Abstract
Proper splicing of pre-mRNA is required for protein synthesis and therefore is a fundamental cellular function. The discovery of a variety of somatic spliceosomal mutations in hematologic malignancies, including myeloid neoplasms and chronic lymphocytic leukemia has pointed to a new leukemogenic pathway involving spliceosomal dysfunction. Theoretically, spliceosomal mutations can lead to activation of incorrect splice sites, intron retention or aberrant alternative splicing occurring in patterns generated by mutations of individual spliceosomal proteins. Such events can produce a defective balance between protein isoforms leading to functional consequences including defective regulation of proliferation and differentiation. The observed pattern of occurrence of highly specific missense mutations coupled with the lack of nonsense mutations and deletions, implies a gain-of-function or better gain-of-dysfunction mechanism. Incorrect splicing of downstream genes such as tumor suppressor genes may result in haploinsufficient expression through nonsense mediated mRNA decay. Thus sliceosomal mutations may, depending on the pattern of affected proteins, lead to similar functional effects on tumor suppressor genes as chromosomal deletions, epigenetic silencing or inactivating/hypomorphic mutations. The prognostic value of the most common mutations and their phenotypic association in the clinical setting is currently being investigated. It is likely that spliceosomal mutations may indicate sensitivity to spliceosome inhibitors applied in the form of a synthetic lethal approach. This manuscript discusses the most current aspects of spliceosomal research in the context of hematologic malignancies.
Molecular pathogenesis of myeloid malignancies: novel somatic mutations
Application of array-based technologies has led to discovery new chromosomal lesions, including micro-deletions, -duplications and somatic uniparental disomy (UPD) associated with myeloid malignancies. New tumor-specific somatic mutations were discovered through rational searches based on more precise delineation of minimally affected chromosomal regions and, more recently, by application of unbiased high-throughput sequencing approaches. Extensions of these technologies led also to a better appreciation of epigenetic changes in the leukemic genome, including cytosine methylation, hydroxymethylation and carboxylation. Newly discovered mutations can be sub-classified into categories such as those contributing to epigenetic instability, mutations in receptor kinases or those involved in apoptotic or differentiation pathways. The recently discovered mutations in spliceosomal protein genes constitute of a novel class of genomic lesions and define an entirely new pathogenic pathway of leukemogenesis.
Spliceosomal function or dysfunction
Spliceosomes and gene expression
During the process of gene expression in eukaryotic cells, pre-mRNA, which contains both introns and exons, undergoes splicing of introns and ligation of exons, a fundamental process needed to form mature mRNA transcripts (Wahl MC. 2009; Ward AJ and Cooper TA et al, 2010; Fig. 1). Recent evidence suggests that most human genes are spliced in two or more patterns to produce mRNAs encoding proteins with altered sequence in a process known as alternative splicing. These alternative splicing events are cell type specific and highly regulated. Spliceosomes are intracellular protein/RNA complexes that catalyze the splicing reaction. The structure of spliceosomes is complex: they contain over 150 distinct proteins and 5 small nuclear (sn) RNAs. As splicing proceeds, the formation of the active spliceosome involves an ordered, stepwise assembly of discrete particles on the pre-mRNA substrate. Early steps involve recognition of the 5’ and 3’ exon/intron junctions through a number of protein-RNA and RNA-RNA interactions. Later steps bring the two complexes together and construct the active spliceosomal complex. The early steps appear to be the site of most alternative splicing regulation (Wang ET et al, 2008).
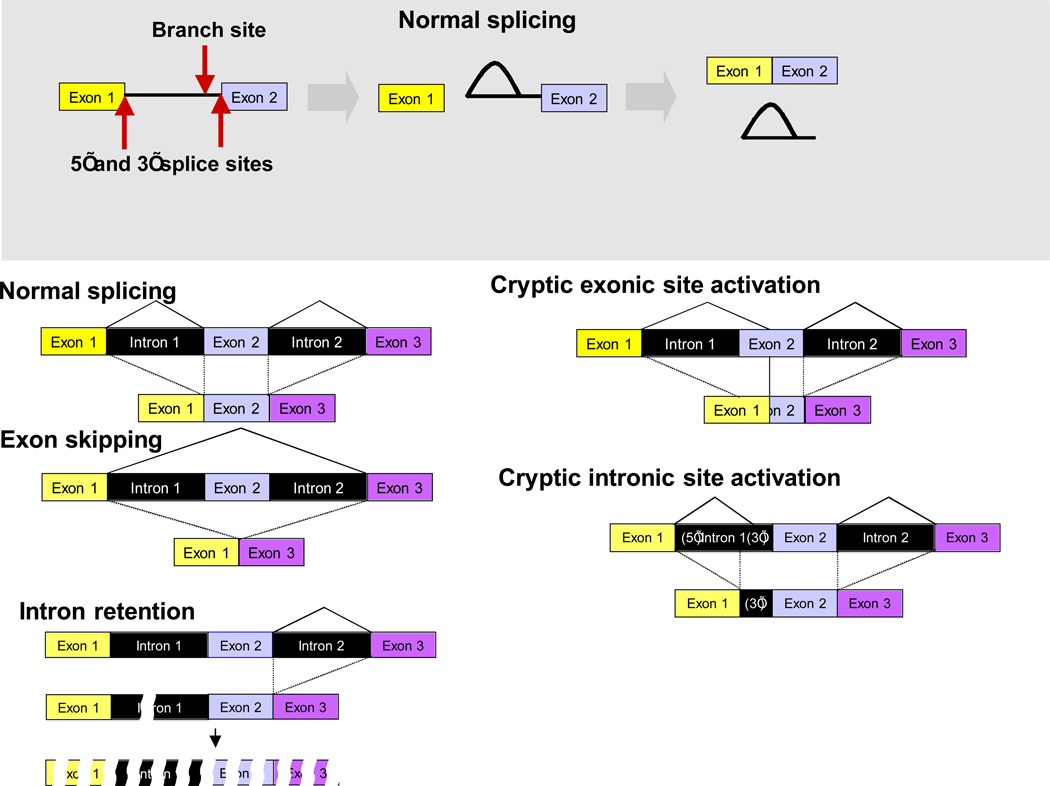
Figure 1. Normal and aberrant pre-mRNA splicing.
