Abstract
OBJECTIVE
An elevated insulin resistance index (homeostasis model assessment of insulin resistance [HOMA-IR]) is more commonly seen in the Mexican American population than in European populations. We report quantitative ancestral effects within a Mexican American population, and we correlate ancestral components with HOMA-IR.
RESEARCH DESIGN AND METHODS
We performed ancestral analysis in 1,551 participants of the Cameron County Hispanic Cohort by genotyping 103 ancestry-informative markers (AIMs). These AIMs allow determination of the percentage (0–100%) ancestry from three major continental populations, i.e., European, African, and Amerindian.
RESULTS
We observed that predominantly Amerindian ancestral components were associated with increased HOMA-IR (β = 0.124, P = 1.64 × 10−7). The correlation was more significant in males (Amerindian β = 0.165, P = 5.08 × 10−7) than in females (Amerindian β = 0.079, P = 0.019).
CONCLUSIONS
This unique study design demonstrates how genomic markers for quantitative ancestral information can be used in admixed populations to predict phenotypic traits such as insulin resistance.
Ethnicity has been suggested as a factor affecting the susceptibility to insulin resistance and related chronic diseases (1,2). However, genetic admixture and major confounding by environmental heterogeneity among human populations complicates the assessment of the role of ethnicity. The opportunity to address these issues was presented to us by our community-recruited Cameron County Hispanic Cohort (CCHC). In this cohort, participants are randomly selected from the adult Mexican American population of a small city: Brownsville, Cameron County in South Texas (3). Cultural and lifestyle homogeneity avoids compounding factors in a way that would be problematic in a major city or across a large geographic area. All the CCHC participants are self-identified Mexican Americans, a rapidly growing minority population known to be genetically admixed with European, African, and Native Amerindian ancestries (1). Elevated homeostasis model assessment of insulin resistance (HOMA-IR) index is commonly seen in this population (4). In the current study, our objective was to assess the ancestral effect on insulin sensitivity by measuring the correlation between quantitative ancestral status of our population and fasting HOMA-IR levels using genomic information from ancestral component analysis.
RESEARCH DESIGN AND METHODS
Subjects
This study investigated 1,551 randomly recruited individuals in the CCHC (3). Sampling bias was corrected based on Census 2000 data to account for age, sex, tract/block, and household clustering.
Genotyping
We genotyped 103 continental ancestry-informative markers (AIMs) for Mexican Americans identified by Kosoy et al. (5), using the Sequenom iPLEX assay (Sequenom, Cambridge, MA). The genotyping call rates of the 103 AIM single-nucleotide polymorphisms were between 95.4 and ∼100%, with a median of 99.9%. For the purpose of quality control, 93 DNA samples were genotyped in duplicate. The concordance of duplicate genotypes was 100%.
Data analysis
We used the Eigensoft Software (version 2) for principal component analysis (PCA) (6) and conducted the ancestral analysis using Admixture 1.1 algorithm (7,8). Ancestral components of each CCHC participant were expressed as percentage units (%) referring to each of three major continental populations, i.e., European, African, and Amerindian. The African and European genotyping data were obtained from the HapMap project (http://hapmap.ncbi.nlm.nih.gov), and genotyping data of 105 Amerindians were obtained from the study by Kosoy et al. (5)
RESULTS
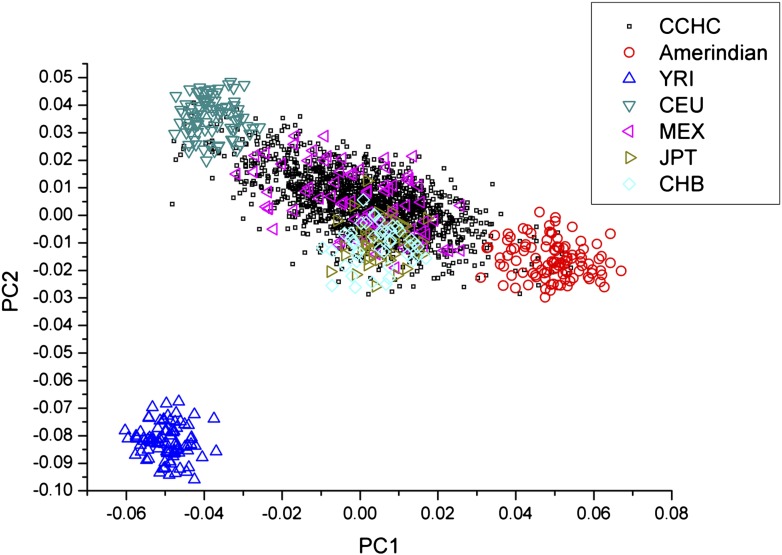
