Abstract
Gene editing using single-stranded oligonucleotides (ODNs) can be used to reverse or create a single base mutation in mammalian cells. This approach could be used to treat genetic diseases caused, at least in part, by a nucleotide substitution. The technique could also be used as a tool to establish single base polymorphisms at multiple sites and thus aid in creating cell lines that can be used to define the basis for drug resistance in human cells. A troubling outcome of the gene-editing reaction is the effect on normal growth of cells that have undergone nucleotide exchange. In this work, we attempt to overcome this reduced proliferation phenotype by changing the method by which the ODN is introduced into the target cell. Using a series of assays that measure gene editing, DNA damage response, and cell viability, we report that chemically modified ODNs, the most active form of ODN for gene editing, can be used successfully if introduced into the cell by the method of nucleofection. Unlike electroporation, which has been used as the standard mode of ODN delivery, one new result is that nucleofection does not induce a dramatic loss of viability within the first 24 hours after the start of gene editing. In addition, and importantly, ODNs introduced to the cell by nucleofection do not activate the DNA damage response pathway as dramatically as ODNs introduced by electroporation. These 2 novel findings are encouraging for the application of gene editing in other systems. However, reduced proliferation phenotype is still observed when the population of corrected cells is monitored out to 8 days, and thus, delivery by nucleofection does not solve the proliferation problem encountered by cells bearing an edited gene.
Introduction
Gene editing using single-stranded oligonucleotides (ssODNs) is one of several approaches being used to correct or create single base changes in chromosomal genes (Liu, et al., 2003; Hu et al., 2005; Urnov et al., 2010; Khan et al., 2010; Wood et al., 2011). In this process, the ssODN binds in homologous register with a complementary target site except for one (predesigned) mispaired base at the center of the aligned strands. The mismatched base pair is resolved in such a way that the nucleotide in the chromosome is swapped out and a complementary (to the ssODN) base is inserted. This nucleotide exchange reaction is facilitated by endogenous enzymes functioning in the DNA repair, recombination and/or replication pathways (Brachman and Kmiec, 2005; Huen et al., 2006; Wang et al., 2006).
While the mechanism and regulation of ssODN-directed gene editing is partially understood and continues to be elucidated (Andersen et al., 2002; Parekh-Olmedo et al., 2005), the frequency with which the editing reaction takes place is still relatively low (1%–3%). This is true for all editing techniques, and therefore experimental protocols must evolve more fully if these approaches are to become a viable therapeutic or research tool. One way to evolve novel approaches is to recognize and more fully characterize barriers to successful implementation. Our lab and others have identified several of the hurdles that continue to impede the pathway toward clinical application (Olsen et al., 2005a; Olsen et al., 2005b; Engstrom and Kmiec, 2007; Olsen et al., 2009; Bonner and Kmiec, 2009; Bonner et al., 2012). Among the most challenging barrier is one in which cells, modified genetically by gene editing, exhibit a slow growth response after targeting, a phenomenon we call reduced proliferation phenotype (RPP). Some groups have argued with merit that the phosphorothioate modifications of the workhorse 72-mer ODN induce a toxic response in the cell and cause the cell to become refractory to growth (Andrieu-Soler et al., 2005; Papaioannou et al., 2009; Aarts and te Riele, 2010). However, the removal of these protective groups depresses the correction efficiency significantly (Papaioannou et al., 2009; Bonner et al., 2012), making it more improbable that such a low population of corrected cells will have a realistic downstream effect. Furthermore, any type of ssODN induces some degree of a DNA damage response in the cell, and it is likely that this activation is at the heart of the RPP.
A fundamental part of ssODN-driven gene editing is the procedure used to introduce the ssODN into the cell. In our hands, electroporation serves to both deliver enough ssODN to divert detectable levels of gene correction and to enable survival at a level at which the cells can be measured robustly (Engstrom et al., 2009). But it is a balance, and in theory if one could improve cell viability without compromising correction levels, then gene editing could become more widely applicable. Coupled with this challenge is the RPP, which might also be related to the negative impact that electroporation has on cells in general. To this end, we have been examining alternative strategies of ODN delivery that can both preserve viability and execute nucleotide exchange at a reasonable level.
In this study, we examine gene editing through a reaction in which the ssODN has been introduced into the cell by a different form of delivery: nucleofection. A mutated enhanced green fluorescent protein (eGFP) gene, which has been used as the standard in many model systems, served as the genetic target since it allows for phenotypic measurement of gene-editing activity. Herein, we report that using nucleofection does not avoid or reverse RPP even though the level of DNA damage pathway activation appears to be reduced and viability improved. Consistent with previous observation, this DNA damage response is again ignited, but at a lower level than the response we observe when the ssODNs are introduced by electroporation. Thus, nucleofection provides a new improved method of transfection for gene delivery technologies that have reduced collateral DNA damage to targeted cells.
Methods and Materials
Cell line and culture conditions
HCT116 cells were acquired from American Type Cell Culture (Manassas, VA). The integrated HCT116-19 cell line was created by integrating a plasmid enhanced green fluorescent protein (pEGFP)-N3 vector (Clontech, Palo Alto, CA) containing a mutated eGFP gene. The mutated eGFP gene has a nonsense mutation at position +67, resulting in a nonfunctional eGFP protein. For these experiments, HCT116(-19) cells were cultured in McCoy's 5A modified medium (Thermo Scientific, Pittsburgh, PA) supplemented with 10% fetal bovine serum, 2mM l-glutamine, and 1% penicillin/streptomycin. Cells were maintained at 37°C and 5% CO2.