A. Pre-mRNA splicing is a two step process catalyzed by spliceosomes and specified by three RNA sequence elements called the 5’ splice site, the 3’ splice site and the branch site. In the first step of the reaction, the 2’ hydroxyl group of the adenosine residue at the branch site attacks the 5’ splice site to form a bi-partite intermediate consisting of the upstream exon and a lariat molecule containing the downstream exon joined to the intron. In the second step, the upstream exon attacks the 3’ splice to yield the ligated exons and the excised intron in lariat form.
B. The splicing pattern of genes can vary due to normal alternative splicing choices or due to mutations of the cis-acting RNA sequence elements or the trans-acting splicing factors. The various outcomes of alternative or pathological splicing events are diagramed. In most cases, misplicing will lead to degradation of the aberrant mRNA either in the nucleus or through nonsense mediated decay in the cytoplasm.
Many organisms including humans have two mutually exclusive types of pre-mRNA introns which are spliced by two distinct spliceosomes. The major type of intron, called U2-dependent, accounts for 99.7% of human introns while the minor type, called U12-dependent, numbers around 800 in humans (Will CL, Lührmann R. 2005; Wachtel C, Manley JL. 2009). These U12-dependent introns are found in around 800 genes along with major class introns. Many of these genes code for essential cellular proteins. The two spliceosomes differ in four of the five snRNAs but share most of the proteins involved in splicing. The different snRNAs recognize the splice site sequences through base pairing interactions accounting for the different specificities of the two spliceosomes and the different consensus sequences of the two classes of intronic splice sites (Wahl MC. 2009; Wachtel C, Manley JL. 2009).
Of importance is that the process of splicing is further regulated by chemical modifications. Various spliceosomal proteins undergo acetylation and are targets for histone acetylases and deacetylases (Kuhn AN, et al, 2009). Similarly, SR-domain containing proteins are substrates SLK like serine kinases; phophorylation appears to promote assembly and activate the splicing process (Prasad J et al, 2009; Muraki M et al, 2004; Pilch B et al, 2004).
Splicosomal genes affected by somatic mutations
Recently, several groups have reported results of either whole genome or exome high throughput sequencing of tumor samples from patients with several types of myeloid neoplasia and chronic lymphocytic leukemia (Visconte V et al, 2011; Makishima H et al, 2012; Yoshida K et al, 2011; Graubert TA et al, 2011; Papaemmanuil E et al, 2011; Rossi D et al, 2011; Wang L et al 2011, Quesada V et al, 2011). A striking feature of these studies is that they revealed frequent somatic mutations in a number of spliceosomal proteins. The most commonly mutated spliceosomal factor genes include (Tab. 1, Fig. 3):
Table 1.
Spliceosomal mutations
| Genes | Mutations | References | Diseases |
|---|---|---|---|
| SF3B1 | Multiple | Visconte et al., Yoshida et al., Papaemmanuil et al., Makishima at al. Rossi at al, Quesada at al | RARS, RCMD-RS RARSt, CLL |
| U2AF1 | Multiple | Yoshida et al. Graubert at al, Makishima at al | MDS, CMML, sAML, AML |
| SRSF2 | Multiple | Yoshida et al. Makishima at al | MDS, CMML |
| ZRSR2 | Multiple | Yoshida et al. Makishima at al | MDS, CMML |
| LUC7L2 | Single | Makishima at al | sAML |
| PRPF8 | Multiple | Makishima at al | sAML |
| U2AF2 | Multiple | Yoshida et al. | MDS |
|
SF1 SAP130 |
Multiple Single |
Yoshida et al. TCGA* |
MDS AML |
| SFRS6 | Single | TCGA* | AML |
| SON | Single | TCGA* | AML |
| U2AF26 | Single | TCGA* | AML |
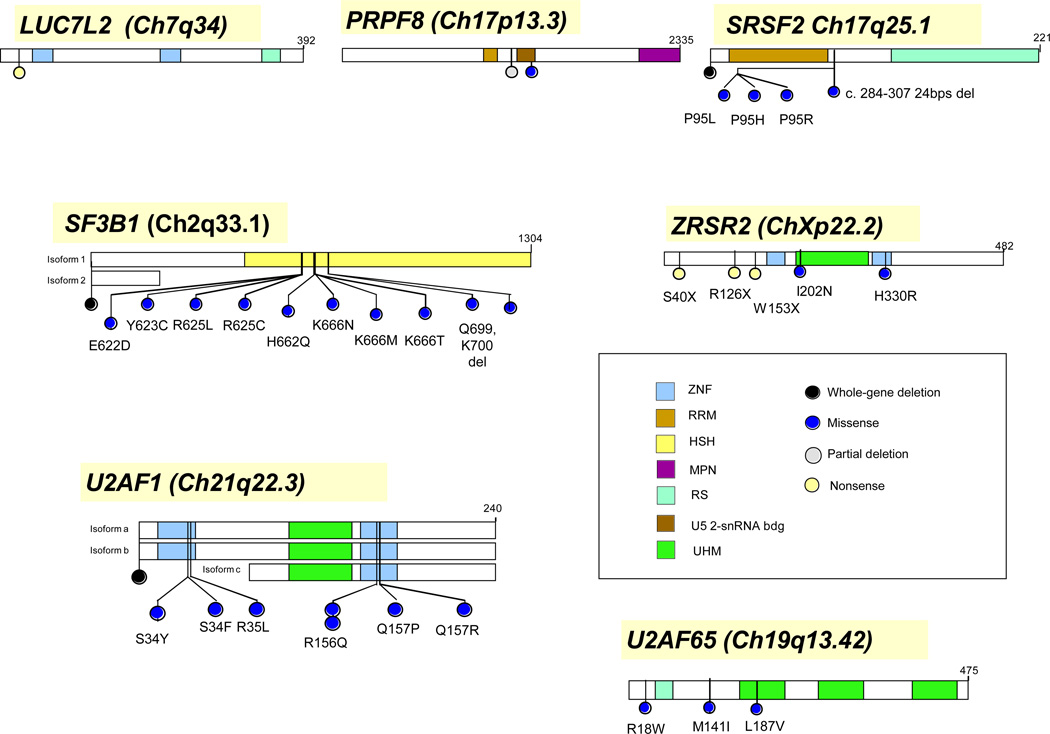
Figure 3. Genomic localization of exemplary spliceosomal mutations.
