Abstract
Kinetochores are complex macromolecular assemblies that link chromosomes to the mitotic spindle, mediate forces for chromosome motion, and generate the checkpoint signal delaying anaphase onset until all chromosomes are incorporated into the spindle. Proper execution of these functions depends on precise interactions between kinetochores and microtubules. While the molecular composition of the kinetochore is well described, structural organization of this organelle at the molecular and atomic levels is just beginning to emerge. Recent structural studies across scales suggest that kinetochores should not be viewed as rigid static scaffolds. Instead, these organelles exhibit a surprising degree of flexibility that enables rapid adaptations to various types of interactions with the mitotic spindle.
Introduction
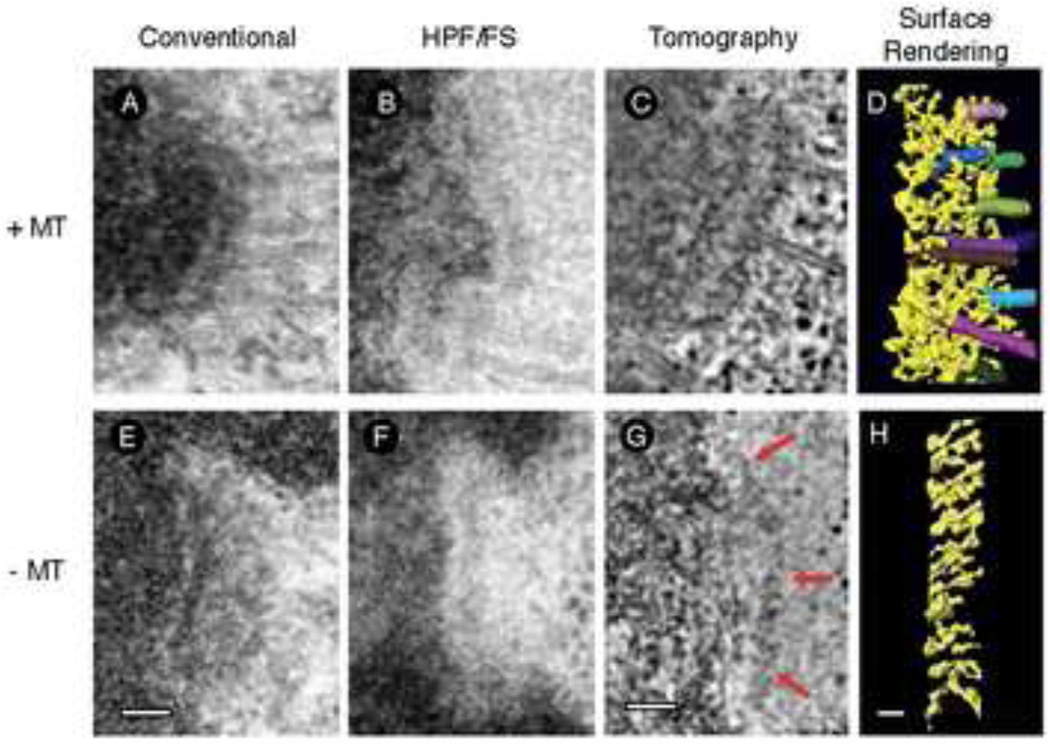
Sublime is perhaps the most appropriate adjective to characterize the kinetochore. The importance of this organelle is unquestionable, but its appearance is so subtle that it took the advent of electron microscopy (EM) to visualize this organelle [1]. In glutaraldehyde-fixed EM samples, kinetochores comprise three morphological domains: a dense inner plate, translucent middle layer, and a dense outer plate that supports a fibrous corona in the absence of spindle microtubule attachment (Fig 1A and E). Paradoxically, introduction of milder fixation procedures such as high-pressure freezing revealed that relatively sharp outer plate of the kinetochore is an artifact induced by the collapse of the labile network of fine fibers comprising the kinetochore outer layer [2] (Fig 1B–D and F–H). A better-preserved kinetochore appears even less structured.
Figure 1. Progressive views of the kinetochore.