Nucleofection of HCT116-19 cells and experimental approach
HCT116-19 cells were seeded at 2.0×106 cells in a 100-mm dish and synchronized with 6 μM aphidicolin for 24 hours prior to targeting. Cells were released for 4 hours prior to harvesting and targeting by washing with phosphate buffered saline (PBS) and adding complete growth media. Prior to targeting, the specific nucleofector solution, SF solution (Lonza, Walkersville, MD), was warmed to room temperature. Complete growth media was added to the 48-well recovery plate and maintained warm by placing in an incubator at 37°C. Cells were harvested by trypsinization followed by centrifugation. Cells were resuspended in warmed SF solution at a concentration of 100,000 cells per 20 μL. Resuspended cells were mixed with the specific concentration of ssODN in a 96-well plate; 20μL was added to a well of the 16-well nucleocuvette and transfected using the 4D-Nucleofector System (Lonza) with program EH-100. Transfected cells were quenched by adding 80 μL of warmed complete growth media and transferred to the previously prepared recovery plates. Plates were recovered at 37°C for between 24 and 192 hours prior to analysis by flow cytometry (for gene-editing activity), DNA damage pathway activation, and viability.
HCT116-19 cells were seeded in a 100-mm dish and synchronized with 6 μM aphidicolin for 24 hours prior to ssODN introduction. Cells were released for 4 hours prior to harvesting and targeting by washing with PBS and adding complete growth media. Cells were resuspended at 1.0×106 cells/mL, and 100 μL was added to 4 mm gap cuvette (BioExpress, Kaysville, UT). For electroporation, the ssODN was added at a concentration of 6 μM (unless otherwise noted) and electroporated (250V, LV, 13-ms pulse length, 2 pulses, 1-second interval) using a BTX ElectroSquare Porator ECM 830 (BTX Instrument Division, Holliston, MA). The cells were recovered in a 48-well plate in complete growth media at 37°C for 24 hours, 48 hours, and/or 72 hours prior to analysis by flow cytometry for viability.
Viability assays
Cell viability was measured by the Guava Viacount Reagent (Millipore, Temecula, CA). Briefly, the Guava Viacount reagent is a mixture of 2 nuclear DNA stains that can differentiate between live cells, dead cells, and cellular debris based on the permeabilities of the different DNA stains. Prior to cell staining, the cells were harvested by trypsinization, washed once with 1×PBS –/–, mixed in a ratio of 1:1 with Guava Viacount reagent and incubated for 5 minutes at room temperature. Analysis was performed on a Guava EasyCyte 5HT Flow Cytometer (Millipore).
Fluorescence of eGFP was measured by fluorescent activated cell sorting (FACS) analysis using a Guava EasyCyte 5HT Flow Cytometer (Millipore). Cells were harvested by trypsinization, washed once with 1×PBS and resuspended in buffer (0.5% bovine serum albumin, 2mM ethylenediaminetetraacetic acid, 2μg/mL propidium iodide in PBS –/–). eGFP fluorescence was calculated in 2 ways: the percentage of the total live eGFP positive population over the total live population and the percentage of the total eGFP positive population over the total cell population. Error bars are produced from 3 sets of data points generated over 3 separate experiments using normal calculation for standard deviation.
Analysis of DNA damage activity
Cells were treated with the FlowCellect Multi-Color DNA Damage Kit (Millipore), which allows for the detection of phosphorylated ataxia talangiectasia mutated (ATM), structural maintenance of chromosomes-1 (SMC-1), and Histone H2A.X. It uses 3 fluorescently labeled antibodies optimized for analysis using flow cytometry. The kit was developed using the DNA damaging reagent Etoposide in HeLa cells as the model system, but the kit can be used with other cell lines to measure the effect of DNA damage through the ATM-dependent pathway. Briefly, cells were harvested by trypsinization, washed with 1× wash buffer, fixed for 20 minutes on ice, washed again with wash buffer, and permeabilized for 20 minutes on ice. After permeabilization, 200,000 cells were transferred to a V-bottom 96-well plate, washed with assay buffer, and then resuspended in 85 μL of assay buffer. The three antibodies (pH2A.X, pATM, and pSMC-1) were added (5 μL) to the cells and allowed to incubate for 1 hour at room temperature. After incubation, the cells were washed with assay buffer and then resuspended in 200 μL of assay buffer and analyzed on a Guava EasyCyte 5HT Flow Cytometer (Millipore). Data are then presented in a scatter plot depicting cells responding positively to a specific antibody.
Results
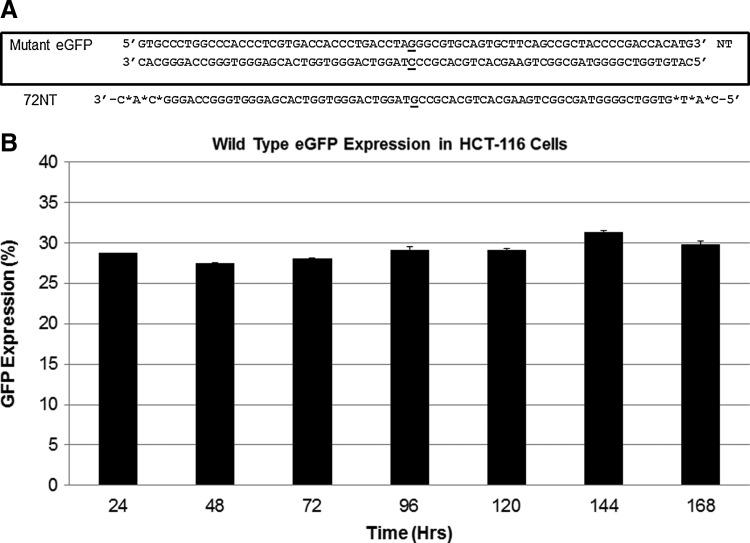
HCT116(-19) cells host a single integrated copy of the mutated eGFP gene that upon correction leads to the expression of a green fluorescent phenotype (Engstrom and Kmiec, 2008). Rescue of the point mutation is directed by single-stranded oligonucleotides (ssODNs) that bind to the mutant gene in the chromosome and facilitate nucleotide exchange in the DNA sequence (Andersen et al., 2002; Liu at al., 2003). The target sequence of the gene is illustrated in Fig. 1A, while the ssODN (72NT) used in the G→C nucleotide exchange reaction is listed below the target sequence. 72NT is a 72-base-long ssODN that contains phosphorothioate linkages (*) between the terminal 3 bases and is complimentary to the nontranscribed strand of the eGFP gene. It is designed to create a specific mismatched base pair within the eGFP gene (underlined).
