Abstract
EGF-R [EGF (epidermal growth factor) receptor] ligands can promote or inhibit cell growth. The biological outcome of receptor activation is dictated, at least in part, by ligand-specified patterns of endocytic trafficking. EGF-R trafficking downstream of the ligands EGF and TGF-α (transforming growth factor-α) has been investigated extensively. However, less is known about EGF-R fates induced by the ligands BTC (betacellulin) and AR (amphiregulin). We undertook comparative analyses to identify ligand-specific molecular events that regulate EGF-R trafficking and degradation. EGF (17 nM) and BTC (8.5 nM) induced significant EGF-R degradation, with or without ectopic expression of the ubiquitin ligase Cbl. Human recombinant AR (17 nM) failed to affect receptor degradation in either case. Notably, levels of ligand-induced EGF-R ubiquitination did not correlate strictly with receptor degradation. Dose–response experiments revealed that AR at a saturating concentration was a partial agonist at the EGF-R, with approx. 40 % efficacy (relative to EGF) at inducing receptor tyrosine phosphorylation, ubiquitination and association with Cbl. EGF-R down-regulation and degradation also were compromised upon cell stimulation with AR (136 nM). These outcomes correlated with decreased degradation of the Cbl substrate and internalization inhibitor hSprouty2. Downstream of the hSprouty2 checkpoint in AR-stimulated cells, Cbl-free EGF-R was incorporated into endosomes from which Cbl–EGF-R complexes were excluded. Our results suggest that the AR-specific EGF-R fate results from decreased hSprouty2 degradation and reduced Cbl recruitment to underphosphorylated EGF-R, two effects that impair EGF-R trafficking to lysosomes.
Keywords: amphiregulin, betacellulin, Cbl, epidermal growth factor receptor, trafficking, ubiquitination
INTRODUCTION
Activated EGF-R [EGF (epidermal growth factor) receptor] elicits cellular signals that are essential for proper mammalian growth and development. These signals are induced by a family of ligands that includes EGF, AR (amphiregulin), TGF-α (transforming growth factor-α), heparin-binding EGF, BTC (betacellulin) and epiregulin. Despite being encoded by distinct genes, the ligands are structurally similar. Each contains a signature motif comprising six cysteine residues that form a three-loop secondary structure via intramolecular bonds. Each ligand can independently bind and activate the EGFR.
EGF family ligands play both overlapping and distinct roles in development. Overlapping roles for EGF family ligands have been observed in the nervous system, prostate gland and gastrointestinal tract development [1– 4]. The co-operative activities of several ligands contribute to mammary gland development [1].
Distinct biological outcomes result from the depletion of specific EGF family ligands. Murine knockout of the ligand TGF-α shows impaired hair follicle development, eye abnormalities and growth defects [1,5–7]. Loss of EGF leads to deficiencies in prostate and central and peripheral nervous system development [8]. AR, in contrast, has been implicated in the development and maintenance of the lung, kidney, liver, skin and mammary glands [1,9–13].
EGF family ligands also have both distinct and overlapping roles in tumour development. Ligand overexpression was observed in most of the tested human cancers [14]. Analyses of tumours, cell lines and transgenic mice have demonstrated that the impact of each ligand on tumour development is unique [15]. In the case of normal human mammary and breast cancer cells, AR enhances cell motility and invasiveness, relative to EGF, by differentially controlling signalling through NF-κB (nuclear factor κB) to IL-1 (interleukin-1) [16,17]. Different tumour types tend to overexpress different ligand subsets, and over-expression of specific combinations of ligands and receptors is a common theme [18].
Several studies have suggested a molecular basis for ligand-specific EGF-R signalling. The ligand AR has been reported to possess significantly reduced receptor-binding affinity, relative to EGF and TGF-α [19–22]. AR’s reduced receptor-binding affinity correlates with decreased EGF-R phosphorylation and altered EGF-R signalling [20,23,24]. Gilmore et al. [25] very recently reported that AR and EGF differ greatly in their induction of EGF-R tyrosine phosphorylation on residues 845, 1045, 1068, 1148 and 1173; both ligands activate Tyr992. The relative hypophosphorylation of receptor tyrosine residues downstream of AR could provide a mechanistic explanation for ligand-specific EGF-R signalling [25]. However, no report to date has shown whether or how AR-associated EGF-R hypophosphorylation affects receptor trafficking.
The non-redundant activities of AR and other EGF family ligands may result from discrete patterns of EGFR trafficking and signalling. Ligand-specific receptor fates can be considered in the context of EGF-R activation by the prototype ligand, EGF. Receptor activation by EGF induces EGF-R dimerization and the autophosphorylation of tyrosine residues within the receptor’s cytoplasmic tail. The phosphorylated tyrosine residues comprise binding sites for SH2 domain (Src homology 2 domain)-containing proteins, which mediate multiple interactions that facilitate receptor trafficking and signalling. Among these SH2 domain-containing regulatory proteins is the E3/ubiquitin ligase Cbl [26]. Cbl’s tyrosine kinase-binding domain is a variant SH2 domain that binds directly to Tyr1045-phosphorylated EGF-R [27]. Cbl also contains a linker region, RING (really interesting new gene) finger domain and RING finger tail that together are sufficient for Cbl-enhanced EGF-R ubiquitination [28].
EGFR ubiquitination has been shown in multiple systems to increase receptor degradation in lysosomes [26 –30]. For optimal EGF-R degradation, Cbl-mediated ubiquitination of distinct substrates is important. At the cell surface, Cbl ubiquitinates its own negative regulator, Sprouty2 (hSpry2, where h is human), in a ligand-dependent manner [31]. This results in the proteosomal degradation of hSpry2 [32]. hSpry2 degradation restores Cbl’s ability to bind EGF-R and enhance receptor ubiquitination and degradation [32].
Ubiquitination also plays a role later in the degradative pathway [27,33]. Ubiquitinated cargoes, including the activated EGFR, are recognized and targeted for incorporation into multivesicular bodies by binding to the UIM (ubiquitin-interacting motif) of Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) [33]. Hrs itself is ubiquitinated, and the dual modifications of Hrs tyrosine phosphorylation and ubiquitination are associated with Hrs degradation and Cbl-enhanced EGF-R degradation [34].
Utilizing EGF-induced receptor trafficking and fate as prototypes for ligand-induced receptor regulation, we undertook a systematic comparison of the three EGF family ligands AR, BTC and EGF. We examined their impact on EGF-R post-translational modifications and/or degradation. Both EGF and BTC were strong inducers of EGF-R degradation; AR was not. Our results suggest that the differential receptor fate induced by AR results from impaired hSpry2 degradation and the reduced recruitment of Cbl to EGF-R, two effects that divert activated receptors from the degradative pathway.