(A) – (D), with microtubules attached, (E) – (H) without microtubules attached. (A) and (E) Classical views of the kinetochore after conventional chemical fixation methods. (B and F) The kinetochore is less distinct after improved structural preservation using high-pressure freezing and freeze substitution, demonstrating that the structure is more open and loosely organized than originally thought. (CD, G–H). Structural details are revealed using electron tomography to obtain 3D images of kinetochores preserved by high-pressure freezing and freeze-substitution. (C), (D), (G), and (H), adapted from [6] with permission.
The kinetochore’s apparent simplicity is deceptive as this biochemically complex organelle contains more than 80–100 specific proteins that are recruited to the kinetochore in a complicated semi-hierarchical manner [reviewed in 3]. At the core is the Constitutive Centromere Associated Network (CCAN) of proteins that specifies the site of kinetochore assembly on chromosomes and is thought to provide a base to assemble the outer kinetochore. The outer domain forms about the KMN network consisting of the KNL1 protein and the Mis12C and Ndc80C heterotetramic complexes (where C indicates complex to distinguish from the Mis12 and Ndc80 proteins). The KMN network is responsible for “load bearing” attachment to the spindle microtubules [4;5]. Together the CCAN, KMN network, and proteins such as CENP-F and Zwint, provide a scaffold for a number of motor, checkpoint, and regulatory components that dynamically associate with the kinetochore. Such molecular complexity reflects diverse chemical interactions as the kinetochore performs three fundamental roles in mitosis: 1) Attach chromosomes to the spindle; 2) Mediate forces that move the chromosomes; 3) Ensure that mitotic exit does not occur prematurely before each and every chromosome is incorporated into the spindle. Obviously, these multifaceted functions require that kinetochore organization be adaptable to the rapidly changing environment of the mitotic spindle. The structural basis for this adaptability is beginning to emerge from recent live-cell observations, biochemistry, and structural studies at multiple scales. This review summarizes these recent developments, with an emphasis on vertebrate kinetochores.
Structure and function of the outer domain
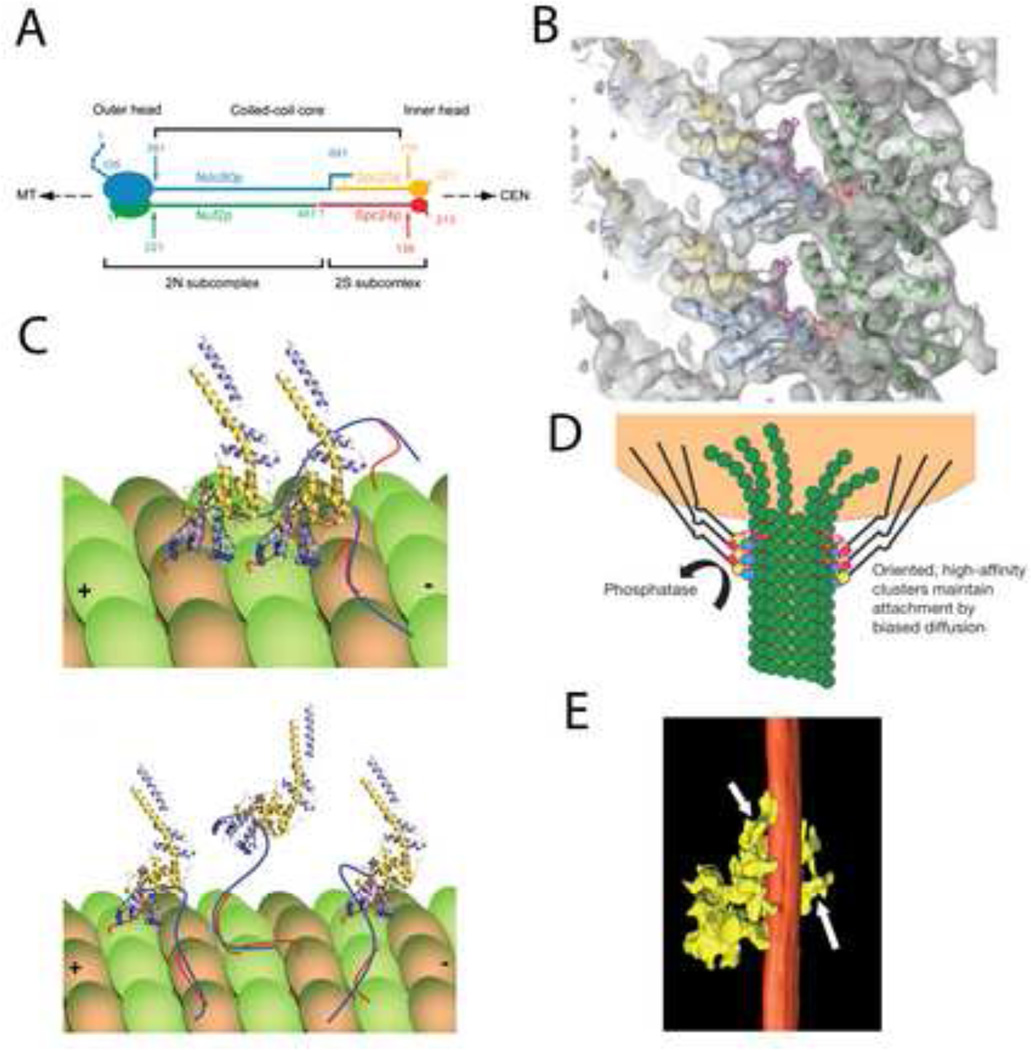