FIG. 1.
Gene editing model system and expression of enhanced green fluorescent protein (eGFP). (A) The mutant sequence of eGFP integrated in HCT116-19 with the nucleotide targeted for change is listed. The DNA oligonucleotide (ODN) is 72NT, a 72-mer with 3 phosphorothioate linkages on each of the termini. Bold indicates the targeted codon, with asterisks indicating the position of phosphorothioate linkages. (B) HCT116-19 cells (1×106) containing a single integrated copy of the wild-type eGFP gene were plated and the expression of eGFP was monitored over the course of 168 hours. The percentage of cells expressing eGFP was calculated using fluorescent activated cell sorting (FACS) analysis.
The general protocol for carrying out gene editing on the mutated eGFP gene in HCT116–19 cells among others has been widely reported (Brachman and Kmiec, 2005; Hu et al., 2005). Here we utilize cells that have been synchronized in early S phase for 24 hours and released for 4 hours prior to the introduction of the ssODN by either electroporation or (later) nucleofection. After a designated time, the cells are processed for FACS and the number of green fluorescent cells quantified. In most cases, we calculate correction efficiency by dividing the number of eGFP+ live cells by the total number of live cells, unless otherwise noted. Previous results had shown the phosphorothioate-modified ssODNs introduced into the cell by electroporation induce an apparent loss of correction efficiency in these HCT116 cells (Ferrara and Kmiec, 2006; Olsen et al., 2009). This reduction of corrected cell percentage is due in large part to the establishment of RPP in modified cells and the continuous growth of unmodified cells. This response is likely due to the activation of the DNA damage response pathway (Ferrara and Kmeic, 2004; Bonner et al., 2012). The level of eGFP expression does not become reduced over time in these cells. In a simple control experiment, we utilized an HCT116-19 cell line that contains a single copy of the wild-type eGFP gene to test whether sustained expression of eGFP alone, in our system, could explain the gradual reduction over time. As shown in Fig. 1B, eGFP expression remains steady throughout a long time course and the percentage of cells expression eGFP remains constant. Thus, we do not believe that continual expression of eGFP accounts for the reduced proliferation phenotype reported previously (Ferrara and Kmiec, 2006; Olsen et al., 2009; Bonner et al., 2012.
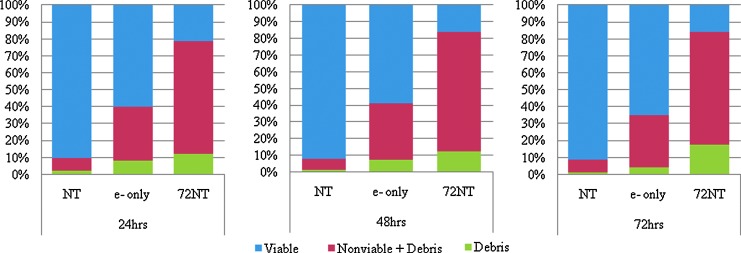
We examined viability of cells under a variety of conditions pertinent to the gene-editing reaction. In particular, we chose conditions that support the highest level of gene editing. Cells were electroporated in the presence or absence of 72NT and viability measured by the Guava ViaCount Assay (Millipore). The Guava ViaCount Assay analyzes cells based on the permeabilities of 2 DNA binding dyes. One of the dyes only stains nucleated cells, while the other stains dying cells. Debris is excluded based on the negative staining from the 2 dyes. Cells that undergo successful gene editing events are likely those that receive a threshold level of targeting ssODN (Drury and Kmiec, 2003; Ferrara et al., 2004). Ironically, this threshold may be high, and reaching or exceeding it may actually result in a certain level of cellular toxicity. As shown in Fig. 2, a significant drop in cell viability occurs within 24, 48 and 72 hours post electroporation. This drop is exacerbated by the presence of the ssODN, which may indicate a type of electrochemical toxicity through the generation of anions. Hence, the reaction conditions using electroporation that are most conducive for correction lead to a reduction in cell viability, results that likely have an impact on long term sustainability of gene-edited cells.
FIG. 2.
Viability analyses after electroporation of 72NT into HCT116 cells. HCT116-19 cells were synchronized with 6 μM aphidicolin for 24 hours, released for 4 hours, and targeted by electroporation with 6 μM 72NT. Cells were allowed to recover for 24 hours (left panel), 48 hours (center panel), and 72 hours (right panel) and then analyzed using the ViaCount reagent by FACS. Viable cells are seen in blue, debris is seen in green, and the nonviable plus debris is depicted in red. E-only refers to cells that are electroporated without ODN, and NT refers to cells not treated in the absence of electroporation.