EXPERIMENTAL
Plasmids
The pAlterMAX-EGF-R, pAlterMAX-HACbl, pcDNA3GFP-Cbl-wt, pcDNA3GFP-Cbl-N and expression constructs have been described [28]. pCMV-FLAG-hSprouty2 has been described in [35]. pcDNA3GFP-Cbl-Y371F was generated through oligonucleotide-directed mutagenesis of pAlterMAX-HACbl by using primer 5′-GCCCATCTCACAGAATAATTCATATTG-3′, with subsequent fragment exchange of mutant sequences into pcDNA3GFP-Cbl WT (wild-type). The plasmid pAlterMAX-EGF-R Y1045F was produced through oligonucleotide-directed mutagenesis, by using primer 5′-GGGTCTGAGCTGAATCGCTGCAAG-3′ and pAlterMAX-EGF-R, followed by fragment exchange.
Antibodies
Anti-EEA1 (early endosome antigen 1) (no. 610456; murine IgG1) antibody was purchased from BD Biosciences. Anti-Cbl [sc-170; (C-15) rabbit polyclonal], anti-EGFR [sc-120; (528) murine IgG2a] and anti-Syk [sc-1240; (4D10) murine IgG2a] antibodies were purchased from Santa Cruz Biotechnology. Anti-GFP (green fluorescent protein) antibody (Ab290; rabbit polyclonal) was acquired from Abcam. Anti-ubiquitin antibody (NCL-UBIQ; rabbit polyclonal; Novocastra Laboratories) was purchased from Vector Laboratories. Anti-EGFR antibody (no. 06-129; sheep polyclonal for immunoblotting) and anti-phosphotyrosine antibody [no. 05-321; (4G10) murine IgG2b] were obtained from Upstate Biotechnology. Peroxidase-conjugated anti-FLAG antibody [A 8592; (M2) murine IgG1] was procured from Sigma. Anti-class I antibody [(W6/32) murine IgG2a] was generated by the University of Iowa Hybridoma Facility, using a hybridoma obtained from the A.T.C.C. (R)-Phycoerythrin-conjugated F(ab′)2 fragment of anti-mouse IgG (H + L) [no. 115-116-146; goat polyclonal] was purchased from Jackson ImmunoResearch Laboratories. Peroxidase-conjugated anti-mouse IgG (no. 55563) secondary antibody and peroxidase-conjugated Protein A (no. 55901) were obtained from ICN Pharmaceuticals. Additional peroxidase-conjugated Protein A (no. 32400) was purchased from Pierce Biotechnology. Peroxidase-conjugated anti-sheep IgG, H + L (no. 402100; rabbit polyclonal), was acquired from Calbiochem–Novabiochem. Alexa Fluor® 555-conjugated anti-mouse IgG2a (A21137; goat polyclonal) and Alexa Fluor® 647-conjugated anti-mouse IgG1 (A21240; goat polyclonal) antibodies were purchased from Invitrogen.
Cells
HEK-293 cells (human embryonic kidney cells) were maintained in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 10 %(v/v) FBS (fetal bovine serum), 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 100 units/ml penicillin–streptomycin and 20 mM Hepes (Gibco BRL). Cells were cultured in an atmosphere with 5 %CO2 at 37 °C.
COS-7 cells were maintained in DMEM containing 10 % FBS, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 100 units/ml penicillin–streptomycin and 20 mM Hepes (Gibco BRL). The cells were cultured at 37 °C in an atmosphere of 5 % CO2.
Transient transfection
HEK-293 cells were transiently transfected using a modification of the calcium phosphate precipitation method [35] and the amounts of DNA indicated in the Figure legends. DNA precipitates were removed from the cells after a 14–16 h incubation period at 37 °C in an atmosphere of 5 %CO2.
COS-7 cells grown to 80–90 % confluence on 100-mm tissue culture dishes received lipid-mediated transfection reagent (Lipofectamine™ 2000; Invitrogen). At 4 h after transfection, media were replaced with 10 % FBS-supplemented DMEM as described above. At 24 h post-transfection, cells were plated on glass coverslips at a density of 1.25 × 104 cells/cm2.
Cell stimulation and treatment
Approx. 40–48 h after transient transfection, the cells were serum-starved (0.5 % FBS in DMEM) for 4–6 h to facilitate the accumulation of inactive EGFRs at the cell surface. The cells were then stimulated with 100 ng/ml EGF (17 nM; Sigma), indicated concentrations of AR (R&D Systems; 98-amino-acid-long form) or 79 ng/ml BTC (8.5 nM; R&D Systems) for the times shown.
To prepare cell lysates, Triton X-100 lysis buffer [50 mM Tris, pH 7.5, 150 mM sodium chloride, 0.5 % Triton X-100 (Fluka), 1 mM sodium orthovanadate, 10 mM sodium fluoride, 1 mM PMSF, 0.07 trypsin inhibitor units of aprotinin/ml and 1 μg/ml each of leupeptin, pepstatin, antipain and chymostatin (Sigma)] was used as described in [28,35]. Lysate protein concentrations were determined using the Bio-Rad Protein Assay reagent (Hercules, CA, U.S.A.) with BSA as the protein standard.
Immunoprecipitation and immunoblotting
Procedures for immunoprecipitation and immunoblotting were described previously [28]. Cell lysate proteins and immunoprecipitates were resolved by SDS/PAGE. Proteins were transferred on to PVDF membranes (Immobilon-P; Millipore) in Caps buffer {10 mM Caps [3-(cyclohexylamino)propane-1-sulfonic acid], pH 11 (Sigma), 10 % (v/v) methanol and 0.01 % SDS (Bio-Rad Laboratories)}. Prior to immunoblotting, the filters were blocked with 2 % (w/v) non-fat dried skimmed milk powder or gelatin (Bio-Rad Laboratories) in 10 mM Tris (pH 8.0), 150 mM NaCl and 0.025 % sodium azide. Proteins immobilized on PVDF were detected using the antibodies indicated, followed by detection with Western Lightning™ Chemiluminescence Reagent Plus (PerkinElmer Life Sciences). The PVDF membranes were stripped for reprobing. To detect ubiquitinated EGF-R, the filters were rinsed thoroughly in water after protein transfer and autoclaved in water for 10 min, followed by blocking and immunoblotting as described in [28]. ScionImage was used for quantitative comparison of immunoblot signals.
Down-regulation assays
At 48 h post-transfection, live cells were harvested from culture plates at 4 °C in 0.5 mM EDTA. The cells were immunostained prior to flow cytometry analysis, as described in [28,35]. In brief, cell samples were processed in triplicate for surface labelling using isotype-matched murine IgG2a anti-EGFR, anti-Syk (negative control) and anti-class I (positive control) antibodies and (R)-phycoerythrin-conjugated goat anti-mouse secondary antibody. A Becton Dickinson FACScan with CellQuest Pro software was used to acquire and analyse data from 5000 GFP-positive cells from each sample pool. Readouts reflected the specific and non-specific binding of each antibody. For each cell population, the specific EGFR MFI (mean fluorescence intensity) was obtained by subtracting the pool’s anti-Syk MFI value (non-specific immunostaining) from its anti-EGF-R MFI value.