The outer layer of the kinetochore is a fibrous meshwork functioning as the principal site of microtubule attachment (Fig.1C and D) [6;7]. A major molecular player in this region is the highly conserved KMN network, whose Ndc80C constituent binds to the outer wall of microtubules [8;9]. Yeast Ndc80C is a 56 nm long “dumbbell” arranged with the globular N-terminal heads of the Ndc80/Nuf2 and Spc24/25 heterodimers at opposite ends of the complex (Fig. 2A) [10]. The two heterodimers are connected by their C-terminal tails. Negative-stain EM demonstrated that the complex is not completely linear due to a flexible hinge in the coiled-coil region of Ndc80C [11].
Figure 2. Ndc80C attachment to the microtubule surface.
(A) Molecular arrangement of the Ndc80 heterotetramer as deduced by immune labeling combined with metal shadow EM. Adapted from [10] with permission. (B) Atomic models of the Ndc80/Nuf2 head of Ndc80C and tubulin, fitted to the cryo-EM map with Ndc80 blue, Nuf2 gold, and tubulin green. Adapted from [17] with permission. (C) Model for vertebrate Ndc80/Nuf2 interaction with microtubules. Ndc80 blue, Nuf2 gold, unstructured tail of Ndc80 dark blue, C-terminal tail of tubulin red. Upper panel shows a model with the Ndc80 tail acting to pack adjacent Ndc80C on to the tubulin lattice. Lower panel shows a model with the Ndc80 tail acting as a tether to keep Ndc80 attached during periods when the calponin “toe” is not bound. Adapted from [19] with permission. (D) Biased diffusion model for Ndc80 capturing the energy of microtubule disassembly. Cooperative binding results in clusters that remain bound even when some molecules of the cluster lose their attachment. Adapted from [17] with permission. (E) Clusters of kinetochore microtubule interactions observed in situ by electron tomography. The surface rendering shows a microtubule (red) with several connections from the outer kinetochore (yellow). Adapted from [6] with permission.
Co-sedimentation binding experiments with purified components indicate that the globular head of the Ndc80/Nuf2 heterodimer binds microtubules, while Spc24/25 localizes the complex at the kinetochore [8]. This finding was corroborated by x-ray crystallography demonstrating that both the Ndc80 [12] and Nuf2 [13] heads contain a calponin-homology domain that is implicated in microtubule attachment of plus-end tracking proteins. However, point mutation studies demonstrate that it is the N-terminal 80 amino acids of Ndc80 that are critical for attachment [14;15]. This unstructured tail is highly basic and is thought to interact with the acidic c-terminal tail on the surface of tubulin as “ionic spaghetti” (i.e., the general ionic bonding of unstructured chains of two proteins rather than bonding between specific residues in each protein). The strength of this binding is reduced by phosphorylation from the kinase Aurora B [14], which in turn is spatially regulated [16].