Exemplary spliceosomal genes affected by somatic mutations. Selected mutations and their locations were shown. Not all mutations described for each of the genes is shown.
SF3B1
Perhaps the most frequently detected spliceosomal factor mutations were found in the SF3B1 gene on chromosome 2q33.1. This gene codes for subunit 1 of the splicing factor 3b protein complex (SF3b) which is involved in the early stages of spliceosomal assembly. The SF3b complex is part of the functional form of the U2 snRNP that binds to the branch site near the 3’ end of introns and helps to specify the site of splicing. Functionally, it cross-link to a 25-nucleotide region in the pre-mRNA located immediately upstream of the intronic branch site. SF3B1 is itself subject to alternative splicing resulting in the production of various different protein isoforms. SF3B1 also plays a role in the minor U12-dependent spliceosome. Inhibition of splicing with spliceostatin A, a compound that targets the SF3B1 protein, or knock down of SF3B1 expression has been shown to alter the fidelity of branch site recognition by U2 snRNP leading to specific alterations in the alternative splicing patterns of many but not all genes (Corrionero et al., 2011). Spliceostatin A and similar compounds inhibit cell proliferation particularly of transformed cell lines suggesting that modulation of SF3B1 activity by mutations could be oncogenic while also providing a target for therepy.
U2AF1 and U2AF26
These genes are located on chromosome 21q22 and 19q13.12, respectively and code for related proteins that play a role in the early steps of 3’ splice site recognition as mutually exclusive partners of the U2AF2 protein. Both proteins belong to the serine/arginine (SR)-rich family of splicing regulatory factors and bind to both the terminal RNA nucleotides of the intron and the U2AF2 protein. This complex then recruits the U2 snRNP to the branch site region of the intron. These proteins also play a role in both constitutive and regulated RNA splicing by directly mediating interactions between the U2AF2 protein and other splicing regulators such as SRSF1 and SRSF2 (see below). Similar to SF3B1, knock down of U2AF1 did not block splicing of all introns but rather had large effects on the alternative splicing of a subset of genes leading to cell cycle arrest (Pacheco TR, et al., 2006).
SRSF1 and SRSF2
These genes are located on chromosome 17q22 and chromosome 17q25.2 respectively. While SRSF2 was found frequently mutated especially in CMML, SRSF1 mutations were less frequent. Both proteins belong to the SR splicing regulatory factor family. Each of these factors contains an RNA recognition motif (RRM) for binding RNA and an RS domain for binding other proteins. The RS domain is rich in serine and arginine residues and facilitates interaction between different SR splicing factors. These proteins bind to splicing regulatory sequence elements in pre-mRNA transcripts and to components of the spliceosome, and can either activate or repress splicing depending on the location of the pre-mRNA binding site. The proteins’ ability to activate splicing is regulated by phosphorylation and interactions with other splicing factor associated proteins. SRSF1 and SRSF2 play a role in preventing exon skipping, ensuring the accuracy of splicing and regulating alternative splicing. SRSF1 has an additional non-splicing function in the maintainance of genome integrity (Li X and Manley JL, 2005)
ZRSR2
This gene is located on chromosome Xp22.2. The corresponding protein associates with the U2AF heterodimer discussed above, which is required for the recognition of a functional 3' splice site in pre-mRNA splicing, and thus may play a role in network interactions during spliceosome assembly. ZRSR2 selectively interacts with the 3'-splice site of U2- and U12-dependent pre-mRNAs and promotes different steps in U2- and U12-dependent intron splicing (Shen H et al., 2010).
Less common mutations in various other genes for proteins with roles in the U2-dependent and U12-dependent spliceosomes have also been found. The number of mutated genes increases as larger sets of whole genome/exome sequenced cases are assembled (Fig. 1).