Immunofluorescence
COS-7 cells were transiently transfected as described above. Cell samples were harvested and processed for immunostaining and immunofluorescence. Briefly, plates were placed on ice, rinsed in cold TBS [Tris-buffered saline; 140 mM NaCl (Fisher Scientific), 2.7 mM KCl (Fisher Scientific) and 25 mM Tris base (Research Products International)], fixed in fixing solution [3 % (w/v) paraformaldehyde (Electron Microscopy Sciences)/2 % (w/v) sucrose (Amresco) in PBS] and rinsed twice in PBS. Cells were then permeabilized by the addition of cold 0.05 % Triton X-100 permeabilization reagent [0.05 % Triton X-100 (Fluka), 20 mM Hepes (Gibco BRL), 3 mM MgCl2, 10 % sucrose and 1 % sodium chloride (Fisher Scientific)]. The cells were rinsed twice in cold PBS and then blocked in 1 %BSA (Amresco) in PBS. After blocking, the coverslips were rinsed in PBS and placed in primary antibodies diluted in blocking solution. This incubation was followed by four rinses in PBS and incubation with the appropriate secondary antibodies diluted in blocking solution. Coverslips were then rinsed and mounted on to microscope slips in Vectashield mounting solution (Vector Laboratories). Images were viewed and captured using a Zeiss 510 confocal microscope.
RESULTS
EGF-R ubiquitination levels do not correlate strictly with the extent of receptor degradation
To compare the EGF family ligands AR, BTC and EGF, we evaluated their impact on EGF-R ubiquitination, tyrosine phosphorylation and degradation. We also determined whether Cbl overexpression would enhance EGF-R degradation following cell stimulation with BTC and AR, thereby implicating Cbl as a regulator of EGF-R fate downstream of these ligands. HEK-293 cells expressing EGF-R and either GFP or GFP–Cbl WT were treated with AR, EGF or BTC for 0–90 min. This time course allowed for the observation of both receptor stimulation and degradation. The concentrations of EGF (17 nM) and BTC (8.5 nM) used here achieved maximal levels of ligand-specific EGF-R phosphorylation, down-regulation and degradation (results not shown). The presence of ubiquitinated receptors throughout the 90-min time course reflected the continuous stimulation and modification of a subset of cell-surface receptors over the ligand incubation period.
As shown in Figure 1, BTC and EGF induced significant receptor degradation in the absence of ectopic Cbl expression, while AR did not (Figure 1A, compare lanes 4, 7 and 10; Figure 1B, white bars). Ectopic expression of GFP–Cbl WT enhanced EGF-R ubiquitination for all of the ligands analysed (compare lanes 2–4 with 12–14, 5–7 with 15–17 and 8–10 with 18–20). Notably, the enhancement of EGF-R ubiquitination correlated with statistically significant enhancement of EGF-R degradation only in the case of EGF-stimulated cells (Figure 1B, compare matched white and grey bars).
Figure 1. Equimolar concentrations of AR and EGF induce different levels of EGF-R ubiquitination, phosphorylation and degradation.
HEK-293 cells were transiently transfected with cDNA encoding EGF-R (0.05 μg) and either GFP (3 μg) or GFP–Cbl WT (4 μg). At 48 h post-transfection, cells were serum-starved and incubated without (0 nM) or with AR (17 nM), BTC (8.5 nM) or EGF (17 nM) for 10, 40 or 90 min. Cell lysate proteins and immunoprecipitates (IP) were gel-resolved, transferred on to PVDF and immunoblotted (IB) with the antibodies indicated. (A) Immunoblotting results from a representative experiment. Top arrow: ubiquitinated EGF-R; middle arrow, non-ubiquitinated EGF-R; bottom arrow, GFP–Cbl; *, phosphorylated Hrs, a marker for EGF-R complex trafficking to early endosomes. (B) Quantitative analysis of three independent experiments, showing the amount of total cellular EGF-R remaining after receptor stimulation by the ligands (Amphi, amphiregulin). Results are means ± S.D. A two-tailed Student’s t test with α=0.05 determined that GFP–Cbl expression significantly affected EGF-R degradation only in the case of receptors activated by EGF (**).
While EGF-R ubiquitination in BTC-treated, GFP-expressing cells was minimal, receptor degradation was clearly detectable; however, AR treatment of GFP–Cbl WT-expressing cells induced greater EGF-R ubiquitination with less degradation (Figure 1A, top two panels, compare lanes 5–7 with lanes 12–14; Figure 1B). Thus the extent of ligand-induced EGF-R degradation did not correlate strictly with the level of receptor ubiquitination.
Another difference between the ligand-activated receptors was the induced level of EGF-R tyrosine phosphorylation. BTC and EGF effected greater total receptor phosphorylation than did AR (Figure 1A, Lysate, pTyr), either in the presence or absence of ectopically expressed Cbl. For equimolar AR and EGF in the absence of GFP–Cbl, our results were similar to those reported by others [12,23,24]. Differential ligand stimulation also resulted in unequal levels of Cbl phosphorylation (bottom arrow) and Hrs tyrosine phosphorylation (asterisk), which serves as a marker for EGF-R trafficking to early endosomes [36]. The identity of the phospho-Hrs species was confirmed using antibodies specific for phospho-Tyr334 Hrs (results not shown) [34].
Although they were not targeted for degradation, AR-stimulated receptors retained the ability to associate with Cbl in a ligand-dependent manner (Figure 1, lanes 12–14). However, Cbl binding was reduced by approx. 50 %, relative to the levels induced by EGF or BTC (Figure 1A, compare lane 12 with lanes 15 and 18, EGF-R IP, GFP–Cbl IB).
AR is a partial agonist at the EGF-R
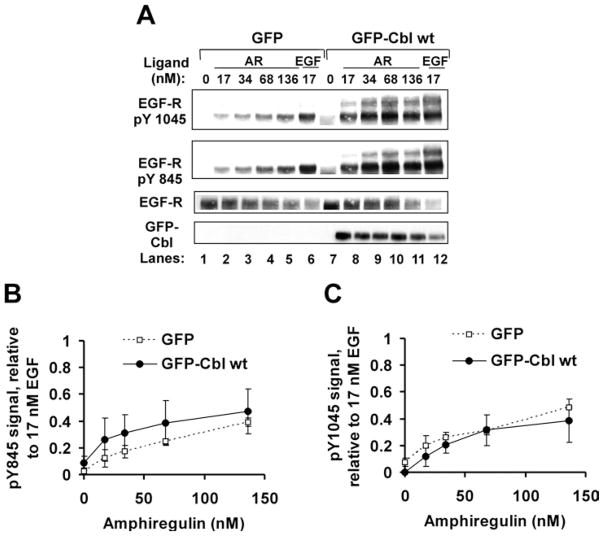
Because AR was clearly less effective than equimolar EGF at activating EGF-R and targeting it for degradation, we performed dose–response experiments to facilitate comparison of the two ligands at doses effecting similar levels of receptor modification. HEK-293 cells expressing EGF-R and either GFP or GFP–Cbl WT were treated for 10 min with 17 nM EGF or AR over the range of 0–136 nM. At the 10 min time point, EGF-induced receptor activation is maximal. Cell lysate proteins and anti-EGF-R immunoprecipitates were prepared and processed for detection of EGF-R activation, as reflected by receptor phosphotyrosine content (Figures 2A and 2B), ubiquitination (Figures 2A and 2C), association with Cbl (Figures 2A and 2D) and site-specific tyrosine phosphorylation (Figures 3A–3C).
