Abstract
In the methylotrophic bacterium Methylobacterium extorquens strain AM1, MxaF, a Ca2+-dependent methanol dehydrogenase (MDH), is the main enzyme catalyzing methanol oxidation during growth on methanol. The genome of strain AM1 contains another MDH gene homologue, xoxF1, whose function in methanol metabolism has remained unclear. In this work, we show that XoxF1 also functions as an MDH and is La3+-dependent. Despite the absence of Ca2+ in the medium strain AM1 was able to grow on methanol in the presence of La3+. Addition of La3+ increased MDH activity but the addition had no effect on mxaF or xoxF1 expression level. We purified MDH from strain AM1 grown on methanol in the presence of La3+, and its N-terminal amino acid sequence corresponded to that of XoxF1. The enzyme contained La3+ as a cofactor. The ΔmxaF mutant strain could not grow on methanol in the presence of Ca2+, but was able to grow after supplementation with La3+. Taken together, these results show that XoxF1 participates in methanol metabolism as a La3+-dependent MDH in strain AM1.
Introduction
Methylotrophs are microorganisms with the ability to utilize reduced C1-compounds, such as methane, methanol and methylamine as their sole carbon and energy source. They are ubiquitous in nature, and some of them are well-known plant epiphytes [1], [2]. Among them, the genus Methylobacterium, an aerobic facultative methylotrophic α-proteobacterium, is one of the most abundant bacterial genera in the phyllosphere [3]–[5], with a titer between 104 and 107 colony-forming units (CFU) per gram fresh weight of plant material [6]. Over the past few decades, considerable work has been done on the methylotrophy of Methylobacterium and their symbiosis with plants, as Methylobacterium can metabolize the methanol released by plants and may also grow on other plant-derived carbon compounds [7]–[9]. M. extorquens strain AM1 serves as an important model organism for studying methylotrophy in bacteria [10], [11], and the genome sequence of the strain is available [12].
In the methylotrophic metabolism of Methylobacterium, methanol is first oxidized to formaldehyde via methanol dehydrogenase (MDH) in the periplasm [13], [14]. MDH is a heterotetrameric protein (α2β2) consisting of two 66-kDa large subunits (MxaF) and two small 8.5-kDa subunits (MxaI) [15], and contains Ca2+ and pyrroloquinoline quinone (PQQ) as a prosthetic group in the active site [15], [16]. MxaF and MxaI are encoded by mxaFI genes located in the large mxa gene cluster [17], and both are essential for growth on methanol, as the loss of these genes in strain AM1 eliminates virtually all methanol dehydrogenase activity [18], [19].
The genome of strain AM1 contains several homologs of MxaF, one of which is named XoxF1 [20]. XoxF1 is predicted to be a PQQ-dependent periplasmic MDH exhibiting 50% sequence identity to MxaF. Recently, Schmidt et al. reported that XoxF1 was found to be strongly expressed in bacterial phyllosphere communities [1], and that the xoxF1-deleted strain was less competitive than the wild-type during colonization in the phyllosphere, although XoxF1 had low MDH activity in strain AM1 [21]. Skovran et al. showed that the double mutant of both xoxF homologs (xoxF1 and xoxF2) was unable to grow on methanol and that the expression of the two-component regulatory systems MxcQE and MxbDM required for activation of the mxa genes is repressed in the double mutant strain [22]. From these facts, it is clear that XoxF functions in the regulation of methanol metabolism, but its catalytic function as an MDH has not been clear.
In our previous work, we showed that lanthanum (La), cerium (Ce), and praseodymium (Pr), all of which are belong to the rare earth elements (REE), increased MDH activity in cell extracts of M. radiotolerans and the non-methylotrophic bacteria Bradyrhizobium sp. [23], [24]. Moreover, the MDHs purified from the cells grown in media containing these metal ions corresponded to XoxF1, while the MDH purified from Ca2+-grown cells corresponded to MxaFI [23], [24]. These results indicate that the MDHs dependent on La3+, Ce3+ or Pr3+ are products of xoxF and that these ions may have important physiological roles in C1 metabolism.
The REEs are a group of 17 elements, specifically, 15 lantanoids plus Sc and Y, and are widely dispersed among many primary and secondary minerals, such as phosphates, carbonates, fluorides, and silicates, especially pegmatites, granites, and related metamorphic and igneous rocks [25]. They are regarded as “the vitamins of modern industry”, since many of them are utilized in a wide range of industrial products such as glass, catalysts, alloys, ceramics, and magnets. As for their effects on life forms, the REEs have not been characterized as either essential or strongly toxic elements in the environment [26], although some have negative effects as inhibitors of several enzymes and proteins [27]–[30], and some exert positive effects as growth promoters for various crops [27].