Consequences of spliceosomal dysfunction at the molecular level
It is notable that most of the splicing factors discussed below are known to act during early exon recognition steps (Figs. 1 & 2). In addition, where this data is known, knockdown of these factors in cells leads to specific alterations in splicing patterns for a subset of genes rather than global affects on most genes. Nevertheless, other mutated splicing factors act later in the splicing pathway and may affect splicing more broadly. The occurrence of mutations of spliceosomal proteins involved in various stages of this process suggests that the entirety of splicing is vulnerable to the acquisition of molecular defects with consequences in disease acquisition and progression Ward AJ, Cooper TA et al, 2010).
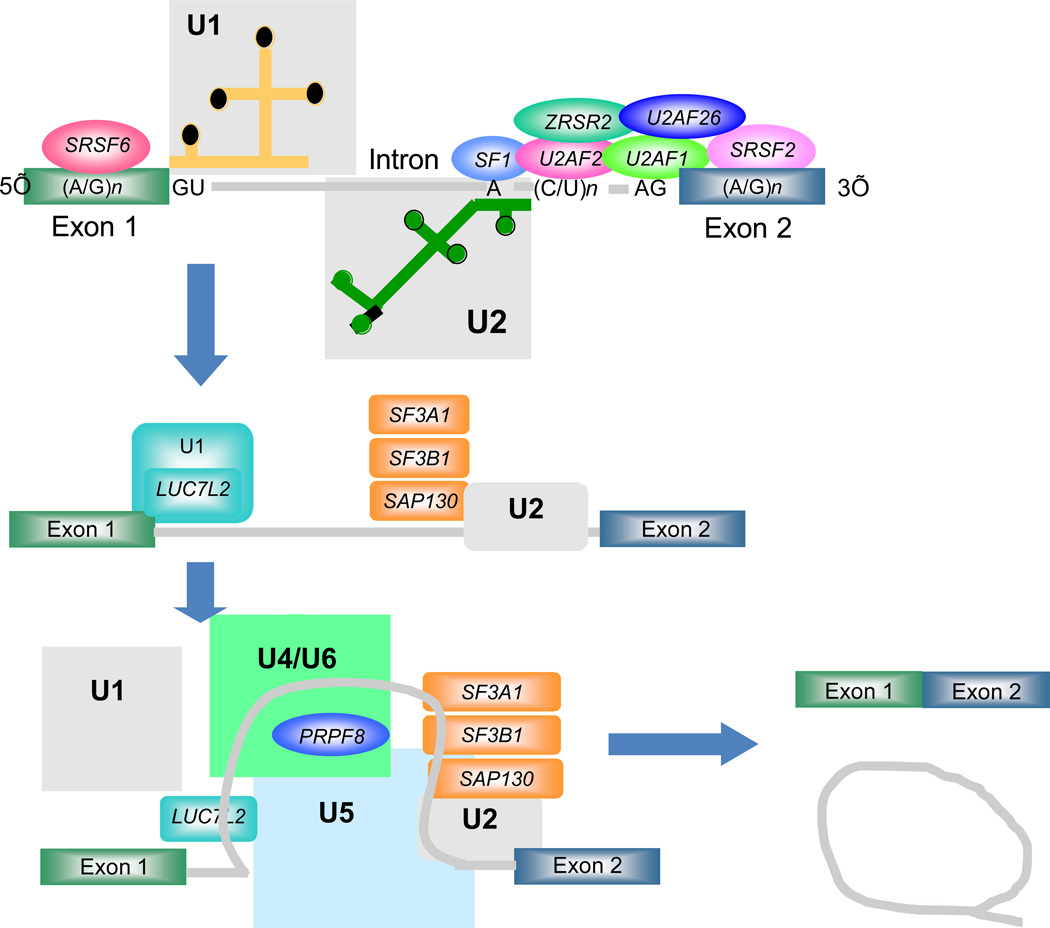
Figure 2. The roles of the various factors mutated in myeloid malignancies in the assembly and function of the spliceosome.
In the major or U2-dependent spliceosome shown here, the 5’ splice site is recognized by the U1 snRNA-protein particle (snRNP) while the branch site is recognized by the U2 snRNP aided by a complex of proteins at the 3’ splice site. The fully bound form of U2 snRNP also associates with the SF3a and SF3b complexes that contain three factors subject to mutations as discussed here. In a later step of the spliceosome formation pathway, a tri-snRNP complex composed of U4, U5 and U6 snRNPs joins the spliceosome bringing with them additional factors subject to mutations such as PRPF8. Further rearrangements of the spliceosome lead to catalysis of the splicing reaction and the production of the spliced product mRNA and the excised intron.
At the molecular level, it is unclear whether the spliceosomal protein mutations encountered in leukemia result in loss of function or gain of function. On the one hand, most of mutations identified to date are missense and affect rather invariant positions in SF3B1, SRSF2, U2AF1 and other spliceosomal proteins (Yoshida K et al, 2011; Makishima H et al, 2012). Similarly, no nonsense or stop codon mutations have been found. These findings argue that the spliceosomal protein mutations lead to gain of function. On the other hand, it would be expected for classical gain of function mutations that one would encounter mutations in a homozygous configuration through somatic UPD (as is observed for many other mutations such as FTL3, JAK2 or CBL) (Makishima H et al, 2011). To this point, however, homozygous or biallelic mutations in spliceosomal proteins have not been described. It may well be that a homozygous mutant cell is non-viable just as the homozygous knock out of SF3B1, for example, is embryonic lethal (Isono K et al, 2005). Nevertheless, the heterozygous state does lead to identifiable alterations in splicing patterns. Similarly, knockdown of U2AF1 expression results in defective splicing of exemplary indicator genes (Fu Y et al, 2011). An increase of inappropriate splicing may ultimately lead to defective expression and thereby indirectly to the loss of function of key downstream target genes. It may thus be most appropriate to refer these mutations as change of function/neomorphic mutations.