Figure 2. Human recombinant AR is a partial agonist at the EGF-R with approx. 40 % efficacy for total EGF-R tyrosine phosphorylation, ubiquitination and co-precipitation of Cbl.
HEK-293 cells were transiently transfected with cDNA encoding EGF-R (0.05 μg) and either GFP (3 μg) or GFP–Cbl WT (4 μg). The cells were serum-starved and then harvested without further treatment (0 nM ligand) or following treatment for 10 min with the indicated ligands at the concentration shown. (A) Immunoblotting results from a representative experiment. Anti-EGFR immunoprecipitates and cell lysate samples were resolved by SDS/PAGE, transferred on to PVDF and immunoblotted with the antibodies shown. Asterisks (*) mark the position of ubiquitinated EGF-R. (B–D) Quantitative analysis of three independent experiments. Results are means ± S.D. (B) Total induced EGF-R tyrosine phosphorylation (pY) reaches a plateau upon cell stimulation with approx. 34 nM AR. The phosphotyrosine content of immunoprecipitated receptors was normalized to EGF-R levels. (C) Total induced EGF-R ubiquitination reaches a plateau upon cell stimulation with approx. 68 nM AR. The ubiquitin content of immunoprecipitated receptors was normalized to the total amount of EGF-R present. (D) Cbl co-precipitation (co-IP) with EGF-R reaches a plateau upon cell stimulation with approx. 68 nM AR. The amount of EGF-R-associated Cbl was normalized to the total amount of EGF-R present.
Figure 3. AR is a partial agonist for site-specific EGF-R tyrosine phosphorylation.
HEK-293 cells were transiently transfected with cDNA encoding EGF-R (0.05 μg) and GFP–Cbl WT (4 μg) or GFP (3 μg). At 48 h post-transfection, cells were serum-starved and incubated without (0) or with EGF (17 nM) or AR (17–136 nM) for 10 min. Cell lysate proteins (100 μg per lane) were gel-resolved, transferred on to PVDF membrane and immunoblotted with the indicated antibodies. (A) Immunoblotting results from a representative experiment. (B, C) Quantitative analysis of phospho-Tyr845 and phospho-Tyr1045 levels in treated cells. In each case, the phosphotyrosine signal was normalized to the corresponding EGF-R level. Results are means ± S.D. for three independent experiments, except in (C), ● (two independent experiments).
For each activation readout, the dose–response curve for increasing amounts of AR reached a plateau at approx. 40 % of the level achieved by EGF. We concluded that AR is a partial agonist at the EGF-R with 40 % efficacy, relative to EGF, for the induction of EGF-R tyrosine phosphorylation, ubiquitination and binding to Cbl.
The limited site-specific tyrosine phosphorylation of EGF-R Tyr1045 was particularly noteworthy, as this modified residue is the recruitment site for Cbl’s tyrosine kinase-binding domain [27]. Impaired phosphorylation of this site following cell activation with AR might explain, at least in part, the observed decrease in EGF-R–Cbl interactions (Figures 2A and 2D).
AR treatment leads to impaired EGF-R down-regulation and degradation
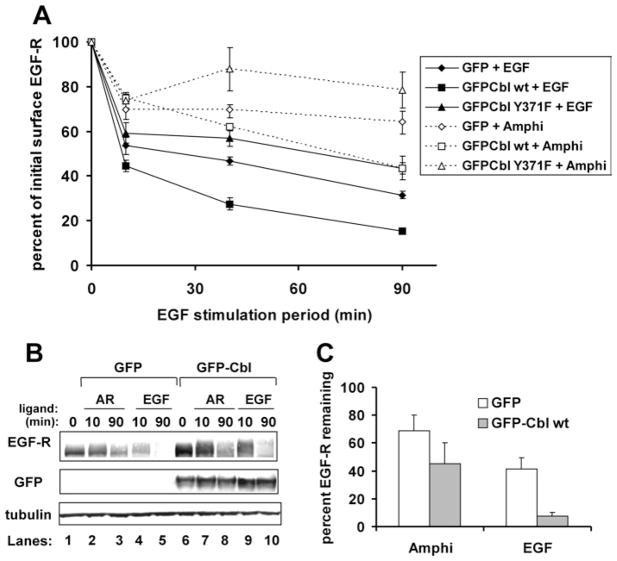
As saturating AR induced modest EGF-R phosphorylation, ubiquitination and Cbl recruitment to the receptor, we wished to determine its impact on EGF-R down-regulation. Here, down-regulation refers to the loss of receptors from the cell surface. We and others previously showed that EGF-induced EGF-R down-regulation and/or degradation is enhanced by ectopic expression of WT Cbl, but inhibited by expression of the E3-defective Cbl mutant Y371F [26,28,34,37]. These overexpression studies correctly implicated Cbl as an endogenous regulator of EGF-induced receptor down-regulation and degradation. In order to determine whether Cbl modulates AR-induced EGF-R down-regulation, HEK-293 cells were transfected to express EGF-R in combination with GFP, GFP–Cbl WT or dominant-negative GFP–Cbl Y371F. Serum-starved cells were incubated over a 90-min time course with or without saturating levels of ligand (17 nM EGF and 136 nM AR). The relative capacity of AR and EGF to effect receptor down-regulation in the absence (GFP) or presence of ectopically expressed Cbl proteins was reflected by their associated down-regulation curves (Figure 4A).
Figure 4. Relative to EGF treatment, AR treatment impairs EGFR down-regulation and degradation.
HEK-293 cells were transiently transfected with cDNA encoding EGF-R WT (0.05 μg) and GFP (3.2 μg), GFP–Cbl WT (4 μg) or GFP–Cbl Y371F (4 μg). (A) Down-regulation assay. At 48 h post-transfection, the cells were serum-starved and incubated at 37 °C without (0 min) or with EGF (17 nM) or AR (136 nM) for 10, 40 or 90 min. Cells were harvested intact on ice, plated in triplicate and stained with anti-EGF-R, anti-Syk (isotype-matched negative control) or anti-MHC Class I (isotype-matched positive control) antibodies. This was followed by incubation with phycoerythrin-conjugated secondary antibody and paraformaldehyde fixation. The surface phycoerythrin signals were analysed by flow cytometry for 5000 GFP-positive cells per sample. Surface receptor levels for each sample were determined by subtracting the mean fluorescence index of the anti-Syk-stained cells from the MFI of the matched anti-EGF-R-stained cells. For each transfection condition, the surface EGFR levels are expressed as a percentage of the EGF-R signal of unstimulated, matched transfection plates. Results are means ± S.D. for three independent experiments. (B) EGF-R degradation assay. A 100 μg cell lysate protein sample was loaded per lane, gel-resolved, transferred on to PVDF and immunoblotted as indicated. Results shown are from a representative experiment employing 17 nM EGF and 136 nM AR. (C) Quantitative analysis of three independent degradation experiments, showing the average amount of total cellular EGF-R remaining after 90 min of receptor stimulation by ligand. Results are means ± S.D.
