency, and selectivity of the most useful PAD inhibitors.
We have also recently described the development of a fluorescence polarization ABPP (fluopolABPP) based high-throughput screen that was used to screen 2000 compounds from the NIH Validation Set. Streptonigrin was the most potent compound identified from the screen and it showed improved potency and selectivity for PAD4 in vitro and improved in vivo potency in HL-60 granulocytes and MCF7 cells versus Cl-amidine.31 Interestingly, streptonigrin is a known anti-cancer agent, suggesting that its efficacy as an anti-neoplastic agent is due in part to its ability to inhibit PAD4 activity. Although it should be noted that this compound is also thought to inhibit tumor cell growth via its ability to inhibit DNA replication, interfere with cellular respiration, and disrupt cell replication.57,58
Conclusionsand Future Perspectives
Although it is now clear that the PADs play a role in a host of inflammatory disorders, their specific roles in both normal and pathological processes are not known. As such, it is difficult to rationalize how PAD inhibitors work to ameliorate disease severity in such a broad spectrum of diseases. For example, PAD4 has been shown to generate citrullinated proteins, and subsequently ACPAs, in both humans and mouse models of RA,43,49,50 suggesting that Cl-amidine or another PAD inhibitor may work by reducing the numbers and types of citrullinated proteins, and, as consequence, inhibit ACPA generation. While this explanation may hold true, in whole or in part, for RA, it cannot explain Cl-amidine efficacy in other disease models where ACPAs are not produced (e.g. colitis and neural regeneration). In at least a subset of these and other diseases PAD inhibitors may exert their effects via their ability to inhibit NET formation.38,41 Since dysregulated NET activity is associated with both ulcerative colitis and lupus, and likely additional immune disorders, PAD inhibition may also represent a method to treat diseases linked to abnormal NET formation.
Alternatively, PAD inhibitors may exert their beneficial effects by promoting cell cycle arrest and/or apoptosis of activated inflammatory cells, which appears to occur in the DSS model of colitis.42 Although this effect is likely mediated in part by effects on p53 (Cl-amidine increases p53 expression in CD45 positive immune cells), PAD inhibitors may also activate the expression of additional genes required for cell cycle arrest or apoptosis. This hypothesis seems reasonable considering that Cl-amidine (and F-amidine) triggers the differentiation and apoptosis of multiple cancer cell lines that are p53+/+ and p53−/− (e.g., HL60, HT29, TK6, and U2-OS cells).42,59–61 Given these findings, coupled to the fact that multiple PADs are overexpressed in several types of cancer, it is tempting to speculate that PAD inhibitors may also show therapeutic utility as anti-neoplastic agents. Consistent with this possibility Cl-amidine and YW3-56 decrease the growth of tumor xenografts (unpublished data and ref. 56). As described above, efficacy may be due in part to effects on p53-dependent gene transcription as has been observed in Cl-amidine treated U2-OS osteosarcoma cells, where Cl-amidine induced the expression of p53 and several downstream target genes including the cyclin dependent kinase inhibitor p21, GADD45, and the proapoptotic protein PUMA.61 PAD inhibition may also prevent cell growth by downregulating c-Fos expression because Coonrod and colleagues have recently demonstrated that PAD4 deiminates Elk-1 and this PTM promotes its phosphorylation, histone H4 acetylation, and c-Fos transcription. In addition to these effects, PAD inhibition may also modulate protein kinase signaling by modifying kinase consensus sequences, as discussed in a recent review.62
Finally, while our understanding of PAD biology is increasing, especially since our introduction of the first generation PAD inhibitors F- and Cl-amidine, much remains to be learned and this will require new chemical tools, including isozyme selective inhibitors, activity-based protein profiling methods, and molecular tools for studying citrullination. The development of PAD-selective inhibitors is especially important because it is unclear which PAD(s) is important for anyone of the aforementioned diseases, and mouse knockouts may not faithfully depict the roles of individual isozymes in a disease because of a failure in central tolerance due to the compensatory effects of other isozymes. Additionally, new inhibitors are needed that overcome some of the drawbacks of current PAD inhibitors (e.g., poor selectivity, bioavailability, relatively low potency, and relatively promiscuous reactivity for the chloro-acetamidine bearing compounds). The demonstration that the related enzyme dimethylarginine dimethylaminohydrolase can be inactivated by 4-halopyridines63 suggests that the development of irreversible inhibitors with novel warheads that show reduced reactivity is possible and likely to be a successful avenue of future research. Finally, the development of chemical tools for the PADs will inevitably provide invaluable insight into both the normal roles of the PADs and their roles in a host of diseases. Subsequently, as our knowledge of disease related PAD activity expands, so will our need for better enzyme inhibitors and molecular tools, generating an exciting and rewarding field of research.