There are several possible effects on downstream gene expression that could result from mutations in the spliceosome components (Fig. 2). SF3B1 and U2AF1 are proteins that help in the recognition of 3’ splice sites in many genes (Wahl MC et al, 2009; Fu Y et al, 2011). SRSF2 is a member of a class of splicing modulators that recognize sequence elements within exons and recruit other splicing factors to nearby splice sites (Shepard PJ, Hertel KJ et al, 2009). There is evidence from specific knock down experiments that different introns differ in their dependence on these factors for accurate splicing. Failure to correctly specify the 3’ splice sites of introns could lead to several defects in gene expression. First, a mutant factor such as SF3B1 or U2AF1 could form inactive splicing complexes at the correct 3’ splice site leading to retention of the entire intron, activation of nearby cryptic splice sites or exon skipping (Fig. 2). Each of these results could produce unstable RNAs either due to nuclear degradation of intron-containing transcripts or cytoplasmic nonsense mediated decay (NMD) due to frame shift-induced stop codons. A similar set of consequences can be imagined for mutations in factors required for 5’ splice site recognition or the later steps of splicing. In general, it is likely that mutations result in altered recognition of sequences coding for 3’, 5’ splice or branch sites. As a consequence binding sites can be missed or alternative sites recognized leading to misplicing that is sequence-specific and thereby generating a pattern of defects distinct for each mutation.
Alterative splicing involves the recognition of cis acting RNA regulatory sequences within the pre-mRNA by trans acting RNA binding proteins that can either recruit splicing factors to adjacent splice sites or block splicing by a number of different mechanisms. A feature of alternatively spliced introns is that they generally have weak splice site sequences presumably to make then dependent on factor binding (Wang ET et al, 2008). The SF3B1 and U2AF1 proteins are believed to be some of the downstream target splicing factors that interact with the regulatory proteins such as SRSF2. Thus, mutations in these factors are likely to cause alterations in the alternative splicing of at least a subset of genes. This has, in fact, been observed upon knock down of these factors in cultured cells (Fu Y et al, 2011).
Spliceosomal dysfunction and consequences for clonal dominance
Irrespective of the basic mechanistic consequences of spliceosomal mutations, the main conceptual questions to be answered in the next years will be: i) how do spliceosomal protein mutations lead to the clonal growth advantage of mutant cells leading to malignant outgrowth (Fig. 4); ii) whether spliceosome mutations have facilitating secondary features or are of ancestral or founding nature; and iii) how different patterns of misplicing translate into distinct phenotypic features i.e., which are the critical downstream target genes and how are they affected.
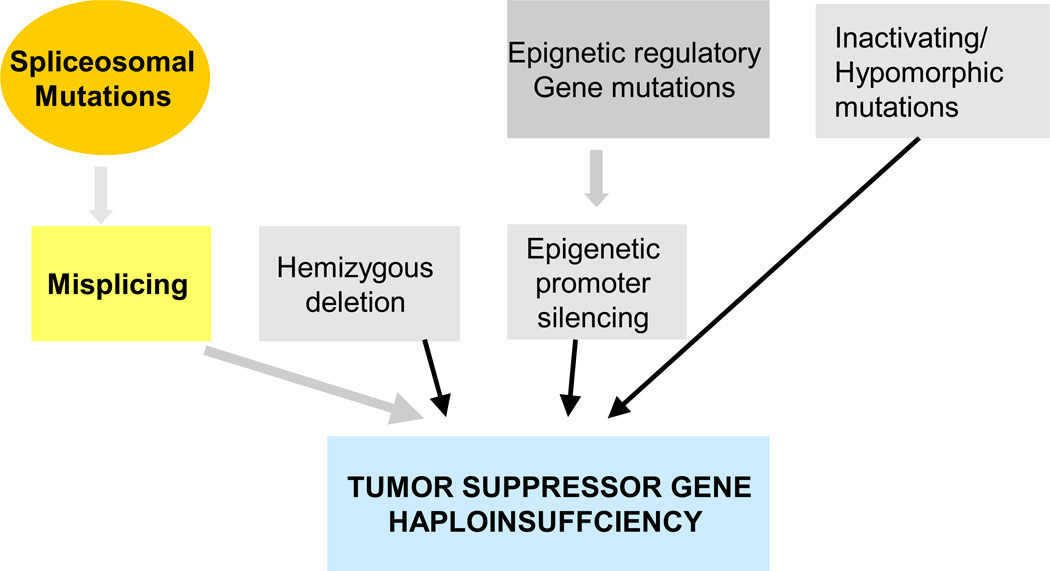
Figure 4. Prooncogenic consequences of spliceosomal mutations.
Spliceosomal mutations may lead to misplicing and ultimately decreased expression of intact TSG resulting in clonal evolution/ selection advantage. Various mechanisms may directly and indirectly lead to a similar result. Similar indirect effects of spliceosomal mutations may lead to alteration in other proteins including those involved in differentiation or other cellular functions.