AR was a poorer inducer of EGF-R down-regulation, regardless of the GFP or GFP–Cbl protein expressed (compare broken lines with solid lines in Figure 4A). Ectopic expression of GFP–Cbl WT enhanced both AR- and EGF-induced EGF-R down-regulation relative to the respective GFP controls, indicating a regulatory role for Cbl in both processes. However, AR treatment maintained higher cell-surface EGF-R levels than were seen in EGF-treated cells expressing the dominant-negative Cbl mutant Y371F (Figure 4A, compare all broken lines with the solid line with triangles). These results indicated that, relative to EGF, AR is impaired for inducing Cbl-modulated EGF-R down-regulation.
It was possible that these differences in down-regulation did not translate to differences in EGF-R degradation. To test this, HEK-293 cells expressing EGF-R, in combination with GFP or GFP–Cbl WT, were stimulated with AR (136 nM) or EGF (17 nM) over a 90-min time course. Lysate proteins from the cells were evaluated for total EGF-R content by immunoblotting. As shown in Figures 4(B) and 4(C), EGF induced significantly greater receptor degradation than did saturating amounts of AR, either in the presence or absence of ectopically expressed Cbl. We concluded that AR is compromised for effecting both EGF-R down-regulation and degradation.
Sprouty2 degradation is impaired in AR-treated cells
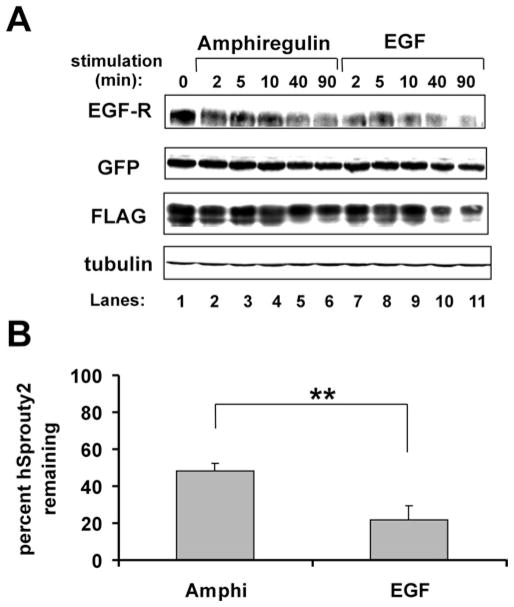
The inability of AR to induce EGF-R down-regulation to the same extent as EGF suggested that AR might impair EGF-R internalization or enhance the recycling of internalized receptors. hSpry2 is a known suppressor of Cbl-enhanced EGF-R internalization [38]. We hypothesized that AR treatment reduces hSpry2 degradation, relative to EGF treatment, thereby decreasing receptor internalization.
To test this idea, HEK-293 cells expressing EGF-R, GFP–Cbl WT and FLAG–hSpry2 were treated with either AR or EGF and then were evaluated for hSpry2 degradation (Figure 5). In concurrence with our previous observations [35], hSpry2 degradation was readily detectable following cell stimulation with EGF (Figure 5A, lanes 7–11). AR treatment induced less hSpry2 degradation over the 90-min time course of stimulation (Figure 5A, lanes 2–6, and Figure 5B). We concluded that AR and EGF can differentially regulate hSprouty2 at the EGF-R internalization checkpoint. This may explain, at least in part, the different impacts of AR and EGF on EGF-R down-regulation.
Figure 5. hSpry2 degradation is impaired in AR-treated cells.
HEK-293 cells were transiently transfected with cDNA encoding EGF-R (0.05 μg), GFP–Cbl WT (4 μg) and FLAG–hSprouty2 (2 μg). At 48 h post-transfection, cells were serum-starved and incubated without (0 min) or with AR (136 nM) or EGF (17 nM) for 2, 5, 10, 40 or 90 min. (A) Immunoblotting results from a representative experiment. A 100 μg cell lysate protein sample was loaded per lane, gel-resolved, transferred on to PVDF and immunoblotted as indicated. (B) Quantitative analysis of three independent degradation experiments, showing the average amount of total cellular hSprouty2 remaining after 90 min of receptor stimulation by ligand. Results are means ± S.D. A two-tailed Student’s t test with α=0.05 determined that a significantly different amount of hSprouty2 degradation was effected by AR compared with EGF (**).
AR treatment fails to induce EGF-R and Cbl co-localization on endosomes
Defects in hSpry2 degradation and EGF-R down-regulation suggested that AR-activated receptors might not be internalized efficiently. To investigate this possibility, subcellular protein co-localization studies were undertaken. GFP–Cbl-expressing COS-7 cells, which contain significant amounts of endogenous EGF-R and exhibit a flatter morphology than HEK-293 cells, were used for these experiments. The cells were serum-starved, treated for various time periods with AR or EGF, fixed and then immunostained to permit visualization of GFP–Cbl WT and EGF-R (Figure 6).
Figure 6. AR treatment leads to the formation of EGF-R-positive vesicles that lack associated GFP–Cbl.

COS-7 cells were transiently transfected with cDNA encoding GFP–Cbl WT (4 μg). At 48 h post-transfection, cells were serum-starved and incubated without ligand (0 min) or with AR (136 nM) or EGF (17 nM) for 5 or 25 min. Following stimulation, the cells were paraformaldehyde-fixed, permeabilized and stained with anti-EGF-R antibody. Scale bar, 20 μm. Images shown are representative results from one experiment. Three independent experiments were performed, with evaluation of more than 20 cells per ligand condition per experiment.
As observed previously [28], Cbl and EGF-R co-localized on the endocytic vesicles of EGF-treated cells over an extended time course of receptor activation (compare Figures 6a and 6b with Figures 6g–6j). The progressive enlargement and perinuclear accumulation of EGF-R–Cbl-positive vesicles were evident as the protein complexes transited the endocytic pathway. Vesicles containing both EGF-R and Cbl also carried the early endosome marker EEA1 (results not shown).
AR treatment resulted in the formation of relatively few, small EGF-R-positive punctae after 5 min of cell stimulation (Figure 6c). Co-staining for EEA1 revealed that some of these structures were early endosomes (results not shown). Thus a saturating concentration of AR enabled a subset of stimulated receptors to traffic to early endosomes. However, GFP–Cbl did not co-localize with endosomal EGF-R (Figure 6d). Instead, GFP–Cbl remained largely cytosolic. This observation was consistent with our protein biochemistry results, which revealed reduced EGF-R–Cbl complex formation early after cell stimulation with AR (Figure 2).