Altering the mode of delivery of the end-modified ODNs
As described above, the preferred method of ODN delivery for gene editing has been electroporation (Parekh-Olmedo et al., 2005). In our hands, this process delivers sufficient quantities of ssODN into the cell and the nucleus to enable gene-editing activity; however, it also results in a degree of collateral damage to the targeted cell (see Fig. 2). A simple alternative for avoiding such negative impact on cellular metabolism and viability might be to alter the mode of delivery. The goal of this change in protocol is to establish conditions that would attain and then maintain an acceptable level of correction efficiency (∼0.5%) over time with a concurrent improvement in cell viability. Nucleofection is reportedly a gentler means of DNA delivery, enabling, under proper conditions, a greater partitioning of DNA into the nucleus as compared with other delivery modes. In addition, cell viability has been found to be substantially increased with nucleofection as compared to electroporation (Gresch et al., 2004).
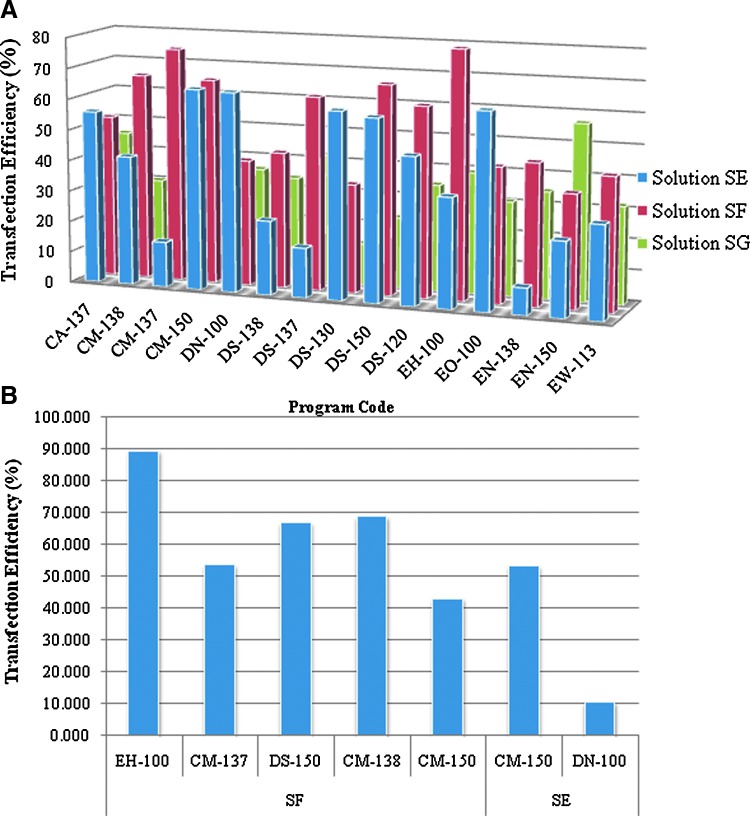
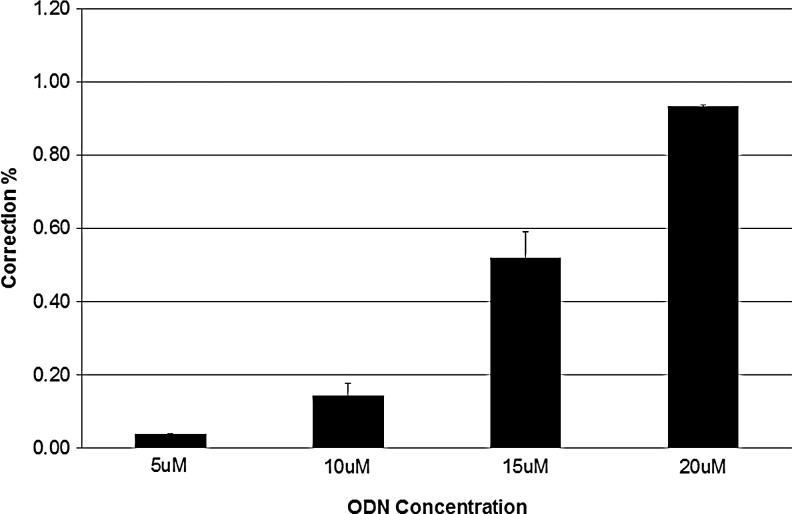
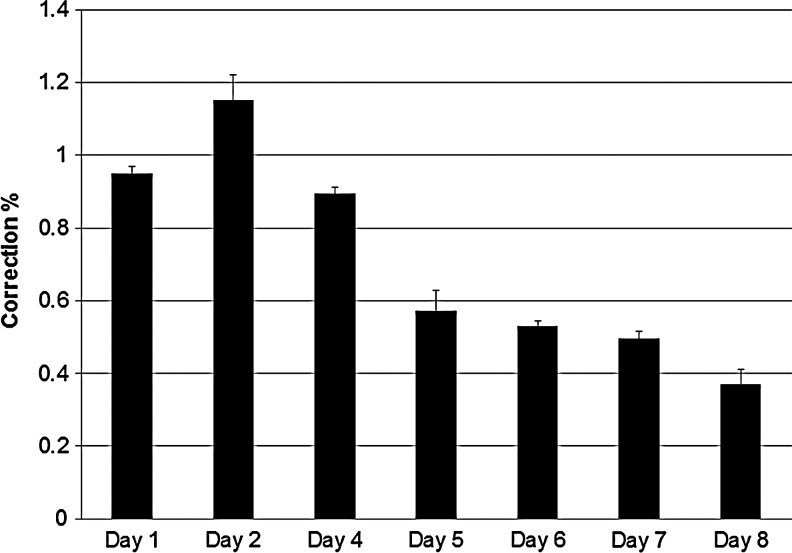
First, we tested a wide variety of nucleofection conditions with the aim of ascertaining the most optimal delivery mode for ssODN. These include a combination of pulses coupled to specialized buffer reagents as provided by Lonza, Inc. A fluorescently Alexa488-tagged 72NT ssODN was introduced into cells, and uptake scored 24 hours later by FACS. As shown in Fig. 3A, a wide variety of conditions support the delivery of ssODNs into the cells with one condition appearing optimal—although several conditions produce comparable results. Due to the manufacturer's restrictions, we cannot define these conditions further, nor do we know the composition of the buffer/delivery reagents. Conditions described as EH-100 for the standard pulse were then used to carry out a gene-editing reaction within a 5–20 μM final concentration range. After 24 hours of recovery, we measured eGFP fluorescence levels by FACS (Fig. 4). There is clearly a high level of gene editing in cells where the ssODN is introduced by nucleofection. Whereas 6μM of ssODN results in approximately 1% correction efficiency when electroporation is used (Olsen et al., 2009; Bonner et al., 2012), here approximately 1% is achieved with 20 μM 72NT; a dose response is also seen with significant levels of gene-editing activity observed between 10 μM and 20 μM. Here, nucleofection enables a significant level of gene editing but requires double the concentration of ssODN to achieve a comparable level of correction. We took the amount of ssODN that directed the highest level of correction and repeated the experiment except that we followed the cells out to 8 days. At daily intervals, correction efficiency was measured by FACS, and the results are presented in Fig. 5. Once again, as seen when electroporation is used to deliver the ssODN, a progressive loss/dilution of corrected cell percentage is seen over time. These results align with the concept of the reduced proliferation phenotype described above.