In this study, using M. extorquens strain AM1 as a model organism to investigate REEs-dependent methylotrophy, we set out (i) to see whether La3+ is involved in methylotrophic growth of the strain, (ii) to assess whether the strain has REE-dependent MDH activity, (iii) to identify the gene encoding REE-dependent MDH, and (iv) to validate the role of XoxF1 and La3+ in methanol metabolism. Our results suggest that XoxF1 is a La3+-dependent functional MDH that may participate in methanol metabolism.
Results
M. extorquens strain AM1 has a methanol-oxidation system independent of Ca2+
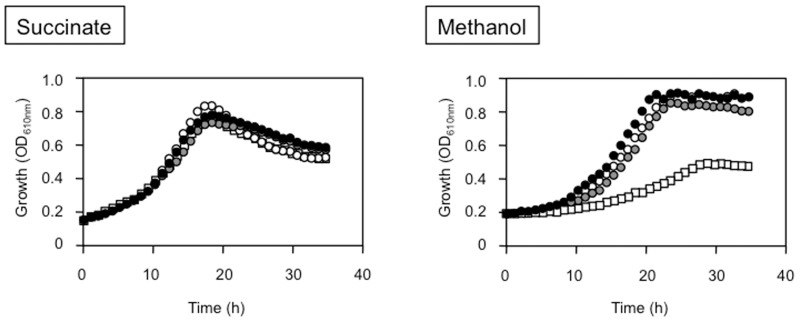
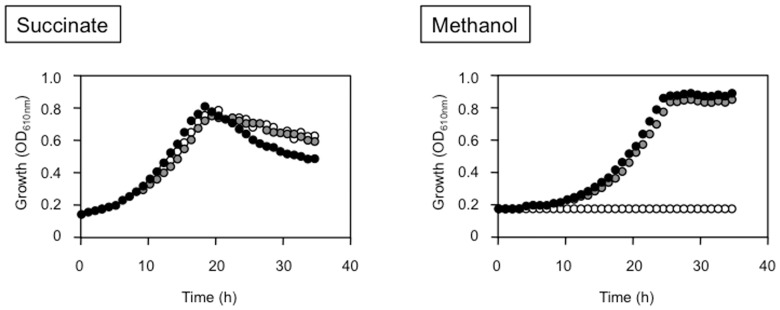
Although MDH activity in Methylobacterium species has been shown to depend on Ca2+ [14], the growth of these strains on methanol without Ca2+ has never been examined. In our previous work, we showed that some REEs increased MDH activity in M. radiotolerans and the non-methylotrophic Bradyrhizobium sp. [23], [24]. These facts suggest that REEs may have some roles as activators or inducers of MDH. Thus, we examined whether M. extorquens strain AM1 could grow on methanol in the presence of La3+ instead of Ca2+. As shown in Fig. 1, strain AM1 could grow normally in methanol/Ca2+ medium. In methanol medium without Ca2+ and La3+, the strain showed very slow growth, because the medium contained a small amount of Ca2+ (0.867 µM). In methanol media containing La3+ and not Ca2+, the strain grew as well as it did in methanol/Ca2+ medium, and the addition of La3+ to methanol/Ca2+ medium had no effect on the growth of strain AM1 (Fig. 1). On the other hand, strain AM1 did not show any growth defect in succinate media even without Ca2+ and La3+. Ca3+ and La3+ have an important role in methanol metabolism but not in succinate metabolism, and strain AM1 has a novel methanol-metabolic pathway that depends on La3+ and not Ca2+.
Figure 1. Growth of the wild-type strain AM1 on methanol or succinate media supplemented with Ca2+ or/and La3+.
The growth of strain AM1 on methanol or succinate media supplemented with Ca2+ (black circle), with La3+ (gray circle), with Ca2++La3+ (white circle) and without Ca2+ and La3+ (white square). The concentration of Ca2+ and La3+ in the medium are 30 µM each. Points on the graphs depict average data from three biological replicates.