While a combination of biochemical and x-ray crystallographic approaches were able to identify the critical domains in Ndc80 for microtubule attachment, the 8.6 Angstrom cryo- EM map was required to determine how these domains interact with microtubules [17]. Docking the atomic models of Ndc80/Nuf2 heads and tubulin in to the Cryo-EM map demonstrates that the calponin homology domain forms a “toe” that fits at the interface of each tubulin monomer (Ndc80 binds to both alpha and beta tubulin) (Fig. 2B). Binding by the toe region is sensitive to conformation with strong preference for the straight protofilaments associated with MT assembly. Consequently, Ndc80C attachment renders MT more resistant to disassembly in the cold and could explain the effect of kinetochore attachment on the conformation of microtubule plus ends [7;17;18]. These reconstructions fail to visualize the disordered N-terminal tail, which electron tomography images of microtubules bound with subsaturating amounts of Ndc80C indicate provides the majority of microtubule affinity, as noted above.
The importance of the unstructured tail is further substantiated by Tooley and colleagues who used a knockdown/ rescue system to demonstrate the importance of a key lysine residue in the toe region for kinetochore-MT attachment in vivo [19]. The study identified two additional lysines and confirmed the importance of the unstructured tail. This led to a three-point contact model for Ndc80 binding with the unstructured tail functioning as a tether (Fig. 2C). Charge is important for these interactions because lysine can be replace by arginine, but not a by a neutral or acidic amino acid.
Sub-saturation binding assays show cooperativity between adjacent Ndc80Cs resulting in a high frequency of clusters of 3 or 4 contact points with microtubules [13;17]. Cooperative binding enables clusters to stay attached when some molecules in the clusters detach, suggesting a mechanism for Ndc80C to follow and capture the energy release by disassembling microtubules in order to power chromosome motion (Fig 2D) [17;19]. The physiological relevance of cooperative binding is also supported by the frequent detection of clusters of kinetochore attachments to microtubule walls in situ [6] (Fig. 2E). The N-terminal tail of Ndc80 is required for cooperative binding, implying that this region aids the kinetochore in following disassembling microtubules [17].
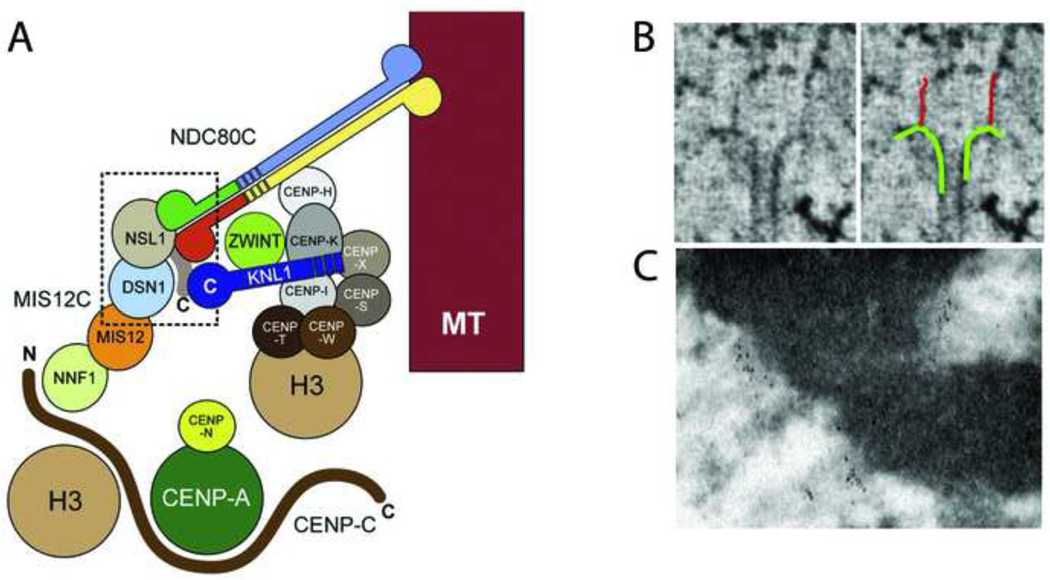
Less is known about structure and functional mechanism of other members of the KMN network, but recent negative stain EM reconstructions show that bacterially expressed Mis12C is a 22-nm long linear array of four globular proteins [20]. Binding, crosslinking, and in vitro EM studies identified the C-terminal tail of NSL1 as the binding site for Spc24/25 and KNL1 at one end of the Mis12C (Fig. 3A). NNF1, which is presumably at the other end of Mis12C, interacts with CENP-C suggesting that Mis12 is the link between Ndc80C and