FIG. 3.
Optimization of ODN delivery into HCT116 cells using nucleofection. HCT116-19 cells were transfected with an Alexa Fluor 488–tagged ODN in appropriate nucleofector solution (solution SE, SF, or SG). (A) For each nucleofector solution, 15 programs were tested (x-axis). Cells were allowed to recover in a 24-well plate with complete growth media for 24 hours, and the transfection efficiency was measured by FACS. Transfection efficiency was measured as the percentage of viable Alexa Flour 488 positive cells to the total viable population as measured by FACS analysis. (B) The top 7 programs were chosen and HCT116-19 cells were transfected with the Alexa Flour 488 ODN to determine the best solution/program combination to use with the HCT116 cell line.
FIG. 4.
Gene editing activity in cells with nucleofection as a delivery mode. HCT116-19 cells were synchronized with 6 μM aphidicolin for 24 hours, released for 4 hours, and then targeted by nucleofection at 5 μM, 10 μM, 15 μM, and 20 μM concentrations of 72NT. Cells were analyzed using FACS 24 hours after targeting. Correction efficiency was calculated as the number of live eGFP positive cells divided by number of total live cells in the population.
FIG. 5.
Levels of gene editing over an extended timeframe. HCT116-19 cells were synchronized with 6μ M aphidicolin for 24 hours, released for 4 hours, and targeted using nucleofection with 20 μM 72NT. Cells were allowed to recover for 1, 2, 4, 5, 6, 7, or 8 days prior to FACS analysis. Correction efficiency was determined as number of eGFP positive cells divided by total number of live cells in the population.
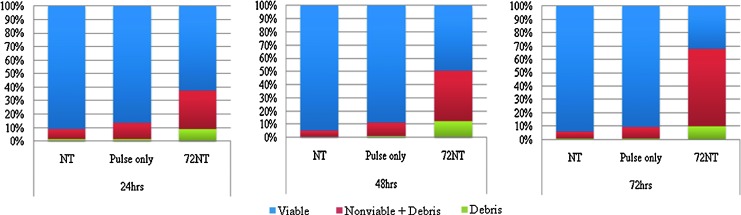
We examined viability over the critical 24- to 72-hour period on a cell population treated with 20 μM 72NT, introduced by nucleofection. As seen in Fig. 7, nucleofection has a less damaging effect on cell viability (compare <with?>pulse-only data to data in Fig. 2). But the addition of 72NT increases cell death and reduces viability in a fashion similar to that of electroporation, but to lesser degree. Nucleofection appears to not impact cell viability as negatively as electroporation immediately after the gene-editing reaction, as the number of cells surviving nucleofection is twice as high at 24 and 48 hours than cells treated by electroporation, and 33% higher at 72 hours. Thus, this form of ODN delivery, even with the terminally modified 72 NT, appears to improve viability yet does not help to overcome the detrimental effects to proliferation of electroporated cells.
FIG. 7.
Viability analyses after nucleofection of 72NT into HCT116 cells. HCT116-19 cells were synchronized with 6 μM aphidicolin for 24 hours, released for 4 hours, and targeted by nucleofection with 20 μM 72NT. Cells were allowed to recover for 24 hours (left panel), 48 hours (center panel) and 72 hours (right panel) and then analyzed using the ViaCount reagent by FACS. Viable cells are seen in blue, debris is seen in green, and the nonviable plus debris is depicted in red. Pulse only refers to cells that are nucleofected without ODN; NT refers to cells that were not treated by electroporation or ODN.