XoxF1 is a functional La3+-dependent MDH
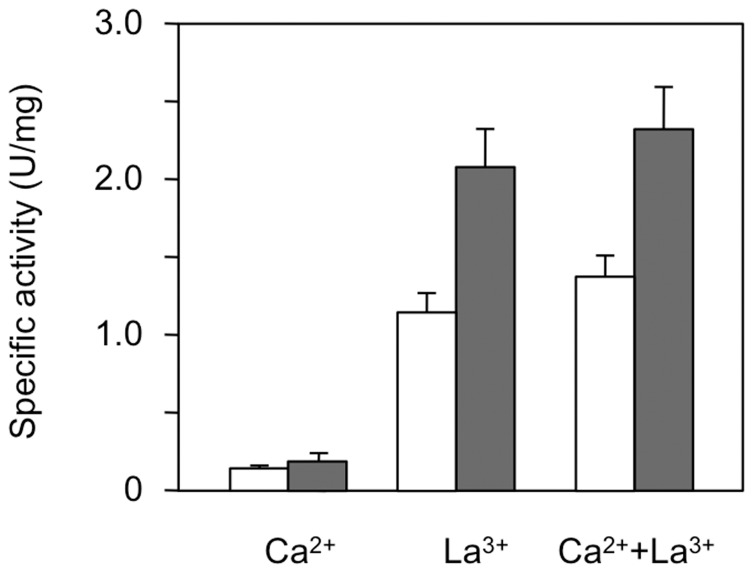
The growth defect of strain AM1 in the methanol medium without Ca2+ was restored by the addition of La3+ to the medium. Next, in order to see whether MDH activity was induced by La3+, we measured MDH activity of strain AM1 grown on media containing La3+. When strain AM1 was grown in methanol media, MDH activity in the cell-free extract was ten times greater in methanol/La3++Ca2+ medium than in methanol/Ca2+ medium, and cells grown in methanol/La3+ medium showed levels of MDH activity similar to those in cells grown in methanol/La3++Ca2+ medium (Fig. 2). Cells grown on the succinate media also had enough MDH activity more than half of the activity in the methanol-grown cells, and the MDH activity induced on the succinate/La3+ medium was higher than that induced on the succinate/Ca2+ medium, as well as the methanol grown cells. There are two possible explanations for this positive effect of La3+: one is that La3+ enhances MDH gene(s) expression and the other is that La3+ activates MDH protein.
Figure 2. MDH activity in strain AM1 grown on the methanol and succinate media with Ca2+ and/or La3+.
Cells were grown aerobically in succinate (white bar) and methanol (gray bar) media with 30 µM La3+ and/or Ca2+ at 30°C. Results are shown as means with standard deviations (n = 3).
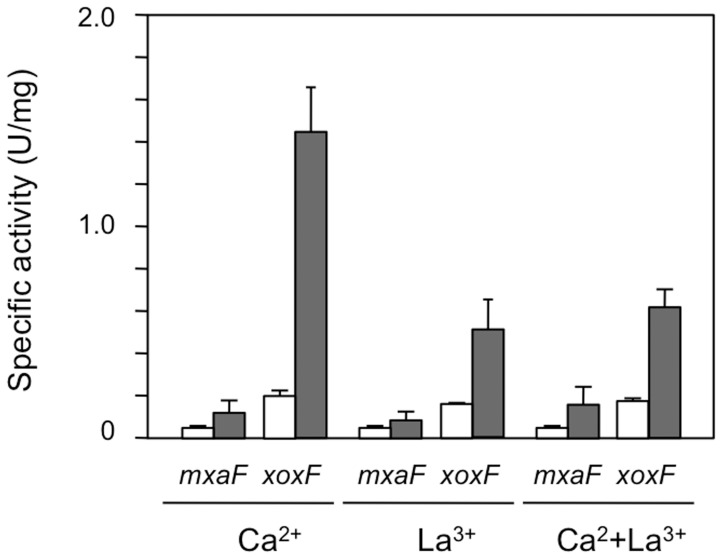
To determine whether La3+ enhances MDH gene(s) expression, we quantified the gene expression levels of mxaF and xoxF1 using cells harboring the xylE reporter gene regulated by the predicted promoter regions, which are 220- and 227-bp upstream sequences of the MxaF and XoxF1 genes, respectively. The reporter activities regulated by the mxaF and xoxF1 promoters were detected in all cells grown on methanol or succinate, and the xoxF1 promoter of the cells grown on methanol/Ca2+ medium exhibited the highest expression activity (Fig. 3). The activities of both promoters on the methanol grown cells exhibited always higher than those on the succinate grown cells (Fig. 3). Moreover, expression activity of the xoxF1 promoter was always greater than that of the mxaF promoter on any media, irrespective of the presence of La3+ and/or Ca2+. XylE activity was not detected in cells harboring the promoterless control plasmid pCM130, irrespective of the carbon sources, as reported previously [31]. These results show that the increase in MDH activity caused by La3+ is due not to an increased expression of MDH genes but rather to post-translational activation of MDH.
Figure 3. Expression of mxaF-xylE and xoxF1-xylE promoter fusions in M. extorquens strain AM1.
The mxaF and xoxF1 promoter activities were determined by measuring catechol 2,3-dioxygenase activity (XylE) in crude cell extracts of strains grown in methanol (black bar) or succinate (white bar) media containing 30 µM La3+ and/or Ca2+. Results are shown as means with standard deviations (n = 3).