At the 25-min time point in AR-stimulated cells, co-localization of GFP–Cbl and EGF-R on endosomes remained minimal (Figures 6e and 6f). Cbl-positive membrane ruffles were prominent in nearly all of the cells (results not shown). Others have reported that the elevation of cellular hSpry2 levels leads to the accumulation of hSpry2 and Cbl in membrane ruffles [39]. The detection of Cbl in AR-induced membrane ruffles complemented our protein biochemistry results, which showed reduced hSpry2 degradation following cell treatment with AR (Figure 5). Collectively, our results indicated that the AR-induced defect in EGF-R endocytic trafficking could be attributed to several ligand-specific outcomes, including (i) the failure of Cbl to degrade hSpry2 efficiently and (ii) the failure of Cbl to associate with hypophosphorylated EGF-R on early endosomes.
DISCUSSION
EGF family ligands can induce distinct biochemical and biological outcomes. The ligands exhibit different potencies for inducing receptor modification and cell growth. This may be due, in part, to their different receptor-binding affinities. Others have shown that AR binds the EGF-R with much lower affinity than does EGF, BTC or TGF-α [19,21]. Theoretically, the reduced EGF-R tyrosine phosphorylation detected in AR-stimulated cells [12,23–25] might arise solely from the ligand’s low receptor-binding affinity.
Our results address this issue. We report that human recombinant AR is a partial agonist at the EGF-R, with approx. 40 % efficacy relative to EGF for inducing receptor tyrosine phosphorylation, ubiquitination, association with Cbl and degradation. Our dose–response experiments revealed that superphysiological concentrations of AR [12] could not effect EGF-R modification to the extent seen with EGF at a high but physiological concentration (Figures 2 and 3). Because increasing amounts of AR failed to shift efficacy readouts beyond their plateau levels, we conclude that low receptor-binding affinity is not the sole factor limiting the efficacy of AR. This conclusion assumes that cell-surface EGFRs are comparably saturated in the presence of 17 nM EGF and 136 nM AR.
The differential efficacy of EGF-R ligands is reminiscent of the different efficacies of ErbB4 ligands. Riese and coworkers showed that neuregulin NRG2β is a potent activator of ErbB4 tyrosine phosphorylation, while NRG2α is not [40]. NRG2β and NRG2α are of high and low efficacy respectively for ErbB4 phosphorylation and IL-3-independent cell growth in BaF3 cells expressing the receptor [40]. Structure–function studies revealed that amino acids in the C-terminus of NRG2β are critical determinants of efficacy, probably through their interaction with ErbB4 [41]. The AR structural features associated with low efficacy of EGF-R activation have not been mapped, but investigations employing EGF–AR chimeras may be useful to define the basis for AR’s function as a partial agonist at the EGF-R.
The endocytic trafficking of ligand-activated EGF-R has been investigated extensively for EGF and TGF-α. However, little is known about EGF-R trafficking induced by the ligands BTC and AR. Our study is the first to identify molecular events, other than receptor phosphorylation, that may influence AR-induced receptor fate. Compared with EGF, AR effects less Sprouty2 degradation (Figure 5) and reduced phosphorylation of EGF-R Tyr1045 (Figures 3A and 3C), which is the binding site for Cbl’s tyrosine kinase-binding domain. One or both of these mechanisms may be responsible for decreased Cbl–EGF-R complex formation in AR-stimulated cells (Figures 2A and 2D). AR-stimulated EGF-R can be internalized, but without associated Cbl (Figure 6). We recently showed that Cbl regulates Hrs and EGF-R fate at the level of endosomal sorting [34], and others have suggested that Cbl–EGF-R complexes must remain intact throughout endocytosis for EGF-R degradation to occur [42–44]. Therefore the AR-associated defect in EGF-R degradation may be due to the failure to recruit Cbl to Hrs-regulated sorting sites on EGF-R-positive endosomes.
It is possible that Cbl-free, AR-activated receptors are internalized and then dissociate from AR early in the process of endocytosis. Previous work by others indicates that such a pH-dependent regulation occurs in the case of TGF-α-stimulated receptors, leading to receptor inactivation and enhanced receptor recycling [43]. However, this has not yet been demonstrated for AR.
What are the potential biological implications of differential ligand-induced EGF-R trafficking and turnover? EGF-R signalling is modulated throughout the endocytic trafficking pathway. EGF-R at the cell surface [45] and in membrane ruffles [46] activates PLC-γ1 (phospholipase C-γ1), inducing its phosphorylation and phosphatidylinositol 4,5-bisphosphate hydrolysis. Grb2 (growth-factor-receptor-bound protein 2) is an adaptor of the Ras signalling pathway that preferentially binds cell-surface EGF-R [44]. Its partner Shc (Src homology and collagen homology) remains associated with EGF-R as it moves from the surface to late endosomes [44,47,48]. These proteins may be recruited, directly or indirectly, to defined EGF-R phosphotyrosine residues as the receptors undergo endocytosis. Because the various EGF family ligands can induce differential receptor phosphorylation and trafficking, it is predicted that they also will induce differential signalling through the PLC-γ1 and Ras/MAPK (mitogen-activated protein kinase) pathways, with associated differences in cell growth regulation. The work presented in this paper does not address this issue. However, others have identified signalling pathways that are modulated in a ligand-specific manner by EGF family members [12,17,49].
Our investigation raises a final point. The conjugation of mono-ubiquitin to EGF-R can enhance receptor down-regulation and degradation [29,50]. Considering only the protein biochemistry results obtained from GFP–Cbl-overexpressing cells, EGF-R degradation might appear to correlate with EGF-R ubiquitination (Figure 1, Ub IB, compare lanes 12–14 with 15–20). However, a comparison of the BTC-stimulated cells expressing GFP with the AR-stimulated cells expressing GFP–Cbl WT showed that the receptor ubiquitination levels do not correlate strictly with receptor degradation (Figure 1, Ub and EGF-R IB, compare lanes 5–7 with 12–14 respectively). At least two interpretations of this finding are possible. First, BTC activation of EGF-R, in the absence of ectopically expressed Cbl, may lead to receptor down-regulation and degradation through an unusual mechanism that is independent of extensive EGF-R ubiquitination. Alternatively, degradative targeting of EGF-R by several ligands may actually require only modest receptor ubiquitination, along with additional downstream regulation of EGF-R, Cbl and/or the endocytic trafficking machinery. In the latter case, limited EGF-R ubiquitination could be sufficient to tether activated EGF-R–Cbl complexes to the UIM of the endosomal trafficking regulator Hrs. This would facilitate downstream regulatory events, including the phosphorylation and ubiquitination of Hrs. Both post-translational modifications are Cbl-regulated and linked to efficient EGF-R degradation [34]. Hrs tyrosine phosphorylation is particularly attractive as a ligand-gated EGF-R trafficking checkpoint, given that it was poorly induced by receptor activation with AR but more robustly affected by EGF and BTC, two ligands that cause EGF-R degradation (Figure 1, and results not shown). Future investigations will determine how the various EGF family ligands regulate the Hrs checkpoint.