By PCA of the 103 AIMs, the CCHC participants showed a predominant admixture of European and Amerindian ancestries (Fig. 1). The genetic ancestry components of this population are superimposable with a population sample of Mexican Americans in Los Angeles, California, that were studied in the HapMap project (Fig. 1). There was considerable variation in the proportions of ancestral components between individuals: the median European ancestry was 45.8%, with 25% percentile of 35.7% and 75% percentile of 55.1%; the median African ancestry was 11.0%, with 5% percentile of 6.6% and 95% percentile of 15.3%; and the median Amerindian ancestry was 42.9%, with 5% percentile of 33.1% and 95% percentile of 53.0%. The coefficient of variation (CV) of European ancestries was 32.5%, the CV of African ancestries was 59.5%, and the CV of Amerindian ancestries was 37.0%.
Figure 1.
PCA of the population structure of the CCHC participants. CCHC participants show a predominant admixture of European and Amerindian ancestries. Amerindian, Mayan from Chimaltenango Guatemala, Nahua Amerindians from central Mexico, and Quechuan Amerindians from Peru (5); CEU, Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain (CEPH) collection; CHB, Han Chinese in Beijing, China; JPT, Japanese in Tokyo, Japan; MEX, Mexican ancestry in Los Angeles, California; YRI, Yoruba in Ibadan, Nigeria.
To explain high levels of HOMA-IR in this population, we tested the correlation between ancestral components and HOMA-IR levels (Supplementary Table 1). Aging and obesity are known risk factors for insulin resistance (9). Socioeconomic status has also been highlighted as an important factor for the development of insulin resistance (10). After adjusting for age, BMI, household annual income, educational levels, and ancestral components, we found that the Amerindian ancestral component is positively associated with elevated HOMA-IR levels (β = 0.124, P = 1.64 × 10−7), and the European ancestral component (also expressed as a percentage) is negatively associated with HOMA-IR levels (β = −0.111, P = 3.18 × 10−6). The correlation is more significant in males (Amerindian β = 0.165, P = 5.08 × 10−7) than in females (Amerindian β = 0.079, P = 0.019). The adjusted sex effect on HOMA-IR levels has β = 0.062 and P = 5.26 × 10−3.
CONCLUSIONS
In this study, we demonstrated that higher HOMA-IR levels are correlated with Amerindian ancestry in a Mexican American population. Amerindian and European populations have different susceptibilities to many metabolic diseases (2). The considerable variability of European and Amerindian ancestries between Mexican American individuals that we show in this study suggests that individuals may have different genetic susceptibility to diseases. The incomplete understanding of the genetic susceptibility to complex diseases increases the importance of examining ethnicity information to identify people at high risk of a disease.
We estimated the proportion of ancestry from each contributing population (European, African, and Amerindian) using AIMs developed specifically for the Mexican American population by Kosoy et al. (5). Kosoy et al. have shown that a subset of as few as 24 AIMs is sufficient for ancestral analysis in Mexican Americans (5). Considerable variation of ancestral components found in CCHC participants allows us to more precisely assess ancestral effects within one single population with limited environmental confounding factors. The decreased insulin sensitivity that we have shown to be associated with Amerindian ancestry may thus contribute substantially to high rates of insulin resistance syndrome in Mexican Americans (4). We also observed an interesting sex effect from ancestry in the degree of insulin sensitivity, possibly related to the protective effect of estrogen in females (11). Compared with the Mexican American reference population in the HapMap project, the CCHC participants of our study have similar distributions of ancestral components. Therefore, the results of this study are representative of the broader population of Mexican origin.