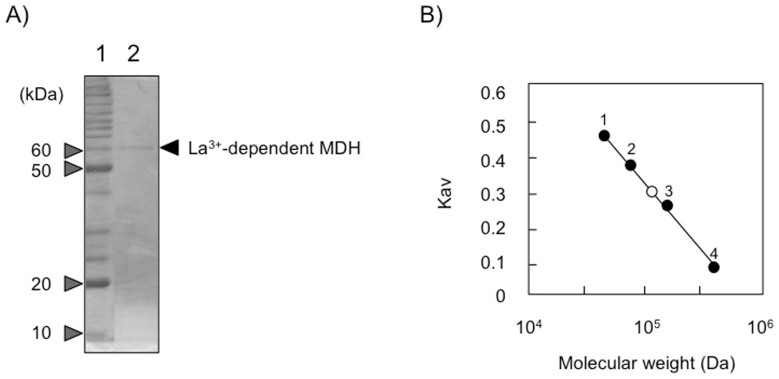
We then purified MDH from strain AM1 cells grown in methanol/La3++Ca2+ medium in order to identify the La3+-dependent MDH and to observe whether MxaF and XoxF are concurrently activated by La3+ and Ca2+ (Table 1). In all the purification steps, we observed only one fraction peak showing MDH activity (data not shown). The purified MDH had a specific activity of 10.0 U/mg of protein. The protein migrated as a single protein band on the SDS-PAGE gel with an apparent molecular mass of 61 kDa. A small protein corresponding to subunit β was not observed (Fig. 4), although the MDH purified from cells grown in methanol/Ca2+ medium showed two bands for α and β subunits (data not shown). Using gel chromatography with a Superdex G-200 GL column, the native molecular weight of the purified protein was estimated to be ca. 117 kDa (Fig. 4B). These results indicated that the purified MDH is a homodimer of only the α subunit. The purified enzyme contained 0.91 atoms of La3+ and 0.39 atoms of Ca2+ per dimer. After treatment with 50 mM EDTA, the La3+ and Ca2+ contents in the enzyme were shown to be 1.24 and 0.10 per dimer, respectively, suggesting that the La3+ is tightly bound to the enzyme. The N-terminal amino acid sequence of the MDH protein was NESVLKGVANPAEQVLQTVD, which was completely identical to 22–41 amino acid residues of the deduced amino acid sequence of the xoxF1 ORF. The predicted cleavage site of the amino acid sequence of XoxF1 as predicted by SignalP version 4.0 [32] was Ala21-Asn22. This cleavage site coincides completely with the N-terminal amino acid sequence of the purified MDH. Taken together, our data show that the xoxF1 gene encodes a functional La3+-dependent MDH and that XoxF1 may have a signal peptide for periplasmic localization.
Table 1. Purification scheme of the La3+-dependent MDH isolated from M. extorquens strain AM1.
| Step | Total activity (Unit) | Specific activity (U/mg) | Purification (fold) | Yield (%) |
| Cell free extract | 46 | 0.62 | 1.0 | 100 |
| PD-10 | 33 | 0.74 | 1.2 | 71 |
| Hi-trap SP HP Sepharose HP | 15 | 14 | 22 | 32 |
| MonoS 5/50 GL | 4.5 | 10 | 17 | 18 |
Figure 4. SDS–PAGE analysis (A) and molecular weight (B) of purified MDH from strain AM1 grown on methanol/Ca2++La3+ medium.
A, Lane 1, marker proteins; lane 2, purified MDH. B, Marker proteins were: 1, ovoalbumin (43 kDa); 2, conalbumin (75 kDa); 3, aldolase (158 kDa); and 4, ferritin (440 kDa).
XoxF1 encodes an essential MDH for the methylotrophy depending on La3+
We next examined the growth behavior and MDH induction patterns of an MxaF-disrupted mutant. In succinate media containing La3+ and/or Ca2+, strain ΔmxaF exhibited normal growth comparable to that of the wild-type strain (Fig. 5). Strain ΔmxaF could not grow on methanol at all in the presence of Ca2+ as previously reported [22], but its growth was restored by supplementation with La3+ even without Ca2+ (Fig. 5).
Figure 5. Phenotypic growth defects in strain ΔmxaF on methanol and succinate media.
Growth on media containing Ca3+ (white circle), La3+ (gray circle), and Ca2++La3+ (black circle). Graphs depict average data from three biological replicates.