the CCAN core [21;22]. Hence the Mis12C-Ndc80C portion of the KNM network could take the appearance of an 80-nm filament linking the inner kinetochore with microtubules, as suggested by electron tomography (Fig. 3B) [18]. However, it is unlikely that connectivity is simply linear because the CCAN proteins CENP-H and CENP-K interact directly with Ndc80C [23;24]. Nevertheless, Mis12C is architecturally positioned at a key intersection between the microtubule binding activity of Ndc80, the outer scaffold function of KNL1, and the inner kinetochore.
Figure 3. Molecular architecture of vertebrate kinetochores.
(A) Model of kinetochore protein interactions based upon structural and binding data. Adapted from [20] with permission. (B) Electron tomography images of long fibrils attaching microtubules to the inner kinetochore in PtK cells. Adapted from [18] with permission. (C) Immuno-EM localization of Ndc80 to the outer kinetochore. Three HeLa cell kinetochores are labeled with the 9G3 antibody to Ndc80 head. Note the linear concentrations of gold particles distal to the chromatin.
The EM and X-ray crystallography studies discussed above revealed many details of the organization of the chromosome-microtubule binding site. Less is known about the spatial distribution of microtubule binding sites within the kinetochore and how the kinetochore structure is affected by various types of microtubule attachments. By nature, EM and X-ray crystallography tend to impose a static view of kinetochores. Yet, evidence is mounting that kinetochores function as flexible, dynamic points of contact between the chromosome and the spindle due to the presence of compliant elements.
Kinetochore compliance: insights and issues
Due to its small size, kinetochore fine detail is obscured by diffraction, making it difficult to learn much about their structure by light microscopy. However, relative positions of individual proteins within the kinetochore can be assessed by measuring the distances between the distributions of proteins labeled with spectrally different fluorophores. This method, commonly referred to as super high resolution co-localization microscopy (SHREC), can partially overcome the limitation imposed by diffraction by assuming that the centers of the point spread functions corresponds to the true positions of the proteins. The main advantage of SHREC is that it can reveal redistribution of components due to transitions in kinetochore architecture. Using SHREC, the relative positions of 16 kinetochore proteins have been established [25] and this molecular map is in good agreement with biochemical and structural data obtained by EM. As expected, the outer domain proteins are more distal to the heterochromatin than CCAN proteins. Immuno-EM studies place Ndc80C in the outer plate (Fig. 3C) [26] and CENP-C in the heterochromatin [27]. This is consistent with the KMN and CCAN networks being located in the outer plate and the distal edge of the heterochromatin in EM images.