A number of genetic variants with key roles in insulin sensitivity have been identified (12,13), including the glucokinase regulatory protein gene (GCKR) variant rs780094, which tags the strongest genetic effect among identified genetic loci of insulin sensitivity (13,14). The GCKR gene variant partially explains lower insulin sensitivity in Amerindians (6.14% of the ancestral effect) (H.-Q.Q., unpublished data). However, a limitation of our current study is the limited number of AIMs genotyped. Admixture mapping using AIMs at high density in this cohort is warranted to further investigate genetic variants contributing to the ancestral effect. Additional study efficiency may be acquired by the combined usage of association mapping (15).
Supplementary Material
Acknowledgments
This work was supported by the National Center on Minority Health and Health Disparities (MD-000170-P20) and by the National Center for Research Resources (Centers for Translational Science Award 1U54-RR-023417-01). H.-Q.Q is supported by intramural funding from The University of Texas School of Public Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
No potential conflicts of interest relevant to this article were reported.
H.-Q.Q. conceived the study, researched the data, and wrote the manuscript. Q.L. researched the data and reviewed and edited the manuscript. Y.L. performed the experiments. C.L.H. provided advice on study design. S.P.F.-H. and J.B.M. developed the CCHC, conceived the study, and wrote the manuscript. H.-Q.Q. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors appreciate Dr. Constantin Polychronakos’s instructive comments on this paper. The authors thank the cohort recruitment team, particularly Rocio Uribe, Elizabeth Braunstein, and Julie Ramirez (The University of Texas School of Public Health, Brownsville Regional Campus [UTSPH-BRC]) and Dr. Anne Rentfro (The University of Texas at Brownsville). The authors also thank Marcela Montemayor (UTSPH-BRC) and other laboratory staff for archiving specimens and performing insulin assays and Christina Villarreal (UTSPH-BRC) for administrative support. The authors thank Valley Baptist Medical Center (Brownsville, TX) for providing space for the Center for Clinical and Translational Science Clinical Research Unit and the community of Brownsville and the participants in the cohort, who so generously participated in this study in their city.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc12-0636/-/DC1.
References
- 1.Manichaikul A, Palmas W, Rodriguez CJ, et al. Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet 2012;8:e1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss KM, Ferrell RF, Hanis CL. A new world syndrome of metabolic diseases with a genetic and evolutionary basis. Am J Phys Anthropol 1984;27(Suppl. 5):153–178 [Google Scholar]
- 3.Fisher-Hoch SP, Rentfro AR, Salinas JJ, et al. Socioeconomic status and prevalence of obesity and diabetes in a Mexican American community, Cameron County, Texas, 2004-2007. Prev Chronic Dis 2010;7:A53. [PMC free article] [PubMed] [Google Scholar]
- 4.Qu HQ, Li Q, Rentfro AR, Fisher-Hoch SP, McCormick JB. The definition of insulin resistance using HOMA-IR for Americans of Mexican descent using machine learning. PLoS ONE 2011;6:e21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 2009;30:69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 7.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009;19:1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou H, Alexander D, Lange K. A quasi-Newton acceleration for high-dimensional optimization algorithms. Stat Comput 2011;21:261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohrt WM, Kirwan JP, Staten MA, Bourey RE, King DS, Holloszy JO. Insulin resistance in aging is related to abdominal obesity. Diabetes 1993;42:273–281 [PubMed] [Google Scholar]
- 10.Lawlor DA, Ebrahim S, Davey Smith G, British Women’s Heart and Health Study Socioeconomic position in childhood and adulthood and insulin resistance: cross sectional survey using data from British Women’s Heart and Health Study. BMJ 2002;325:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geer EB, Shen W. Gender differences in insulin resistance, body composition, and energy balance. Gend Med 2009;6(Suppl. 1):60–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers JC, Elliott P, Zabaneh D, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 2008;40:716–718 [DOI] [PubMed] [Google Scholar]
- 13.Dupuis J, Langenberg C, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beer NL, Tribble ND, McCulloch LJ, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet 2009;18:4081–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Young JH, Fox E, et al. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet 2011;20:2285–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.