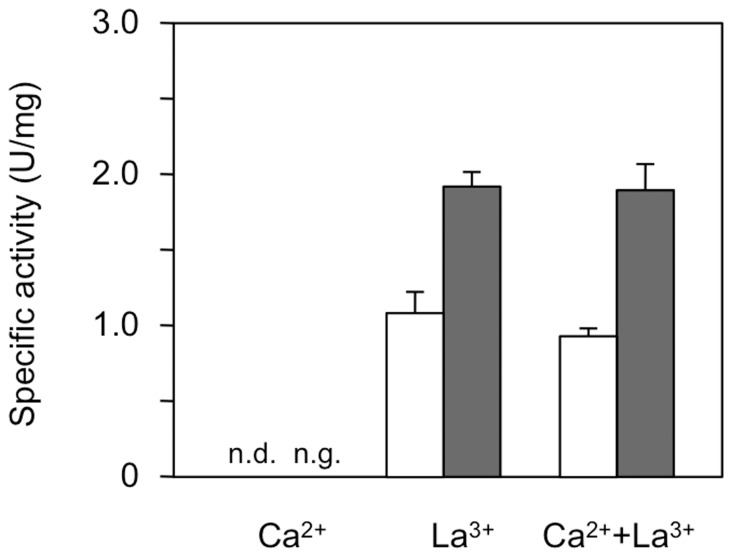
Strain ΔmxaF grown in succinate/Ca2+ medium did not show detectable MDH activity (Fig. 6). Strain ΔmxaF grown in succinate or methanol media containing La3+, however, showed MDH activity comparable to that of the wild-type strain grown in succinate or methanol media containing La3+ (Fig. 6). These results suggest that the XoxF1 is able to function as the MDH in the cells in the presence of La3+, in place of MxaF, which explains the growth of the mutant on methanol in the presence of La3+.
Figure 6. MDH activity in strain ΔmxaF grown on the methanol and succinate media with Ca2+ and/or La3+.
Cells were grown aerobically in succinate (white bar) and methanol (gray bar) media with 30 µM Ca3+, La3+, and Ca2++La3+ at 30°C. n.d.: not detected, and n.g.: no growth. Results are shown as means with standard deviations (n = 3).
Discussion
Methylobacterium species and various other bacteria harbor xoxF, which is homologous to mxaF, encoding the large subunit of MDH. Over the last few years, the function of XoxF has been the subject of controversy. Using strain AM1, Schmidt et al. showed that XoxF1 had low activities of methanol and formaldehyde dehydrogenase activity [21], and the work by Skovran et al. suggested that XoxF1 and XoxF2 might have some roles in the expression of the MDH genes [22]. Nevertheless, the function and physiological role of XoxF in methanol metabolism is not yet completely understood.
In this work, we showed that XoxF1 from strain AM1 functions as a La3+-dependent MDH and has a role in La3+-dependent methanol metabolism of the strain, because (i) purified XoxF1 from the strain grown on methanol in the presence of La3+ contained La3+ with significant MDH activity, (ii) strain AM1 could grow on the methanol/La3+ medium even without Ca2+, and (iii) the growth defect of strain ΔmxaF was completely restored by supplementation with La3+. In our previous work, we have shown that MDHs purified from M. radiotolerans strain NBRC15690 grown in the presence of REEs and non-methylotrophic bacterium Bradyrhizobium sp. strain MAFF211645 have significant MDH activity and that the N-terminal amino acid sequences of both enzymes were identical to those of XoxF homologues [23], [24]. These facts suggest that XoxF1 in strain AM1 has an important physiological role as an MDH in the methanol metabolism in the presence of La3+, and that an REE-dependent methanol-metabolic pathway may be distributed among known methylotrophic bacteria as well as in other non-methylotrophic bacteria containing XoxF homologues.
It has been reported that XoxF1 purified from strain ΔmxaF harboring the pCM80-xoxF-His vector grown on Ca2+-containing medium exhibits low MDH activity (V max value for methanol is 0.015 U/mg) [21]. La3+-dependent XoxF1, however, exhibited significant specific activity for methanol (10.0 U/mg), with levels over fifteen times higher than those in purified Ca2+-induced MxaFI from cells grown on Ca2+-containing medium (0.66 U/mg) (data not shown). Strain ΔmxaF grown in the methanol/Ca2+ medium had little MDH activity despite high expression levels of the xoxF1 gene (Fig. 3). Moreover, Ca2+-induced XoxF1 was a monomer [21], while La3+-containing XoxF1 was a homo-dimer of α-subunits only (Fig. 4). Thus it can be hypothesized that La3+ can facilitate the dimerization of XoxF1 protein, which in the absence of La3+ is an inactive monomeric apo-enzyme. Similarly, since there was no fraction showing any MDH activity except for that containing XoxF1, MxaFI may be inactive in cells grown in the presence of La3+, although their hetero-tetramerization has not been examined (data not shown). It has been reported that Ca2+-dependent enzymes and other proteins, e.g., horseradish peroxidase, might be inhibited by La3+ [27]–[30]. Thus we hypothesize that La3+ may inhibit MxaFI posttranslational activation and/or its activity. Taken together, our work suggests that the posttranslational activation of XoxF1 and that of MxaFI require La3+ and Ca2+, respectively, and that strain AM1 has the ability to generate MDH, either XoxF1 or MxaF, depending on which metal is present, for methanol metabolism.