References
- 1.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Zee BM, Levin RS, DiMaggio PA, Garcia BA. Epigenetics & Chromatin. 2010;3:1–11. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vossenaar ER, Zendman AJW, van Venrooij WJ, Pruijn GJM. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 4.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discovery Dev. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson PR, Fast W. ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 6.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. BMC Cancer. 2009;9:40. doi: 10.1186/1471-2407-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, Murayama S, Asaga H, Toda T, Kumura N, Maruyama N. J Neurosci Res. 2005;80:120–128. doi: 10.1002/jnr.20431. [DOI] [PubMed] [Google Scholar]
- 8.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 9.Moscarello MA, Mastronardi FG, Wood DD. Neurochem Res. 2007;32:251–256. doi: 10.1007/s11064-006-9144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schellekens GA, Jong BAd, Hoogen FHvd, Putte LBvd, Venrooij WJv. J Clin Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Nat Struct Mol Bio. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 12.Cherrington BD, Morency E, Struble AM, Coonrod SA, Wakshlag JJ. PLoS One. 2010;5:e11768. doi: 10.1371/journal.pone.0011768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang B, Shin HY, Choi JK, Nguyen du PT, Jeong BH, Ishigami A, Maruyama N, Carp RI, Kim YS, Choi EK. J Neuropathol Exp Neurol. 2011;70:116–124. doi: 10.1097/NEN.0b013e318207559e. [DOI] [PubMed] [Google Scholar]
- 14.Asaga H, Nakashima K, Senshu T, Ishigami A, Yamada M. J Leukoc Biol. 2001;70:46–51. [PubMed] [Google Scholar]
- 15.Darrah E, Rosen A, Giles JT, Andrade F. Ann Rheum Dis. 2012;71:92–98. doi: 10.1136/ard.2011.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, Serre G. J Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 17.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Proc Natl Acad Sci U S A. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Q, Fast W. J Biol Chem. 2011;286:17069–17078. doi: 10.1074/jbc.M111.230961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Q, Bedford MT, Fast W. Mol Biosyst. 2011;7:2286–2295. doi: 10.1039/c1mb05089c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanikawa C, Espinosa M, Suzuki A, Masuda K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y, Matsuda K. Nat Commun. 2012;3:676. doi: 10.1038/ncomms1676. [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Biochem Biophys Res Comm. 2002;290:979–983. doi: 10.1006/bbrc.2001.6303. [DOI] [PubMed] [Google Scholar]
- 22.Vossenaar ER, Van Venrooij WJ. Arthritis Res Ther. 2004;6:107–111. doi: 10.1186/ar1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 24.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discov Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 25.Arita K, Shimizu T, Hashimoto H, Hidaka Y, Yamada M, Sato M. Proceedings of the National Academy of Sciences. 2006;103:5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones JE, Slack JL, Fang P, Zhang X, Subramanian V, Causey CP, Coonrod SA, Guo M, Thompson PR. ACS Chemical Biology. 2011;7:160–165. doi: 10.1021/cb200258q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Sato M, Thompson PR. Biochemistry. 2006;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Causey CP, Jones JE, Slack JL, Kamei D, Jones LE, Subramanian V, Knuckley B, Ebrahimi P, Chumanevich AA, Luo Y, Hashimoto H, Sato M, Hofseth LJ, Thompson PR. Journal of Medicinal Chemistry. 2011;54:6919–6935. doi: 10.1021/jm2008985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuckley B, Bhatia M, Thompson PR. Biochemistry. 2007;46:6578–6587. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. Chem Bio Chem. 2010;11:161–165. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Knuckley B, Bhatia M, Pellechia PJ, Thompson PR. Journal of the American Chemical Society. 2006;128:14468–14469. doi: 10.1021/ja0656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ke Z, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. J Phys Chem B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X. Arthritis & Rheumatism. 