A lot of the pieces in this puzzle are still missing but there are several theoretical possibilities as to how spliceosomal mutations exert their proleukemogenic properties or contribute to specific phenotypes (as, for example, is seen for SF3B1 mutations and ring sideroblasts in MDS). Splicing defects may lead to degradation of defective mRNA and result in decreased expression of tumor suppressor genes (TSG). For instance, intron retention may result in NMD. Functionally, this result would be equivalent to haploinsufficiency as seen in loss of chromosomal material or to inactivating or hypomorphic mutations in the corresponding TSG (Fig. 4). Consistent with this theory, we have reported misplicing of specific introns in RUNX1 and EZH2 genes (Makishima H et al, 2011), both known to be the target of direct mutations or to be contained within the boundaries of chromosomal deletions. Each type of spliceosomal lesion may affect many target mRNAs and thereby produce a very individual phenotype but one can hypothesize that some of the defectively spliced genes may have a crucial role in leukemogenesis or be essential for specific phenotypic disease features. Potential mechanisms include aberrant alternative splicing which could lead to altered relationship between the isoforms (e.g., pro- versus anti-apoptotic isoforms of Bax family genes) that contribute to prooncogneic potential or exon skipping which can lead to the generation of defective proteins with dominant negative effects (Kim H et al, 2009). Similarly, this down-regulation of SF3B1 induced an increase in the Bcl-x(s) with a concomitant decrease in the Bcl-x(L) splice variants and immunoreactive protein levels, thereby decreasing the Bcl-x(L)/Bcl-x(s) ratio. Specific down-regulation of SF3B1 also inhibited the ability of exogenous ceramide treatment to further induce the activation of the Bcl-x(s) 5' splice site (Massiello A et al 2006).
The issue as to whether spliceosomal mutations are initiating events has also not been answered. In our serial analyses, we have observed the presence of U2AF1 mutations from the very beginning, prior to the unequivocal diagnosis of the malignant process (Abu Kar S et al, 2012; Muramatsu H et al, 2012). However, heterozygous SF3B1 knock out mice that might be presumed to phenocopy the human mutations, do not develop MDS. In fact, these mutations seem to be associated with the phenotypic feature of ring sideroblasts rather than with malignant disease corresponding to the observation that SF3B1 mutations in MDS appear to be associated with good prognosis (Visconte V et al, 2012, Malcovati L et al, 2012). This clinical observation could also argue against their ancestral nature. Spliceosomal mutations may occur in early progenitors. Then, during lineage-specific differentiation, when mRNAs targeted by mutations are expressed, the defects become functional and lead to specific phenotypes. For instance, SF3B1 mutations produce changes in erythroid precursors when present in the context of MDS or SRSF2 mutations are encountered in association with chronic myelomonocytic leukemia (CMML). It is also possible that spliceosomal mutations occur in more differentiated precursor cells. This may be the case in CLL where likely lymphoid-specific genes are affected by misplicing (Rossi D et al, 2011; Wang L et al 2011, Quesada V et al, 2011).
Specific spliceosmal mutations and their clinical implications
Leukemias associated with specific spliceosomal protein mutations
High-throughput sequencing screens of patients with MDS have led the to identification of cases characterized by the presence of somatic mutations in various genes of the spliceosomal machinery (Tab. 1, Fig. 3). Initially, SF3B1 was found to be mutated in patients with refractory anemia (RA), refractory anemia with refractory cytopenia and multilineage dysplasia (RCMD) and refractory anemia with ring sideroblasts (RARS and RCMD-RS) as well as RARS with thrombocytosis (Visconte V et al, 2011; Papaemmanuil E et al, 2011; Yoshida K et al, 2011; Makishima H et al, 2012). It appears that SF3B1 mutations are associated with RS as a phenotypic feature rather than transforming properties. Knockdown of SF3B1 in cell lines does not result in accelerated growth and the heterozygous knock out mouse does not develop MDS and the phenotype may need to be investigated in a conditional knockout setting (Tiu R et al. unpublished results). It is possible that one or more specific mRNAs coding for proteins associated with mitochondrial iron metabolism is/are misspliced. The specific mutations in SF3B1 are quite invariant with mutations of K700 accounting for the majority (Visconte V et al, 2011).
In 3 studies, SF3B1 mutations were found to be present in 5–15% of cases (17% in fludarabine refractory patients). In CLL, the mutagenic event likely affects lymphoid precursor/memory cells and, due to the lack of massive iron utilization machinery that is operative in erythroid cells, iron rings are not present. Similarly, to RARS, SF3B1 mutations in CLL were mostly missense and recurrently targeted 3 hotspots (4codons 662, 666, and 700). CLL SF3B1 mutations were consistently associated with a poor prognosis, fludarabine-refractoriness. While they occurred occurred primarily in CLL with deletions of 11q, they were mutually exclusive with TP53 mutation (Rossi D et al, 2011; Wang L et al, 2011; Quesada V at al, 2011).
In MDS, SF3B1 mutations account for the majority (80%) of cases characterized by increased numbers of RS and affects around 28% of low risk MDS but is less frequently found in advanced from of MDS, including sAML as well as being found in a small number of cases (7%) of CMML (Yoshida K et al, 2011; Makishima H et al, 2011; Abu Kar S et al, 2012; Muramatsu H et al, 2012). Mutation of SF3B1 has been proposed to account for favorable prognosis but multivariate adjustment for other parameters suggests that SF3B1 mutations are not independent predictors of risk (Patnaik MM et al, 2012). However, a relative paucity of SF3B1 mutations suggests that progression to sAML is not a frequent event in low risk MDS with mutant SF3B1.