Acknowledgments
Cells and plasmids used in the present study were gifts from David Spector, Fred Quelle and Hamid Band. We thank John Koland, Barry Kasson and Gina Visser Smit for helpful discussions on this work. Excellent technical support was provided by Jeremy Hoffmann, Andrew Place, Suying Liu and Joe Place. We gratefully acknowledge assistance from the staff of the Roy J. and Lucille A. Carver College of Medicine Central Microscopy Research and Flow Cytometry Facilities. Funding for this work was provided by: Research Scholar Grant RSG-03-046-01 from the American Cancer Society and 1 RO1 CA109685-01A2 from the National Cancer Institute, NIH (National Institutes of Health) (to N. L. L.); institutional training grants NRSA T32 DEO14678-04 and DEO14678-03 [from the NIDCR (National Institute of Dental and Craniofacial Research) to Christopher Squier and the Dows Institute and College of Dentistry at the University of Iowa, for support of K. A. S. and N. L. L. respectively]; and award DAMD17-98-1-8038 from the U.S. Department of Defense Breast Cancer Research Program (to N. L. L.). The content of this information does not necessarily reflect the position or the policy of the U.S. Government, the National Cancer Institute, or the National Institutes of Health, and no official endorsement should be inferred.
Abbreviations used
- AR
amphiregulin
- BTC
betacellulin
- DMEM
Dulbecco’s modified Eagle’s medium
- EEA1
early endosome antigen 1
- EGF
epidermal growth factor
- EGF-R
EGF receptor
- FBS
fetal bovine serum
- HEK-293 cell
human embryonic kidney cell
- Hrs
hepatocyte growth factor-regulated tyrosine kinase substrate
- hSpry2
human Sprouty2 IL, interleukin
- MFI
mean fluorescence intensity
- PLC-γ1
phospholipase C-γ1
- RING
really interesting new gene
- TGF-α
transforming growth factor-α
- UIM
ubiquitin-interacting motif
- WT
wild-type
References
- 1.Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 2.Xian CJ, Li L, Deng YS, Zhao SP, Zhou XF. Lack of effects of transforming growth factor-α gene knockout on peripheral nerve regeneration may result from compensatory mechanisms. Exp Neurol. 2001;172:182–188. doi: 10.1006/exnr.2001.7771. [DOI] [PubMed] [Google Scholar]
- 3.Abbott BD, Lin TM, Rasmussen NT, Albrecht RM, Schmid JE, Peterson RE. Lack of expression of EGF and TGF-α in the fetal mouse alters formation of prostatic epithelial buds and influences the response to TCDD. Toxicol Sci. 2003;76:427–436. doi: 10.1093/toxsci/kfg238. [DOI] [PubMed] [Google Scholar]
- 4.Troyer KL, Luetteke NC, Saxon ML, Qiu TH, Xian CJ, Lee DC. Growth retardation, duodenal lesions, and aberrant ileum architecture in triple null mice lacking EGF, amphiregulin, and TGF-α. Gastroenterology. 2001;121:68–78. doi: 10.1053/gast.2001.25478. [DOI] [PubMed] [Google Scholar]
- 5.Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGFα deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- 6.Mann GB, Fowler KJ, Gabriel A, Nice EC, Williams RL, Dunn AR. Mice with a null mutation of the TGFα gene have abnormal skin architecture, wavy hair, and curly whiskers and often develop corneal inflammation. Cell. 1993;73:249–261. doi: 10.1016/0092-8674(93)90227-h. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz EA, Seroogy KB, Schroeder JA, Russell WE, Evans EP, Riedel RF, Phillips HK, Harrison CA, Lee DC, Luetteke NC. Characterization of the mouse transforming growth factor α gene: its expression during eyelid development and in waved 1 tissues. Cell Growth Differ. 1996;7:1271–1282. [PubMed] [Google Scholar]
- 8.Xian CJ, Zhou XF. EGF family of growth factors: essential roles and functional redundancy in the nerve system. Front Biosci. 2004;9:85–92. doi: 10.2741/1210. [DOI] [PubMed] [Google Scholar]
- 9.Schuger L, Johnson GR, Gilbride K, Plowman GD, Mandel R. Amphiregulin in lung branching morphogenesis: interaction with heparan sulfate proteoglycan modulates cell proliferation. Development. 1996;122:1759–1767. doi: 10.1242/dev.122.6.1759. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai H, Tsukamoto T, Kjelsberg CA, Cantley LG, Nigam SK. EGF receptor ligands are a large fraction of in vitro branching morphogens secreted by embryonic kidney. Am J Physiol. 1997;273:F463–F472. doi: 10.1152/ajprenal.1997.273.3.F463. [DOI] [PubMed] [Google Scholar]
- 11.Berasain C, Garcia-Trevijano ER, Castillo J, Erroba E, Santamaria M, Lee DC, Prieto J, Avila MA. Novel role for amphiregulin in protection from liver injury. J Biol Chem. 2005;280:19012–19020. doi: 10.1074/jbc.M413344200. [DOI] [PubMed] [Google Scholar]
- 12.Kansra S, Stoll SW, Johnson JL, Elder JT. Autocrine extracellular signal-regulated kinase (ERK) activation in normal human keratinocytes: metalloproteinase-mediated release of amphiregulin triggers signaling from ErbB1 to ERK. Mol Biol Cell. 2004;15:4299–4309. doi: 10.1091/mbc.E04-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kansra S, Stoll SW, Johnson JL, Elder JT. Src family kinase inhibitors block amphiregulin-mediated autocrine ErbB signaling in normal human keratinocytes. Mol Pharmacol. 2005;67:1145–1157. doi: 10.1124/mol.104.004689. [DOI] [PubMed] [Google Scholar]
- 14.Normanno N, Bianco C, De Luca A, Salomon DS. The role of EGF-related peptides in tumor growth. Front Biosci. 2001;6:D685–D707. doi: 10.2741/normano. [DOI] [PubMed] [Google Scholar]
- 15.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 16.Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281:37728–37737. doi: 10.1074/jbc.M606532200. [DOI] [PubMed] [Google Scholar]
- 17.Streicher KL, Willmarth NE, Garcia J, Boerner JL, Dewey TG, Ethier SP. Activation of a nuclear factor κB/interleukin-1 positive feedback loop by amphiregulin in human breast cancer cells. Mol Cancer Res. 2007;5:847–861. doi: 10.1158/1541-7786.MCR-06-0427. [DOI] [PubMed] [Google Scholar]
- 18.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Shoyab M, Plowman GD, McDonald VL, Bradley JG, Todaro GJ. Structure and function of human amphiregulin: a member of the epidermal growth factor family. Science. 1989;243:1074–1076. doi: 10.1126/science.2466334. [DOI] [PubMed] [Google Scholar]