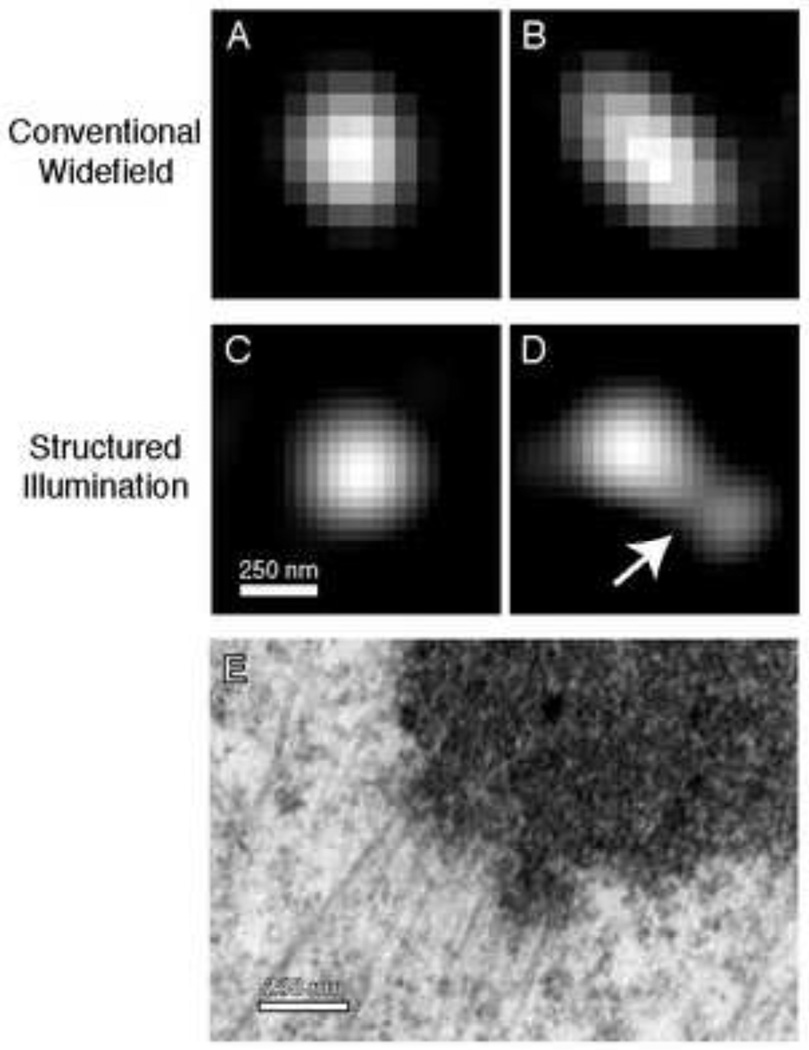
Measurements of distances between two-color labeled kinetochore protein populations in human [28] and Drosophila [29] cells demonstrated that the distance between fluorescent centroids (delta) changes as a result of attachment to dynamic microtubules. Further, outer kinetochore protein centroids are displaced inward toward the centromere upon treatment with Taxol [25]. These data imply that certain components of kinetochores can be stretched. Studies seeking to examine changes within kinetochores will benefit greatly from super resolution techniques which now make it possible to visualize shape and subdomains of labeled kinetochore proteins (Fig. 4). How the shapes vary between protein populations and across time will be crucial to understanding the complexity of kinetochore structure (Fig. 4E) and the response of kinetochores to mechanical force.
Figure 4. Details of kinetochore morphology revealed by super resolution microscopy.
(A) Labeled kinetochore proteins, such as the outer plate component Hec1, appear as round, diffraction limited point spread functions in conventional light microscopy. Methods based on centroid analysis fit the distribution of pixel intensities to a Gaussian function to find the center with sub-pixel accuracy. (B) Even in widefield, some Hec1 foci deviate from a PSF-like distribution of fluorescence, hinting at structural variation. (C and D) Hec1 immunostaining visualized with structured illumination microscopy (SIM). While some kinetochores still appear round, others exhibit a surprising amount of structural detail. In (D), a depression separates two apparent subdomains (arrow). (E) EM image of a kinetochore from a PtK1 cell highlighting the complexity of kinetochore morphology that can exist.
As a significant step in this direction, Suzuki and colleagues used immuno-EM to track the redistribution of inner kinetochore proteins CENP-A, CENP-C, CENP-R, and CENP-T from a rectangular to ellipsoid shape upon microtubule attachment in chicken DT40 cells [30]. Such a transition could shift the inner fluorescence centroid and increase delta measured by LM, but direct LM/EM correlation of the same kinetochores will be needed to firmly establish the relationship between these phenomena. Interestingly, this same study finds that the rectangular shape of gold labeling of outer plate components Ndc80, Mis12, and CENP-E does not change, despite the fact that large scale rearrangements in the outer kinetochore occur have been reported upon microtubule attachment or suppression of plus end dynamics [6;25]. Thus, it is likely that stretch arises from a combination of elastic elements throughout the kinetochore.