In subsequent studies, other spliceosomal protein mutations were identified with U2AF1 and SFSR2 mutations being the most common (Yoshida K et al, 2011; Makishima H et al, 2012; Graubert TA et al, 2011). SRSF2 mutations are affecting the same position (canonical) and have been found in 28–40% % of CMML patients as well as in some sAML cases, possibly derived from SFSR2 (Yoshida K et al, 2011; Makishima H et al, 2012; Abu Kar S et al, 2011). SRSF2 mutations have, to date, not been shown to convey an unfavorable prognosis and can coexist with mutations in TET2, ASXL1 and RUNX1 but not U2AF1. U2AF1 mutations were found in 10% of MDS including more advanced stages as well as sAML but also in up to 20% of patients with CMML. Most of the mutations affect 2 canonical zinc finger domains. Interestingly both SRSF2 and U2AF1 have not been found to be mutated in closely related pediatric leukemias including JMML (Hirabayashi S et al, 2012; Abu Kar S et al, 2012; Muramatsu H et al, 2011). Mutated U2AF1 appears to convey poor prognosis in CMML (Makishima H et al, 2011). While mutually exclusive with SRSF2 mutations, both gene mutations have been found in serial samples from the initial presentation onward and, in cross sectional studies of CMML-2 and sAML, the frequency of these mutations does not differ.
Other splicing factors that are also frequently mutated in myeloid neoplasms include ZRSR2 (URP), SF1, PRPF40B, U2AF2 (U2AF65) and SF3A1 (Yoshida K et al, 2011; Makishima H et al, 2012). These mutations occur in a mutually exclusive pattern and also, with the exception of ZRSR2, were only amino acid substitution mutations often with a high recurrence of individual missense mutations. In the case of ZRSR2, missense, nonsense and frame shift mutations were all detected (Yoshida K et al, 2011).
Therapeutic implications
Should spliceosomal mutations be proven to result in a gain of function or neomorphic phenotype, application of specific spliceosomal inhibitors may allow for targeting of mutant phenotype. So far, some spliceosomal inhibitors have been reported to have significant anti-tumor cell activity in culture (Albert BJ et al, 2007, 2009; Corrionero A et al, 2011). These inhibitors have yet to be tested on tumor cells known to be carrying any of the spliceosomal mutations discussed here. It may also be possible to find inhibitors that are specific to the mutant form of these factors that would restore the normal splicing phenotype. A different mode of action can also be proposed from observation of the mutational pattern in spliceosomal genes. Since most of the mutations are heterozygous, it is possible that homozygous inactivation of spliceosomal genes is lethal. In such a scenario, a spliceosome inhibitor targeted at the specific factor may be more toxic to the heterozygous mutant cells than normal cells. For examples, it seems that lenalidomide acts in a such a manner through selective sensitivity of cells with del(5q).
Conclusions
Genes coding for splicosomal proteins appear to be a frequent targets of somatic mutations in hematologic malignancies, including MDS, MDS/MPN and AML as well as in CLL. It is likely that further sequencing of various cancer genomes will reveal additional genes affected by somatic mutations in leukemia. It is possible that spliceosomal dysfunction caused by these mutations results in specific misplicing patterns in TSG or in genes associated with differentiation processes. The recognition sequences for 3’ and 5’ splicing sites may allow for prediction of target mRNAs prone to defective splicing by specific mutations. The clinical relevance of spliceosomal mutations include their prognostic significance, association with specific morphologic features and potential role as biomarkers for targeted therapies.
Table 2.
Distribution of splicosomal mutations among patients with hematologic malignancies.
| Disease | Estimated mutational frequencies | |||
|---|---|---|---|---|
| SF3B1 | U2AF1 | SRSF2 | ZRSR2 | |
| CLL | 15% | NF | NF | NF |
| Low risk MDS RARS/RCMD-RS |
20–28% 65–73%% |
5–6% 1–3% |
5–8% 5–6% |
1% 5% |
| CMML | 3–5% | 8–15% | 27–40% | 8% |
| RAEB1/2, sAML | 2–5% | 6–16% | 4–6% | 6% |
| AML | 2–3% | 1–8% | 1–3% | 1% |
Literature: NF not found; Literature CLL (Rossi D et al, 2011; Quesada V et al, 2012), MDS and AML (Visconte V et al, 2011; Makishima H et al, 2012; Yoshida K et al, 2011; Graubert TA et al, 2011; Papaemmanuil E et al, 2011). CMML (Yoshida K et al, 2011, Makishima H et al, 2012; Muramatsu H et al, 2011; Abu Kar S et al, 2012), ZRSR2 (Yoshida K et al, 2011)
References
- Abu Kar S, Jankowska A, Makishima H, Visconte V, Jerez A, Sugimoto Y, Muramatsu H, Traina T, Afable M, Guinta K, Tiu RV, Przychodzen B, Dunbar A, Sakaguchi H, O’Keefe Ch, Sekeres MA, Maciejewski JP. Spliceosome gene mutations are also present in the diverse mutational spectrum of chronic myelomonocytic leukemia. Hematologica. 2012 doi: 10.3324/haematol.2012.064048. 2012, submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert BJ, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol Cancer Ther. 2009;8:2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert BJ, Sivaramakrishnan A, Naka T, Czaicki NL, Koide K. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J Am Chem Soc. 2007;129:2648–2659. doi: 10.1021/ja067870m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Masuda A, Ito M, Shinmi J, Ohno K. AG-dependent3'-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res. 2011 May;39(10):4396–4404. doi: 10.1093/nar/gkr026. 