La3+ is one of the REEs, which are relatively abundant in the earth's crust (35 µg/g for La3+, 66 µg/g for Ce3+, and 40 µg/g for Nd3+); in fact, the abundance of Ce3+ is almost equal to those of much more commonly studied elements in the environment, such as Cu and Zn [26]. La3+ exists in all plants examined, with levels of around 0.178–3.1 µg/g dry mass in leaves, which is in the same range as Mn (0.5–15.6 µg/g dry mass) and Fe (1.33–2.5 µg/g dry mass) [25]. Meanwhile, Delmotte et al. have reported that XoxF is highly expressed in bacterial phyllosphere communities in situ, with a prevalence similar to that of MxaF as demonstrated by shotgun proteomics [1]. Therefore, the Methylobacterium species, as major plant epiphytes, would be readily able to access La3+ and Ca2+ on plant surfaces in the natural environment, and it is highly possible that XoxF1 is active on plant leaf surfaces together with MxaFI, because XoxF1 and MxaF are induced by methanol regardless of the presence of La3+ and/or Ca2+.
In this paper, we showed that XoxF1 is a functional MDH that depends on La3+. As far as we know, this is the first report of a metabolic pathway and enzyme dependent on an REE as a cofactor. Recently, XoxF was reported to be involved in a complex regulatory cascade of a MxcQE two-component system [22]. Taking our data together with these reports, it appears that XoxF may play a dual role in both regulation of MDH genes and catalysis of methanol oxidation.
Materials and Methods
Bacterial strains, media, and cultivation
M. extorquens strains and plasmids used in this study are described in Table 2. M. extorquens strains were cultivated in minimal salts (MS) media [33] supplemented with 0.5% methanol or 0.4% succinate as a carbon source. MS medium with 0.5% methanol is referred to as methanol/Ca2+ medium, methanol/Ca2+ medium containing 30 µM LaCl3 is referred to as methanol/Ca2++La3+ medium, and MS medium with 0.5% methanol containing 30 µM LaCl3 instead of CaCl2 is referred to as Methanol/La3+ medium. For the cultivation of strain AM1 to purify La3+-dependent MDH, a 1/10 nutrient medium supplemented with 0.5% methanol and 30 µM LaCl3 was used [23]. In this medium, the content of Ca2+ was 31.8 µM. When appropriate, antibiotics were added at the following concentrations: tetracycline (Tc), 10 µg/ml, and kanamycin (Km), 50 µg/ml.
Table 2. Strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference | |
| Strains | AM1 | Wild type (JCM 2805) | JCM |
| Rifr derivative (wild type) | 18,19 | ||
| ΔmxaF | mxaF::Km | This study | |
| Plasmids | pCM184 | Allelic exchange suicide vector (Kmr Tcr Apr) | 35 |
| pΔmxaF | pCM184 with mxaF upstream and downstream flanks (Kmr Tcr Apr) | This study | |
| pCM130 | Promoter-less xylE fusion vector (Tcr) | 31 | |
| pRM246 | pCM130 with mxaF promoter region (Tcr) | This study | |
| pRM248 | pCM130 with xoxF1 promoter region (Tcr) | This study | |
Cultivation of M. extorquens strains was done in 200 µl of MS media in 96 well round bottom microplates (Asahi Glass Co., Ltd., Chiba, Japan) at 28°C with reciprocal shaking, and growth was monitored by measuring the optical density at 610 nm in the a HiTS BioMicroplate reader (Scinics co, Ltd., Tokyo, Japan).
Escherichia coli strains DH5α and S17-1 [34] were routinely cultivated at 37°C in Luria-Bertani medium. The following antibiotic concentrations were used: tetracycline, 10 µg/ml, kanamycin, 50 µg/ml, and ampicillin, 100 µg/ml.
Construction of null mutants
Null mutants were generated in mxaF using the allelic exchange vector pCM184 [35]. The following primers were used for the amplification of the up- and downstream regions of mxaF: upstream of mxaF, mxaFup-fw (5′-GAATTCTCACGATGCGGCACTTCGGATG-3′) and mxaFup-rv (5′-GAATTCTTCTCGACCTTCCACACCGTCTC-3′); downstream of mxaF, mxaFdn-fw (5′-GTTAACGGATGGACCACCTCGCCAAGGA-3′) and mxaFdn-rv (5′-GAGCTCTTCGCATCTGCCGTC, AGGCAGT-3′). Each PCR fragment was introduced into pCM184. The resulting allelic exchange vectors were introduced into M. extorquens strain AM1 via conjugation using E. coli strain S17-1. Mutations were confirmed by diagnostic PCR.
Construction of promoter fusions
mxaF and xoxF1 promoter fusions with xylE encoding catechol 2,3-dioxygenase were constructed in vector pCM130 [31]. The following primers were used for amplification of the promoter regions of mxaF and xoxF1: mxaF promoter, PmxaF-fw (5′-GGATCCGGTCAAGACGATGCCAATAC-3′) and PmxaF-rv (5′-AAGCTTCTCGGAAGTCATCCGAAGTG-3′); xoxF1 promoter, PxoxF1-fw (5′-GGATCCTTCGTTCAAGCTTCGGTTTC-3′) and PxoxF1-rv (5′-GCATGCTCATGGATTCCTCCGACAAG-3′). The resulting PmxaF-xylE and PxoxF1-xylE fusions were transferred into strain AM1 via conjugation.