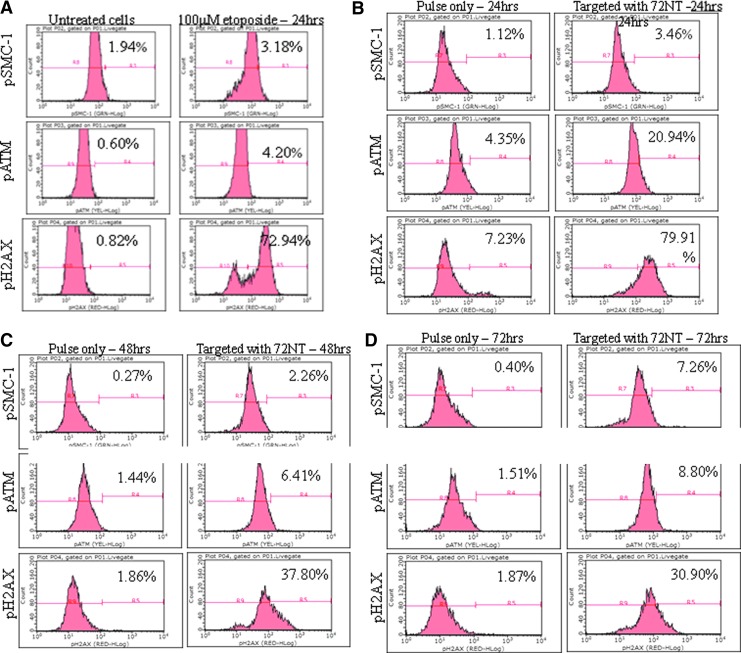
Previously, we reported that several key proteins, SMC1, ATM, and H2AX, become phosphorylated or activated upon electroporation of 72NT (Bonner et al., 2012). It has been suggested that the activation of ATM and its downstream targets (including CHK1 and CHK2) results in cell-cycle stalling (Ferrara and Kmiec, 2006) and fork collapse (Wu et al., 2005; Bonner and Kmiec, 2009; Bonner et al., 2012). We repeated these types of experiments but changed the method of delivery from electroporation to nucleofection to test if the DNA damage response pathway became activated. This type of activation could contribute to RPP. Synchronized cells were released after 24 hours and 72NT was introduced under optimal conditions, those that promote gene editing. In Fig. 6, panel A, cells without any manipulation but after synchronization and release were evaluated. A minor basal level of activation is observed, indicating that treatment with aphidicolin does not induce the DNA damage response pathway in any significant way. In contrast, treatment of released cells with 100 μM etoposide for 24 hours, as predicted, induces a significant rise in all three panels, especially pH2AX. The activation status of SMC1, ATM, and H2AX after nucleofection of 72NT was then analyzed at 24, 48, and 72 hours, respectively, after addition of 72NT (Fig. 6). The results show that H2AX is readily phosphorylated by 24 hours, similar to the response observed when cells are treated with etoposide. A reduction in the population of cells containing activated H2AX is also seen at 48 hours, as predicted from previous observations (Engstrom et al., 2009). pH2AX levels are diminished to some degree but remain significantly high out to 72 hours. The activation response of SMC1 and ATM is significant at 24 hours (particularly ATM) before beginning to diminish at the 48- and 72-hour time points respectively. Taken together, these results indicate that the DNA damage response pathway is in fact also activated early in the gene-editing reaction when 72NT is introduced into the cell by nucleofection.
FIG. 6.
Activation of DNA damage response proteins with nucleofection as the method of delivery. HCT116-19 cells were synchronized with 6 μM aphidicolin for 24 hours, released for 4 hours, and targeted by nucleofection with 20 μm 72NT ODN. (A) The addition of eptoposide (100 μmol) for 24 hours after release serves as a positive control for induction of ataxia talangiectasia mutated (ATM), Histonc H2A.X (H2A.X), and structural maintenance of chromosomes-1 (SMC1) respectively. (B–D) Cells were recovered for 24, 48, and 72 hours (panels B, C, and D respectively) into a 48-well plate. Pulse only refers to cells nucleofected without ssODN. Cells were harvested by trypsinisation, fixed, permeabilized, and stained with phosphorylated Histone H2A.X (pH2A.X), phosphorylated Ataxia telangiectasia mutated (pATM), and phosphorylated structural maintenance of chromosomes-1 (pSMC-1) antibodies; DNA damage analysis was carried out by FACS. Phosphorylation or activation is measured by the shift, to the right, of the histograms. The number in the upper right hand corner refers to the percentage of cells scoring positive for activation, quantified within the gated position.
Discussion
Our observations suggest that ODN-directed gene editing can, in fact, be carried out with nucleofection, attaining correction levels within the range of approximately 0.7% to 1%. Once again, however, an impact on cell proliferation is observed, as reported earlier by others, including Olsen et al. (Olsen et al., 2005 2005a or 2005b) and Ferrara et al. (Ferrara and Kmiec, 2006; Ferrara et al., 2007). This effect seems to manifest after the gene editing reaction, during cell cycle progression and through replication fork stalling; stalled replication forks then collapse and double-stranded DNA breaks emerge (Bonner et al., 2012). The data support a scenario in which the conducive conditions for gene editing are also the ones that result in a decrease in the percentage of corrected cells over time.
This reduced proliferation phenotype may be explained in part by the activation of proteins involved in the DNA damage response pathway (Olsen et al., 2005b; Ferrara and Kmiec, 2006; Huen et al., 2006; Olsen et al., 2009) rather than a pure loss of cell viability. These proteins, in particular ATM, H2AX, and SMC1, initiate a cascade of events that lead to a slowdown of the cell cycle, cells with both ATM and H2AX becoming phosphorylated within the first 24 hours post ODN delivery. This activation has been attributed to the presence of phosphorothioate linkages on the free ends of the ODN (Olsen et al., 2005b; Aarts and te Riele, 2009; Papaioannou et al., 2009; Aarts and te Riele, 2010). And it is true that this effect can be moderated to some degree by changing the position or eliminating the chemical modifications on the ODN altogether. While some stalling takes place whenever single-stranded DNA ends are introduced into the cell, it is possible that various molecular compositions impact the sustainability of gene editing in a cell population more dramatically. In our hands, however, removing or changing the terminal modifications from 72NT results in lower levels of gene editing (Bonner and Kmiec, 2009). In particular, the overall frequency drops (Olsen et al., 2005b; Bertoni et al., 2009; Papaionnou et al., 2009), and thus endpoints that require the clonal isolation of an edited cell, will be difficult to achieve. To this end, we have recently found that the 72NT molecule bearing modified ends induces replication stress by blocking fork movement (Bonner et al., 2012). Thus, it appears prudent to continue searching for alternative protocols, still using 72NT, that minimize RPP.