2011 Epub 2011 Feb 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, McLellan MD, Dooling DJ, Abbott RM, Fulton RS, Schmidt H, Kalicki-Veizer J, O'Laughlin M, Grillot M, Baty J, Heath S, Frater JL, Nasim T, Link DC, Tomasson MH, Westervelt P, DiPersio JF, Mardis ER, Ley TJ, Wilson RK, Walter MJ. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011 Dec 11;44(1):53–57. doi: 10.1038/ng.1031. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Flotho C, Moetter J, Heuser M, Hasle H, Gruhn B, Klingebiel T, Thol F, Schlegelberger B, Baumann I, Strahm B, Stary J, Locatelli F, Zecca M, Bergstraesser E, Dworzak M, van den Heuvel-Eibrink MM, De Moerloose B, Ogawa S, Niemeyer CM, Wlodarski MW. Spliceosomal gene aberrations are rare, coexist with oncogenic mutations, and are unlikely to exert a driver effect in childhood MDS and JMML. Blood. 2012 Jan 11; doi: 10.1182/blood-2011-12-395087. 2012 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Isono K, Mizutani-Koseki Y, Komori T, Schmidt-Zachmann MS, Koseki H. Mammalian polycombmediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes Dev. 2005 Mar 1;19(5):536–541. doi: 10.1101/gad.1284605. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hsieh JJ, Cheng EH. Deadly splicing: Bax becomes Almighty. Mol Cell. 2009 Jan 30;33(2):145–146. doi: 10.1016/j.molcel.2009.01.004. 2009. [DOI] [PubMed] [Google Scholar]
- Kuhn AN, Van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA. 2009;15:153–75. doi: 10.1261/rna.1332609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005 Aug 12;122(3):365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, Przychodzen B, Bupathi M, Guinta K, Afable MG, Sekeres MA, Padgett RA, Tiu RV, Maciejewski JP. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012 Feb 9; doi: 10.1182/blood-2011-12-399774. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Maciejewski JP. Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res. 2011 Jun 15;17(12):3913–3923. doi: 10.1158/1078-0432.CCR-10-2900. 2011 Epub 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5' splice site selection of Bcl-x pre-mRNA. FASEB J. 2006 Aug;20(10):1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- Muraki M, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279:24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- Muramatsu H, Makishima H, Maciejewski JP. Chronic myelomonocytic leukemia and atypical chronic myeloid leukemia: novel pathogenetic lesions. Semin Oncol. 2012 Feb;39(1):67–73. doi: 10.1053/j.seminoncol.2011.11.004. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell. 2006;17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, Godfrey AL, Rapado I, Cvejic A, Rance R, McGee C, Ellis P, Mudie LJ, Stephens PJ, McLaren S, Massie CE, Tarpey PS, Varela I, Nik-Zainal S, Davies HR, Shlien A, Jones D, Raine K, Hinton J, Butler AP, Teague JW, Baxter EJ, Score J, Galli A, Della Porta MG, Travaglino E, Groves M, Tauro S, Munshi NC, Anderson KC, El-Naggar A, Fischer A, Mustonen V, Warren AJ, Cross NC, Green AR, Futreal PA, Stratton MR, Campbell PJ. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011 Oct 13;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik MM, Lasho TL, Hodnefield JM, Knudson RA, Ketterling RP, Garcia-Manero G, Steensma DP, Pardanani A, Hanson CA, Tefferi A. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012 Jan 12;119(2):569–572. doi: 10.1182/blood-2011-09-377994. 2012 Epub 2011 Nov 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch B, Allemand E, Facompre M, Bailly C, Riou JF, Soret J, Tazi J. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 2001;61:6876–6884. [PubMed] [Google Scholar]
- Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, Fangazio M, Vaisitti T, Monti S, Chiaretti S, Guarini A, Del Giudice I, Cerri M, Cresta S, Deambrogi C, Gargiulo E, Gattei V, Forconi F, Bertoni F, Deaglio S, Rabadan R, Pasqualucci L, Foà R, Dalla-Favera R, Gaidano G. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011 Dec 22;118(26):6904–6908. doi: 10.1182/blood-2011-08-373159. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, López-Guerra M, Colomer D, Navarro A, Baumann T, Aymerich M, Rozman M, Delgado J, Giné E, Hernández JM, González-Díaz M, Puente DA, Velasco G, Freije JM, Tubío JM, Royo R, Gelpí JL, Orozco M, Pisano DG, Zamora J, Vázquez M, Valencia A, Himmelbauer H, Bayés M, Heath S, Gut M, Gut I, Estivill X, López-Guillermo A, Puente XS, Campo E, López-Otín C. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011 Dec 11;44(1):47–52. doi: 10.1038/ng.1032. 2011. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. The SR protein family. Genome Biol. 2009;10(10):242. doi: 10.1186/gb-2009-10-10-242. 2009 Epub 2009 Oct 27. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3' splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24:2389–2394. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011 Sep 11;478(7367):64–69. doi: 10.1038/nature10496. 2011. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, O'Keefe C, Rogers HJ, Sekeres MA, Maciejewski JP, Tiu RV. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2011 Sep 2; doi: 10.1038/leu.2011.232. 2011 doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220:152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem. 2005 Aug;386(8):713–724. doi: 10.1515/BC.2005.084. Review. [DOI] [PubMed] [Google Scholar]
- Wachtel C, Manley JL. Splicing of mRNA precursors: the role of RNAs and proteins in catalysis. Mol Biosyst. 2009 Apr;5(4):311–316. doi: 10.1039/b820828j. Epub 2009 Jan 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, Hacohen N, Reed R, Meyerson M, Golub TR, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011 Dec 29;365(26):2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]