2010;62:1630–1640. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slack JL, Jones LE, Bhatia MM, Thompson PR. Biochemistry. 2011;50:3997–4010. doi: 10.1021/bi200309e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lange S, Gögel S, Leung KY, Vernay B, Nicholas AP, Causey CP, Thompson PR, Greene NDE, Ferretti P. Developmental Biology. 2011;355:205–214. doi: 10.1016/j.ydbio.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. The Journal of Experimental Medicine. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohlake P, Whiteley C. Molecular Neurobiology. 2010;41:149–158. doi: 10.1007/s12035-010-8112-x. [DOI] [PubMed] [Google Scholar]
- 40.Neeli I, Khan SN, Radic M. The Journal of Immunology. 2008;180:1895–1902. doi: 10.4049/jimmunol.180.3.1895. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, Allis CD, Coonrod SA. The Journal of Cell Biology. 2009;184:205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2011;300:G929–G938. doi: 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willis VC, Gizinski AM, Banda NK, Causey CP, Knuckley B, Cordova KN, Luo Y, Levitt B, Glogowska M, Chandra P, Kulik L, Robinson WH, Arend WP, Thompson PR, Holers VM. The Journal of Immunology. 2011;186:4396–4404. doi: 10.4049/jimmunol.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan MJ. Nat Rev Rheumatol. 2011;7:691–699. doi: 10.1038/nrrheum.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemmers S, Teijaro JR, Arandjelovic S, Mowen KA. PLoS ONE. 2011;6:e22043. doi: 10.1371/journal.pone.0022043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leffler J, Martin M, Gullstrand B, Tydén H, Lood C, Truedsson L, Bengtsson AA, Blom AM. The Journal of Immunology. 2012;188:3522–3531. doi: 10.4049/jimmunol.1102404. [DOI] [PubMed] [Google Scholar]
- 48.Savchenko AS, Inoue A, Ohashi R, Jiang S, Hasegawa G, Tanaka T, Hamakubo T, Kodama T, Aoyagi Y, Ushiki T, Naito M. Pathology International. 2011;61:290–297. doi: 10.1111/j.1440-1827.2011.02651.x. [DOI] [PubMed] [Google Scholar]
- 49.Lundberg K, Nijenhuis S, Vossenaar ER, Palmblad K, Venrooij WJv, Klareskog L, Zendman AJW, Harris HE. Arthritis Res Ther. 2005;7:R458–R467. doi: 10.1186/ar1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJW, Saxne T, Malmstr V, Venables PJ. Arthritis & Rheumatism. 2008;58:2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki m, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 52.Chen CC, Isomoto H, Narumi Y, Sato K, Oishi Y, Kobayashi T, Yanagihara K, Mizuta Y, Kohno S, Tsukamoto K. Clinical Immunology. 2008;126:165–171. doi: 10.1016/j.clim.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 53.Knuckley B, Luo Y, Thompson PR. Bioorganic & Medicinal Chemistry. 2008;16:739–745. doi: 10.1016/j.bmc.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pritzker LB, Moscarello MA. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1998;1388:154–160. doi: 10.1016/s0167-4838(98)00175-7. [DOI] [PubMed] [Google Scholar]
- 55.Stone EM, Schaller TH, Bianchi H, Person MD, Fast W. Biochemistry. 2005;44:13744–13752. doi: 10.1021/bi051341y. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, Fisher M, Oshaben K, Zhao N, Gu Y, Wang D, Chen G. J Biol Chem. 2012 doi: 10.1074/jbc.M112.375725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolzan AD, Bianchi MS. Mutat Res. 2001;488:25–37. doi: 10.1016/s1383-5742(00)00062-4. [DOI] [PubMed] [Google Scholar]
- 58.Knipp M, Vasak M. Anal Biochem. 2000;286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 59.Slack J, Causey C, Thompson P. Cellular and Molecular Life Sciences. 2011;68:709–720. doi: 10.1007/s00018-010-0480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. J Biol Chem. 2008;283:20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, Gilmour DS, Wang Y. Molecular and Cellular Biology. 2008;28:4745–4758. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rust HL, Thompson PR. ACS Chemical Biology. 2011;6:881–892. doi: 10.1021/cb200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson CM, Monzingo AF, Ke Z, Yoon DW, Linsky TW, Guo H, Robertus JD, Fast W. Journal of the American Chemical Society. 2011;133:10951–10959. doi: 10.1021/ja2033684. [DOI] [PMC free article] [PubMed] [Google Scholar]