In this study we have been able to define a new protocol in which gene editing can be carried out using a new mode of DNA delivery, a method that improves the viability of the cells and this is an important step forward for the technology. These outcomes have been achieved because nucleofection can overcome some of the limitations attributed to electroporation (Gresch et al., 2004). Nucleofection preserves the efficient delivery of large molecules into cells, characteristic of electroporation, while enabling cell viability; our results support these claims. Nucleofection utilizes a specifically regulated series of pulses that does not disable the cell and nuclear membrane beyond repair. Specific reagents are used in the nucleofection process and in cell restoration concurrent with the voltage pulse. Nucleofection also delivers a higher percentage of large molecules such as ODNs into the nucleus (Gresch et al., 2004). The cytotoxic anions, generated by electroporation, are likely buffered by the high ionic strength of the reagents used in the process. It is likely that these high buffering capacity reagents also counteract the cytotoxic nature of the phosphorothioate linkages. Thus, this procedure allows us to utilize the most active ODN, 72 NT while simultaneously reducing cytotoxicity. This observation might have significant value for studies in the mechanism of gene editing in other cell systems or in vivo; for example, cell viability is the paramount feature and is crucial for targeting systems in which replication or differentiation may not be desired, such as progenitor cells. The success of gene editing relies first on cell survival and any reaction component that improves viability is an advance.
While we have been able to improve cell viability during the gene editing process—and move it toward in vivo implementation, the major problem addressed in previous studies—reduced proliferation still remains. In our view, as explained above, the central problem is the activation of the DNA damage response pathway and its downstream influence on checkpoint blockage. Our current studies are aimed at causing a transitory diminution in the levels of proteins involved in this pathway in the hope that the corrected cells can be restored to a normal proliferation phenotype or can avoid this growth retardation altogether. In general, this approach centers on developing conditions or creating environments in which edited cells can grow prior to dilution and clonal isolation while trying to maintain a reasonable level of gene editing activity.
Acknowledgments
We thank members of the Kmiec laboratory for critical evaluation of the data. We are indebted to Ms. Sarah Greene for manuscript preparation and editing. This work was supported by NIH grant 5R01CA089325-12.
Author Disclosure Statement
No competing financial interests exist.
References
- AARTS M. T.E. RIELE H. Parameters of oligonucleotide-mediated gene modification in mouse ES cells. J. Cell Mol. Med. 2009;14(6B):1657–1667. doi: 10.1111/j.1582-4934.2009.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AARTS M. T.E. RIELE H. Subtle gene modification in mouse ES cells: evidence for incorporation of unmodified oligonucleotides without induction of DNA damage. Nucleic Acids Res. 2010;38:6956–6967. doi: 10.1093/nar/gkq589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AARTS M. T.E. RIELE H. Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Therapy. 2011;18:213–219. doi: 10.1038/gt.2010.161. [DOI] [PubMed] [Google Scholar]
- ANDERSEN M.S. SORENSEN C.B. BOLUND L. JENSEN T.G. Mechanisms underlying targeted gene correction using chimeric RNA/DNA and single-stranded DNA oligonucleotides. J. Mol. Med. 2002;80:770–781. doi: 10.1007/s00109-002-0393-8. [DOI] [PubMed] [Google Scholar]
- ANDRIEU-SOLER C. CASAS M. FAUSSAT A.M. GANDOLPHE C. DOAT M. TEMPE D. GIOVANNANGELI C. BEHAR-COHEN F. CONCORDET J.P. Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs. Nucleic Acids Res. 2005;33:3733–3742. doi: 10.1093/nar/gki686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTONI C. RUSTAGI A. RANDO T.A. Enhanced gene repair mediated by methyl-CpG-modified single-stranded oligonucleotides. Nucleic Acids Res. 2009;37:7468–7482. doi: 10.1093/nar/gkp757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONNER M. KMIEC E.B. DNA breakage associated with targeted gene alteration directed by DNA oligonucleotides. Mutat. Res. 2009;669:85–94. doi: 10.1016/j.mrfmmm.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONNER M. STROUSE B. APPELGATE M. LIVINGSTON P. KMIEC E. DNA damage response pathway and replication fork stress during oligonucleotide directed gene editing. Mol. Ther. 2012;1:e18. doi: 10.1038/mtna.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRACHMAN E.E. KMIEC E.B. Gene repair in mammalian cells is stimulated by the elongation of S phase and transient stalling of replication forks. DNA Repair (Amst.) 2005;4:445–457. doi: 10.1016/j.dnarep.2004.11.007. [DOI] [PubMed] [Google Scholar]