- 20.Chung E, Graves-Deal R, Franklin JL, Coffey RJ. Differential effects of amphiregulin and TGF-α on the morphology of MDCK cells. Exp Cell Res. 2005;309:149–460. doi: 10.1016/j.yexcr.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Neelam B, Richter A, Chamberlin SG, Puddicombe SM, Wood L, Murray MB, Nandagopal K, Niyogi SK, Davies DE. Structure–function studies of ligand-induced epidermal growth factor receptor dimerization. Biochemistry. 1998;37:4884–4891. doi: 10.1021/bi972548x. [DOI] [PubMed] [Google Scholar]
- 22.Adam R, Drummond DR, Solic N, Holt SJ, Sharma RP, Chamberlin SG, Davies DE. Modulation of the receptor binding affinity of amphiregulin by modification of its carboxyl terminal tail. Biochim Biophys Acta. 1995;1266:83–90. doi: 10.1016/0167-4889(94)00224-3. [DOI] [PubMed] [Google Scholar]
- 23.Beerli RR, Hynes NE. Epidermal growth factor-related peptides activate distinct subsets of ErbB receptors and differ in their biological activities. J Biol Chem. 1996;271:6071–6076. doi: 10.1074/jbc.271.11.6071. [DOI] [PubMed] [Google Scholar]
- 24.Riese DJ, Kim ED, Elenius K, Buckley S, Klagsbrun M, Plowman GD, Stern DF. The epidermal growth factor receptor couples transforming growth factor-α, heparin-binding epidermal growth factor-like factor, and amphiregulin to Neu, ErbB-3, and ErbB-4. J Biol Chem. 1996;271:20047–20052. doi: 10.1074/jbc.271.33.20047. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore JL, Scott JA, Bouizar Z, Robling A, Pitfield SE, Riese DJ, II, Foley J. Amphiregulin–EGFR signaling regulates PTHrP gene expression in breast cancer cells. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 28.Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Jr, Miyake S, Meissner-Lula N, Hsu VW, Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem. 2000;275:367–377. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- 29.Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- 30.Haglund K, Shimokawa N, Szymkiewicz I, Dikic I. Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc Natl Acad Sci USA. 2002;99:12191–12196. doi: 10.1073/pnas.192462299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubin C, Litvak V, Medvedovsky H, Zwang Y, Lev S, Yarden Y. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr Biol. 2003;13:297–307. doi: 10.1016/s0960-9822(03)00053-8. [DOI] [PubMed] [Google Scholar]
- 32.Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr Biol. 2003;13:308–314. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 33.Urbe S, Sachse M, Row PE, Preisinger C, Barr FA, Strous G, Klumperman J, Clague MJ. The UIM domain of Hrs couples receptor sorting to vesicle formation. J Cell Sci. 2003;116:4169–4179. doi: 10.1242/jcs.00723. [DOI] [PubMed] [Google Scholar]
- 34.Stern KA, Visser Smit GD, Place TL, Winistorfer S, Piper RC, Lill NL. Cbl enhances epidermal growth factor receptor degradation by modulating Hrs tyrosine phosphorylation. Mol Cell Biol. 2007;27:888–898. doi: 10.1128/MCB.02356-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visser GD, Lill NL. The Cbl RING finger C-terminal flank controls epidermal growth factor receptor fate downstream of receptor ubiquitination. Exp Cell Res. 2005;311:281–293. doi: 10.1016/j.yexcr.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 36.Urbe S, Mills IG, Stenmark H, Kitamura N, Clague MJ. Endosomal localization and receptor dynamics determine tyrosine phosphorylation of hepatocyte growth factor-regulated tyrosine kinase substrate. Mol Cell Biol. 2000;20:7685–7692. doi: 10.1128/mcb.20.20.7685-7692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 38.Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. EMBO J. 2002;21:4796–4808. doi: 10.1093/emboj/cdf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong ES, Lim J, Low BC, Chen Q, Guy GR. Evidence for direct interaction between Sprouty and Cbl. J Biol Chem. 2001;276:5866–5875. doi: 10.1074/jbc.M006945200. [DOI] [PubMed] [Google Scholar]
- 40.Hobbs SS, Coffing SL, Le AT, Cameron EM, Williams EE, Andrew M, Blommel EN, Hammer RP, Chang H, Riese DJ., II Neuregulin isoforms exhibit distinct patterns of ErbB family receptor activation. Oncogene. 2002;21:8442–8452. doi: 10.1038/sj.onc.1205960. [DOI] [PubMed] [Google Scholar]
- 41.Hobbs SS, Cameron EM, Hammer RP, Le AT, Gallo RM, Blommel EN, Coffing SL, Chang H, Riese DJ., II Five carboxyl-terminal residues of neuregulin2 are critical for stimulation of signaling by the ErbB4 receptor tyrosine kinase. Oncogene. 2004;23:883–893. doi: 10.1038/sj.onc.1207250. [DOI] [PubMed] [Google Scholar]
- 42.Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- 43.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burke P, Schooler K, Wiley HS. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haugh JM, Schooler K, Wells A, Wiley HS, Lauffenburger DA. Effect of epidermal growth factor receptor internalization on regulation of the phospholipase C-gamma1 signaling pathway. J Biol Chem. 1999;274:8958–8965. doi: 10.1074/jbc.274.13.8958. [DOI] [PubMed] [Google Scholar]
- 46.Diakonova M, Payrastre B, van Velzen AG, Hage WJ, van Bergen en Henegouwen PM, Boonstra J, Cremers FF, Humbel BM. Epidermal growth factor induces rapid and transient association of phospholipase Cγ1 with EGF-receptor and filamentous actin at membrane ruffles of A431 cells. J Cell Sci. 1995;108:2499–2509. doi: 10.1242/jcs.108.6.2499. [DOI] [PubMed] [Google Scholar]
- 47.Haugh JM, Huang AC, Wiley HS, Wells A, Lauffenburger DA. Internalized epidermal growth factor receptors participate in the activation of p21ras in fibroblasts. J Biol Chem. 1999;274:34350–34360. doi: 10.1074/jbc.274.48.34350. [DOI] [PubMed] [Google Scholar]
- 48.Oksvold MP, Skarpen E, Lindeman B, Roos N, Huitfeldt HS. Immunocytochemical localization of Shc and activated EGF receptor in early endosomes after EGF stimulation of HeLa cells. J Histochem Cytochem. 2000;48:21–33. doi: 10.1177/002215540004800103. [DOI] [PubMed] [Google Scholar]
- 49.Shin HS, Lee HJ, Nishida M, Lee MS, Tamura R, Yamashita S, Matsuzawa Y, Lee IK, Koh GY. Betacellulin and amphiregulin induce upregulation of cyclin D1 and DNA synthesis activity through differential signaling pathways in vascular smooth muscle cells. Circ Res. 2003;93:302–310. doi: 10.1161/01.RES.0000086803.64109.9E. [DOI] [PubMed] [Google Scholar]
- 50.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]