Abstract
Dichloroacetamide safeners protect maize (Zea mays L.) against injury from chloroacetanilide and thiocarbamate herbicides. Etiolated maize seedlings have a high-affinity cytosolic-binding site for the safener [3H](R,S)-3-dichloroacetyl-2,2,5-trimethyl-1,3-oxazol-idine ([3H]Saf), and this safener-binding activity (SafBA) is competitively inhibited by the herbicides. The safener-binding protein (SafBP), purified to homogeneity, has a relative molecular weight of 39,000, as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and an isoelectric point of 5.5. Antiserum raised against purified SafBP specifically recognizes a 39-kD protein in etiolated maize and sorghum (Sorghum bicolor L.), which have SafBA, but not in etiolated wheat (Triticum aestivum L.), oat (Avena sativa L.), barley (Hordeum vulgare L.), tobacco (Nicotiana tabacum L.), or Arabidopsis, which lack SafBA. SafBP is most abundant in the coleoptile and scarcest in the leaves, consistent with the distribution of SafBA. SBP1, a cDNA encoding SafBP, was cloned using polymerase chain reaction primers based on purified proteolytic peptides. Extracts of Escherichia coli cells expressing SBP1 have strong [3H]Saf binding, which, like binding to the native maize protein, is competitively inhibited by the safener dichlormid and the herbicides S-ethyl dipropylthiocarbamate, alachlor, and metolachlor. SBP1 is predicted to encode a phenolic O-methyltransferase, but SafBP does not O-methylate catechol or caffeic acid. The acquisition of its encoding gene opens experimental approaches for the evaluation of the role of SafBP in response to the relevant safeners and herbicides.
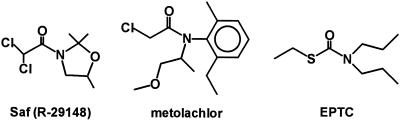
Herbicide safeners, also known as antidotes, are used to protect crops from herbicide injury (Hatzios, 1983, 1989; Fuerst, 1987). One important class of safeners is the dichloroacetamides, such as dichlormid, which are used in combination with thiocarbamate and chloroacetanilide herbicides on maize (Zea mays L.) and sorghum. Examples of these classes of compound are shown in Figure 1. Although it is well established that in maize dichloroacetamide safeners elevate GSH levels and induce novel GSH S-transferases, the mechanism by which they do so is not known (Lay et al., 1975; Fuerst et al., 1993).
Figure 1.
Structures of compounds discussed in the text: Saf, a dichloroacetamide herbicide safener; metolachlor, a chloroacetanilide herbicide; and EPTC, a thiocarbamate herbicide.
Etiolated maize seedlings contain a soluble, high-affinity-binding activity (Kd, 0.12 μm) for a tritiated form of the dichloroacetamide safener Saf (Fig. 1) (Walton and Casida, 1995). Although present in all tissues, SafBA is highest in the coleoptile. There is a good correlation between inhibition of Saf binding in vitro and safener action in vivo among a series of Saf analogs, with dichlormid, a widely used dichloroacetamide safener, being the strongest binding antagonist of any compound tested (IC50, 10 nm). Chloroacetanilide and thiocarbamate herbicides, which dichloroacetamide safeners protect against, are also strong competitive inhibitors of Saf binding; metolachlor and EPTC (Fig. 1) have IC50 values of 110 and 40 nm, respectively (Walton and Casida, 1995). Despite their intensive use in agriculture for more than 30 years, the mode of action of these herbicides is currently unknown.
To begin to address the question of the role of SafBA in the response of maize seedlings to dichloroacetamide safeners and, possibly, to the two classes of herbicides, we have further characterized SafBA. Here we report the purification of SafBP, immunological studies of its tissue and species distribution, and the cloning, sequencing, and expression in Escherichia coli of a cDNA encoding SafBP.
MATERIALS AND METHODS
Genotypes and sources of plant materials are as described by Walton and Casida (1995). The Arabidopsis thaliana ecotype was Columbia and the tobacco (Nicotiana tabacum) cultivar was SR-1. Plants were grown in the dark on wet paper towels for 4 to 7 d.
Binding Assay
Racemic Saf and [3H]Saf (specific activity, 15 Ci/mmol) were prepared according to the method of Latli and Casida (1995). The sources of other chemicals are given by Walton and Casida (1995). [3H]Saf binding was measured by incubating extracts with approximately 400,000 cpm of [3H]Saf in 50 mm Tris-HCl, pH 8.0, in a total volume of 1 mL for 60 min at 22°C. The extracts were then filtered through glass-fiber filters (GF/A, Whatman) that had been soaked in 0.3% (v/v) polyethylenimine and rinsed with water. The radioactivity retained on the filters was measured by scintillation counting (Walton and Casida, 1995).
Purification of SafBP
Coleoptiles of 4-d-old maize (Zea mays L.) seedlings grown in partial darkness on wet paper towels at 23°C (typically 5–10 g fresh weight) were ground in 50 mm Tris-HCl, pH 8.0, plus 0.4 m Suc (Walton and Casida, 1995). After being filtered through cheesecloth, the extracts were treated by adding (NH4)2SO4 to 50% saturation (0.295 g/mL), stirring for 30 min at 4°C, and centrifuging for 15 min at 10,000g. (NH4)2SO4 was again added to the supernatant to a final concentration of 70% saturation (an additional 0.127 g/mL) and after stirring and centrifugation the pellet was redissolved in chromatofocusing start buffer (25 mm piperazine-HCl, pH 6.3) and desalted on a gel-filtration column (PD-10, Pharmacia).