- DRURY M.D. KMIEC E.B. DNA pairing is an important step in the process of targeted nucleotide exchange. Nucleic Acids Res. 2003;31:899–910. doi: 10.1093/nar/gkg171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENGSTROM J.U. KMIEC E.B. Manipulation of cell cycle progression can counteract the apparent loss of correction frequency following oligonucleotide-directed gene repair. BMC Mol. Biol. 2007;8:9. doi: 10.1186/1471-2199-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENGSTROM J.U. KMIEC E.B. DNA Replication, cell cycle progression, and the targeted gene repair reaction. Cell Cycle. 2008;7:1402–1414. doi: 10.4161/cc.7.10.5826. [DOI] [PubMed] [Google Scholar]
- ENGSTROM J.U. SUZUKI T. KMIEC E.B. Regulation of targeted gene repair by intrinsic cellular processes. Bioessays. 2009;31:159–168. doi: 10.1002/bies.200800119. [DOI] [PubMed] [Google Scholar]
- FERRARA L. ENGSTROM J.U. SCHWARTZ T. PAREKH-OLMEDO H. KMIEC E.B. Recovery of cell cycle delay following targeted gene repair by oligonucleotides. DNA Repair (Amst.) 2007;6:1529–1535. doi: 10.1016/j.dnarep.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRARA L. KMIEC E.B. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004;32:5239–5248. doi: 10.1093/nar/gkh822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRARA L. KMIEC EB. Targeted gene repair activates Chk1 and Chk2 and stalls replication in corrected cells. DNA Repair (Amst) 2006;5:422–431. doi: 10.1016/j.dnarep.2005.11.009. [DOI] [PubMed] [Google Scholar]
- FERRARA L. PAREKH-OLMEDO H. KMIEC E. Enhanced oligonucleotide-directed gene targeting in mammalian cells following treatment with DNA damaging agents. Exp. Cell Res. 2004;300:170–179. doi: 10.1016/j.yexcr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- GRESCH O. ENGEL F.B. NESIC D. TRAN T.T. ENGLAND H.M. HICKMAN E.S. KÖRNER I. GAN L. CHEN S. CASTRO-OBREGON S. HAMMERMANN R. WOLF J. MÜLLER-HARTMANN H. NIX M. SIEBENKOTTEN G. KRAUS G. LUN K.L. New non-viral method for gene transfer into primary cells. Methods. 2004;33:151–163. doi: 10.1016/j.ymeth.2003.11.009. [DOI] [PubMed] [Google Scholar]
- HUEN M.S. LI X.T. LU L.Y. WATT R.M. LIU D.P. HUANG J.D. The involvement of replication in single stranded oligonucleotide-mediated gene repair. Nucleic Acids Res. 2006;34:6183–6194. doi: 10.1093/nar/gkl852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU Y. PAREKH-OLMEDO H. DRURY M. SKOGEN M. KMIEC E.B. Reaction parameters of targeted gene repair in mammalian cells. Mol. Biotechnol. 2005;29:197–210. doi: 10.1385/MB:29:3:197. [DOI] [PubMed] [Google Scholar]
- KHAN I.F. HIRATA R.K. WANG P.R. LI Y. KHO J. NELSON A. HUO Y. ZAVALJEVSKI M. WARE C. RUSSELL D.W. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU L. PAREKH-OLMEDO H. KMIEC E.B. The development and regulation of gene repair. Nat. Rev. Genet. 2003;4:679–689. doi: 10.1038/nrg1156. [DOI] [PubMed] [Google Scholar]
- OLSEN P.A. RANDOL M. KRAUSS S. Implications of cell cycle progression on functional sequence correction by short single-stranded DNA oligonucleotides. Gene Ther. 2005a;12:546–551. doi: 10.1038/sj.gt.3302454. [DOI] [PubMed] [Google Scholar]
- OLSEN P.A. RANDOL M. LUNA L. BROWN T. KRAUSS S. Genomic sequence correction by single-stranded DNA oligonucleotides: role of DNA synthesis and chemical modifications of the oligonucleotide ends. J. Gene Med. 2005b;7:1534–1544. doi: 10.1002/jgm.804. [DOI] [PubMed] [Google Scholar]
- OLSEN P.A. SOLHAUG A. BOOTH J.A. GELAZAUSKAITE M. KRAUSS S. Cellular responses to targeted genomic sequence modification using single-stranded oligonucleotides and zinc-finger nucleases. DNA Repair (Amst.) 2009;8:298–308. doi: 10.1016/j.dnarep.2008.11.011. [DOI] [PubMed] [Google Scholar]
- PAPAIOANNOU I. DISTERER P. OWEN J.S. Use of internally nuclease-protected single-strand DNA oligonucleotides and silencing of the mismatch repair protein, MSH2, enhances the replication of corrected cells following gene editing. J. Gene Med. 2009;11:267–274. doi: 10.1002/jgm.1296. [DOI] [PubMed] [Google Scholar]
- PAREKH-OLMEDO H. FERRARA L. BRACHMAN E. KMIEC E.B. Gene therapy progress and prospects: targeted gene repair. Gene Ther. 2005;12:639–646. doi: 10.1038/sj.gt.3302511. [DOI] [PubMed] [Google Scholar]
- RADECKE S. RADECKE F. PETER I. SCHWARZ K. Physical incorporation of a single-stranded oligodeoxynucleotide during targeted repair of a human chromosomal locus. J. Gene Med. 2006;8:217–228. doi: 10.1002/jgm.828. [DOI] [PubMed] [Google Scholar]
- URNOV F. REBAR E. HOLMES M. ZHANG H. GREGORY P. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- WANG Z. ZHOU Z.J. LIU D.P. HUANG J.D. Single-stranded oligonucleotide-mediated gene repair in mammalian cells has a mechanism distinct from homologous recombination repair. Biochem. Biophys. Res. Commun. 2006;350:568–573. doi: 10.1016/j.bbrc.2006.09.078. [DOI] [PubMed] [Google Scholar]
- WOOD A. LO T. ZEITLER B. PICKLE C.S. RALSTON E.J. LEE A.H. AMORA R. MILLER J.C. LEUNG E. MENG X. ZHANG L. REBAR E.J. GREGORY P.D. URNOV F.D. MEYER B.J. Targeted genome editing across species Using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU X.S. XIN L. YIN W.X. SHANG X.Y. LU L. WATT R.M. CHEAH K.S. HUANG J.D. LIU D.P. LIANG C.C. Increased efficiency of oligonucleotide-mediated gene repair through slowing replication fork progression. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2508–2513. doi: 10.1073/pnas.0406991102. [DOI] [PMC free article] [PubMed] [Google Scholar]