Preparation of crude extracts and enzyme assays
Cells were grown on each medium for 36 h, then harvested and resuspended in 20 mM Tris–HCl buffer, pH 8.0. Cells were broken with a 3110BX mini-beadbeater (Biospec Products, Bartlesville, OK, USA) or an Ultrasonic Disruptor UD-201 (Tomy Seiko Co., Ltd., Tokyo, Japan). Cell debris was removed by centrifugation at 12,000×g for 10 min at 4°C.
MDH and catechol dioxygenase (XylE) activities were measured according to the methods of Day and Anthony [36] and Springer et al. [37], respectively. Protein concentration was determined according to the method of Bradford [38] with a protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA) by using bovine serum albumin as the standard.
Purification of La3+-dependent MDH
Cell-free extracts prepared from cells grown on methanol/Ca2++La3+ medium were applied to a PD-10 column (GE Healthcare UK Ltd., Buckinghamshire, UK) pre-equilibrated with 25 mM MES buffer, pH 5.0. The protein was eluted with the same buffer, and applied to a HiTrap Sepharose HP column pre-equilibrated with 25 mM MES buffer, pH 5.5. The protein was eluted with the same buffer containing 1 M NaCl at a flow rate of 1.0 ml/min. The active fractions were pooled and concentrated using an Ultrafree-MC microcentrifuge filter unit with a molecular weight cutoff of 5000 Da (Sigma-Aldrich Co., St. Louis, MO, USA). The purity of the enzyme was confirmed by SDS-PAGE analysis (12.5% polyacrylamide gel). The N-terminal amino acid sequence of the purified protein was determined on a protein sequencer Model 610A (Applied Biosystems, Foster City, CA, USA).
Determination of La3+ and Ca2+ contents of the media and the purified enzyme
The contents of La3+ and Ca2+ in the media were determined using an Agilent 7500cx ICP-MS system (Agilent Technologies, Inc., Santa Clara, CA, USA).
The buffer containing purified enzyme was re-equilibrated in 25 mM Tris-HCl buffer, pH 8.0, using a PD-10 column. The enzyme (2.6 µM) was then incubated with 50 mM EDTA, pH 8.0, at 30°C for 2 h, after which it was desalted and concentrated with an Amicon Ultra-0.5 mL 3 K concentrator (Millipore, Billerica, MA, USA) concentrator and diluted with Milli Q water (Millipore). The La3+ and Ca2+ contents in the enzyme were determined using an ULTIMA 2 ICP-OES spectrometer (Horiba Ltd., Kyoto, Japan).
Acknowledgments
The authors are grateful to Professor Mary E. Lidstrom (University of Washington) for providing R.M. with the plasmid pCM184 for making null mutants and the plasmid pCM130 for making promoter fusions. The authors are also grateful to Y. Izumi (Institute of Plant Science and Resources, Okayama University) for ICP-MS analysis.
Funding Statement
The authors have no support or funding to report.
References
- 1. Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, et al. (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci U S A 106: 16428–16433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kawaguchi K, Yurimoto H, Oku M, Sakai Y (2011) Yeast methylotrophy and autophagy in a methanol-oscillating environment on growing Arabidopsis thaliana leaves. PLoS ONE 6: e25257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, et al. (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6 doi:10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knief C, Frances L, Cantet F, Vorholt JA (2008) Cultivation independent characterization of Methylobacterium populations in the plant phyllosphere by automated ribosomal intergenic spacer analysis. Appl Environ Microbiol 74: 2218–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA (2010) Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J 4: 719–728. [DOI] [PubMed] [Google Scholar]
- 6.Holland MA, Long RLG, Polacco JC (2002) Methylobacterium spp.: Phylloplane bacteria involved in cross-talk with the plant host. In: Lindow SE, Hecht-Poinar EI Elliott VJ, editors. Phyllosphere Microbiology. APS Press. pp. 125–135.