Atomic force microscopy of the reconstituted CENP-T-W complex suggest that these inner kinetochore components could account for the bulk of kinetochore stretch observed by LM [30]. The images showed that CENP-T possesses a long flexible element that enables the separation between N and C-termini to vary from almost 0 nm, to 90 nm when tension is applied. Two complexes within the KMN network at the microtubule binding site also have the potential for flexibility. As discussed above, Mis12C forms a straight 21–23 nm rod based on its hydrodynamic properties and appearance in negative stain [20]. This elongated conformation is likely to represent the structure in Taxol-treated cells based on the fact that SHREC mapping of kinetochore architecture found the length of the complex to be 9 nm in untreated cells and 19 nm in the presence of the drug [25]. This 10 nm of flexibility alone cannot explain stretch detected by LM, which is an order of magnitude larger in human kinetochores. Another possible candidate is the Ndc80 complex, which either has a straight, 57 nm long rod shape, or a bent conformation arising from a kink in the coiled-coil region of the Ndc80 subunit. The resulting kink shortens the complexes’ length by ~15% in negatively stained EM images [11].
While SHREC has been extremely valuable in identifying the phenomena of kinetochore stretch, this approach is limited to detection of relative distances. It cannot distinguish between stretching/folding of molecular complexes vs. reorientation of ensembles within the kinetochore architecture. Therefore, solving the mystery of kinetochore compliance will require integrating data from structural studies and live cell imaging across resolution scales.
Flexibility of kinetochore-spindle interactions
The outer kinetochore is an irregular fibrous mat that appears to function like spider web attaching to the walls of any microtubules in the vicinity, including those that end in a different kinetochore [6]. Hence kinetochores possess the flexibility to interact with both the plus end and the lateral surface of microtubules. Functionally, gliding of kinetochores along the sides of single [31] or bundled microtubules [32] powers chromosome movement to the poles and equator, respectively.
In human cells lateral interactions can support spindle assembly and chromosome congression in the absence of plus-end attachments as shown by simultaneously knocking down the kinesin 14 family member HSET and the Ndc80 complex component Nuf2 [33]. The key to overcoming lack of end-on attachments is CENP-E motor activity to drive congression using lateral interactions. While this study showed that such attachments are sufficient to support spindle assembly in human cells under perturbed conditions, determining the importance of lateral attachments during normal mitosis required technical development to track kinetochores and the spindle poles in 3D throughout mitosis [34] or meiosis [35]. Tracking measurements clearly show that upon initial encounter with microtubules most chromosomes move toward a position approximately equidistant between the two centrosomes, rather than moving toward a pole. This results in a pronounced chromosome ring at the equator in both human somatic cells [34] and mouse oocytes [35], suggesting it is a conserved intermediate stage that facilitates spindle assembly and proper chromosome segregation. The ring stage is characterized by dramatic rotations of the kinetochore-kinetochore axis and frequent loss of inter-kinetochore stretch that are inconsistent with stable end-on attachment of both sisters to oppositely oriented spindle poles. EM images confirm that lateral interactions predominate during the early stages of mitosis. The evidence suggests that many chromosomes initially use labile lateral interactions that are gradually converted to stable end-on attachments at the equator. Yet, some of these fully congressed chromosomes do not possess amphitelic attachment in human cells [34], suggesting that the flexibility to modulate kinetochore-microtubule interaction is maintained much later into mitosis than was previously thought.
Conclusions
Owing to complete genome sequencing and large scale gene inactivation studies in model organisms the list of principal kinetochore components is largely complete. It has become possible to start fitting these molecules into the kinetochore structure revealed by EM and in some cases understanding interactions at the atomic level. At the same time, novel LM techniques reveal that kinetochore structure offers a great deal of flexibility and adaptability. The distribution of individual molecular components within the kinetochore changes rapidly in response to various types of microtubule attachments. The next big challenge will be to correlate these redistributions with changes in the orientation of individual complexes that can only detected in EM. The key to achieving this goal is likely in combining various types of microscopy on a single kinetochore. Correlative LM/EM studies will be needed to uncover the exact sources of compliance within kinetochores and reveal the details of how chromosome interactions with the spindle are fine tuned.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 2.McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gascoigne KE, Cheeseman IM. Kinetochore assembly: if you build it, they will come. Curr.Opin.Cell Biol. 2011;23:102–108. doi: 10.1016/j.ceb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deluca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J.Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–114. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat.Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576.. This paper is an electron tomography study that demonstrates the loose network construction of the outer kinetochore and the variation of microtubule attachment sites. It points out the prominence of lateral attachments and architectural rearrangement upon microtubule attachment.
- 7.VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr.Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 9.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J.Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc.Natl.Acad.Sci.U.S.A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J.Mol.Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat.Struct.Mol.Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 13. Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos RG, Maiolica A, Polka J, De Luca JG, De WP, Salek M, Rappsilber J, Moores CA, Salmon ED, Musacchio A. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020.. The authors constructed and crystallized the Ndc80 Bonsai complex and obtained the atomic structure. A pair of tightly interacting calponin homology domains was identified in the Ndc80 and Nuf2 head domains and cooperative binding was observed.
- 14.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr.Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr.Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome biorientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423.. This cryo-EM study produced a density map of the Ndc80-tubulin interface with sufficient resolution to identify Ndc80 domains key for microtubule binding. The Ndc80 calponin homology domain binds to both the inter- and intra dimer interfaces and is sensitive to microtubule conformation. The unstructured N-terminal tail enables cooperative binding along the microtubule surface, which could be important for chromosome transport.
- 18. McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038.. Electron tomography study quantifying the effect of end-on kinetochore binding on the plus-end conformation of microtubules. The study also describes long fibrils connecting microtubules with the inner kinetochore.
- 19. Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol.Biol.Cell. 2011;22:1217–1226. doi: 10.1091/mbc.E10-07-0626.. A depletion/mutant rescue strategy was used to demonstrate the importance of key positively charged residues in the Ndc80 calponin-homology domain for chromosome alignment in situ. The authors propose a tripartite attachment of the calponin domain with the unstructured tail providing a tether during transient loss of attachment through the calponin homology domain.
- 20. Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, Maiolica A, Stark H, Musacchio A. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J.Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070.. This study uses negative stain electron microscopy and chemical cross-linking methods to show that Mis12C is 22 nm long linear complex connected to Spc24/24 and KNL1 through the NSL1 subunit.
- 21.Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr.Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Screpanti E, De AA, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr.Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikami Y, Hori T, Kimura H, Fukagawa T. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol.Cell Biol. 2005;25:1958–1970. doi: 10.1128/MCB.25.5.1958-1970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol.Biol.Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, Deluca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, Stukenberg PT, Desai A, Salmon ED. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035.. This study used SHREC to determine the relative locations of 16 kinetochore proteins with 5 nm accuracy. Upon treating cells with taxol there was a distinct shift in one set of proteins relative the Ndc80 head.
- 26.Deluca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol.Biol.Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitoh H, Tomkiel J, Cooke CA, Ratrie H, III, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 28.Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J.Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J.Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J.Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050.. The authors use immuno-EM to show that changes occur in the distribution of inner kinetochore proteins. Further analysis with AFM is used to identify structural changes in the CENP-T-W complex.
- 31.Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J.Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cai S, O'Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nat.Cell Biol. 2009;11:832–838. doi: 10.1038/ncb1890.. Depletion of select proteins results in chromosome congression in the absence of end-on kinetochore-microtubule attachments.
- 34. Magidson V, O'Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012.. 4D tracking of centrosomes and single kinetochore pairs was performed to quantitatively analyse the behavior of mitotic components in human cells. A prometaphase ring of chromosomes was found to accelarate search and capture to promote the efficiency of spindle assembly.
- 35. Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals errorprone homologous chromosome biorientation in Mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031.. Tracking of all kinetochores in 3D throughout meiosis in mouse oocytes reveales a prometaphase ring of chromosomes and a high frequency of chromosome misattachment to the spindle, suggesting that the process of spindle assembly is error prone.