- 7. Abanda-Nkpwatt D, Müsch M, Tschiersch J, Boettner M, Schwab W (2006) Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J Exp Bot 57: 4025–4032. [DOI] [PubMed] [Google Scholar]
- 8. Gourion B, Rossignol M, Vorholt JA (2006) A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc Natl Acad Sci U S A 103: 13186–13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sy A, Timmers AC, Knief C, Vorholt JA (2005) Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl Environ Microbiol 71: 7245–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME (2003) Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol 185: 2980–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schrader J, Schilling M, Holtmann D, Sell D, Filho MV, et al. (2009) Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol 27: 107–115. [DOI] [PubMed] [Google Scholar]
- 12. Vuilleumier S, Chistoserdova L, Lee MC, Bringel F, Lajus A, et al. (2009) Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4: e5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anthony C (2000) Methanol dehydrogenase, a PQQ-containing quinoprotein dehydrogenase. Subcell Biochem 35: 73–117. [DOI] [PubMed] [Google Scholar]
- 14. Anthony C, Williams P (2003) The structure and mechanism of methanol dehydrogenase. Biochim Biophys Acta 1647: 18–23. [DOI] [PubMed] [Google Scholar]
- 15. Williams PA, Coates L, Mohammed F, Gill R, Erskine PT, et al. (2005) The atomic resolution structure of methanol dehydrogenase from Methylobacterium extorquens . Acta Crystallogr D Biol Crystallogr 61: 75–79. [DOI] [PubMed] [Google Scholar]
- 16. Anthony C, Zatman LJ (1964) The methanol-oxidizing enzyme of Pseudomonas sp. M 27. Biochem J 92: 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lidstrom ME, Anthony C, Biville F, Gasser F, Goodwin P, et al. (1994) New unified nomenclature for genes involved in the oxidation of methanol in Gram-negative bacteria. FEMS Microbiol Lett 117: 103–106. [DOI] [PubMed] [Google Scholar]
- 18. Nunn DN, Lidstrom ME (1986) Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol 166: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nunn DN, Lidstrom ME (1986) Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J Bacteriol 166: 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chistoserdova L, Lidstrom ME (1997) Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 143: 1729–1736. [DOI] [PubMed] [Google Scholar]
- 21. Schmidt S, Christen P, Kiefer P, Vorholt JA (2010) Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology 156: 2575–2586. [DOI] [PubMed] [Google Scholar]
- 22. Skovran E, Palmer AD, Rountree AM, Good NM, Lidstrom ME (2011) XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol 193: 6032–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fitriyanto NA, Fushimi M, Matsunaga M, Pertiwiningrum A, Iwama T, et al. (2011) Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111: 613–617. [DOI] [PubMed] [Google Scholar]
- 24. Hibi Y, Asai K, Arafuka H, Hamajima M, Iwama T, et al. (2011) Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans . J Biosci Bioeng 111: 547–549. [DOI] [PubMed] [Google Scholar]
- 25. Tyler G, Olsson T (2005) Rare earth elements in forest-floor herbs as related to soil conditions and mineral nutrition. Biol Trace Elem Res 106: 177–191. [DOI] [PubMed] [Google Scholar]
- 26. Tyler G (2004) Rare earth elements in soil and plant systems-a review. Plant Soil 267: 191–206. [Google Scholar]
- 27.Brawn PH, Rathjen AH, Graham RD, Tribe DE (1990) Rare earth elements in biological systems., In: Gschneidner KA Jr, Eyring L, Editors, Handbook on the physics and chemistry of rare earths, vol. 13. North-Holland, Elsevier Science Publishers BV, pp. 423–452.
- 28. Erdmann F, Jung M, Eyrisch S, Lang S, Helms V, et al. (2009) Lanthanum ions inhibit the mammalian Sec61 complex in its channel dynamics and protein transport activity. FEBS Lett 583: 2359–2364. [DOI] [PubMed] [Google Scholar]
- 29. Liu P, Liu Y, Lu Z, Zhu J, Dong J, et al. (2004) Study on biological effect of La3+ on Escherichia coli by atomic force microscopy. J Inorg Biochem 98: 68–72. [DOI] [PubMed] [Google Scholar]
- 30. Wang L, Zhou Q, Lu T, Ding X, Huang X (2010) Molecular and cellular mechanism of the effect of La(III) on horseradish peroxidase. J Biol Inorg Chem 15: 1063–1069. [DOI] [PubMed] [Google Scholar]
- 31. Marx CJ, Lidstrom ME (2001) Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147: 2065–2075. [DOI] [PubMed] [Google Scholar]
- 32. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 33. Harder W, Attwood M, Quayle JR (1973) Methanol assimilation by Hyphomicrobium spp. J Gen Microbiol 78: 155–163. [Google Scholar]
- 34. Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotechnol 1: 784–779. [Google Scholar]
- 35. Marx CJ, Lidstrom ME (2002) Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33: 1062–1067. [DOI] [PubMed] [Google Scholar]
- 36. Day DJ, Anthony C (1990) Methanol dehydrogenase from Methylobacterium extorquens AM1. Methods Enzymol 188: 210–216. [Google Scholar]
- 37. Springer AL, Morris CJ, Lidstrom ME (1997) Molecular analysis of MxbD and MxbM, a putative sensor-regulator pair required for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology 143: 1737–1744. [DOI] [PubMed] [Google Scholar]
- 38. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]