The protein extract was fractionated by chromatofocusing on a Mono P HR5/20 column (Pharmacia) using an HPLC column (Beckman) equipped with a controller and two pumps (model 421A controller and model 114M pumps, Beckman). The elution buffer was 10% (v/v) Polybuffer 74 (Pharmacia) adjusted to pH 4.6 with HCl. Fractions (each 1 mL) with binding activity were pooled and adjusted to 0.1 m KH2PO4, pH 7.0, and 1.7 m (NH4)2SO4. This preparation was then fractionated by hydrophobic-interaction chromatography on an HPLC column (TSK-Phenyl-5-PW, Bio-Rad). Buffer A was 0.1 m KPO4, pH 7.0, plus 1.7 m TSK-(NH4)2SO4, and buffer B was water. The gradient was 100% A to 100% B in 30 min at a flow rate of 1 mL/min.
Fractions (each 1 mL) with binding activity were pooled, desalted on a PD-10 column, and fractionated by anion-exchange chromatography on a TSK-DEAE-5-PW HPLC column. Buffer A was 25 mm Tris-HCl, pH 8.0, and buffer B was 25 mm Tris-HCl plus 0.8 m KCl, pH 8.0. The gradient was 100% A to 100% B in 30 min at a flow rate of 1 mL/min. One-milliliter fractions were collected, and aliquots of each fraction were assayed for SafBA and analyzed by SDS-PAGE (12% [v/v] acrylamide; Scott-Craig et al., 1992). Gels were stained with Coomassie blue R-250. The Mr of the proteins was estimated using prestained standards from GIBCO-BRL. Gel-filtration chromatography was on a Superdex 200H 10/30 column (Pharmacia) eluted with 0.1 m KH2PO4, pH 7.0. The column was equilibrated with gel-filtration standards from Bio-Rad.
The N-terminal sequence could not be obtained from purified SafBP. Internal peptides were generated by digestion with trypsin or endoproteinase Asp-N (both from Sigma) at the Michigan State University Macromolecular Facility (East Lansing). Digestions were with 0.3 μg/mL proteinase in 10% acetonitrile, 0.1 m Tris-HCl, pH 8.2, and 1% reduced Triton X-100 (Sigma), for 24 h at 37°C. Peptides were fractionated on a 0.8- × 250-mm microbore reverse-phase column (LC Packings, San Francisco, CA), packed with 5-μm, 300-Å C18 resin (Vydac, Hesperia, CA). The flow rate was 40 μL/min, and the gradient was 95% solvent A to 50% solvent A in 145 min. Solvent A was 0.1% trifluoroacetic acid and solvent B was 90% (v/v) acetonitrile containing 0.085% trifluoroacetic acid. Sequencing was done by automated Edman degradation.
Antibody Preparation and Immunoblotting
Purified SafBP (50 μg) was homogenized in Titermax adjuvant (CytRx, Norcross, GA) and injected subcutaneously into New Zealand White rabbits. Rabbits were bled after 42 and 56 d without any boost. The crude antiserum could be used to detect SafBP on immunoblots at a 1:16,000 dilution and was routinely used at a 1:5,000 dilution. SDS-PAGE gels were blotted onto 0.2-μm-pore nitrocellulose (Schleicher & Schuell) using a semidry Multiphor II apparatus according to the manufacturer's instructions (LKB, Pharmacia). Bound anti-SafBP antibody was detected using goat anti-rabbit IgG conjugated to alkaline phosphatase (Kirkegaard and Perry Laboratories, Gaithersburg, MD) (Scott-Craig et al., 1992).
Cloning of SBP1 cDNA by PCR
Routine nucleic acid manipulations were carried out as described by Sambrook et al. (1989) unless otherwise specified. The cDNA library, made from poly(A+) RNA extracted from 12-d-old etiolated maize seedlings of inbred B73 in vector λZipLox (GIBCO-BRL), was obtained from Dr. Tim Helentjaris (Pioneer Hi-Bred International, Johnston, IA).
PCR was with DNA extracted from the cDNA library as a template. PCR reactions were carried out in 100-μL final volumes using 0.25 unit of Taq polymerase (Promega), 20 mm Tris, pH 8.9, 50 mm KCl, 0.01% Triton X-100, 1 mm DTT, 1.5 mm MgCl2, 75 μm of each deoxynucleotide triphosphate, 1 μm of each primer, and 100 ng of cDNA library template for 45 cycles (one cycle being 94°C for 1 min, 50°C for 2 min, and 72°C for 3 min). Three pairs of degenerate primers based on the amino acid sequences of three peptides (nos. 5, 6, and 8 in Table I) from purified SafBP were synthesized in both orientations and tried in all combinations.
Table I.
Peptides obtained from SafBP
| Tryptic Peptide Sequence | Deduced Amino Acid Sequence and Position in SBP1 |
|---|---|
| 1. AFADIWFV | 16- AqADIWrl |
| 2. DKQSSE(E)(G)(M)Y(L)ISPV(S)YL | 87- DKQSSEEerYrISPVSYL |
| 3. DVVTSPFEELXGAT(L) | 143- DVVTSPFEELhGATL |
| 4. DHLHGATLFHESMGSL | 150- eeLHGATLFHESMGSL |
| 5. DNFGIEIAMXEF | 181- DNFGIEIAMrEF |
| 6. DGAVKNYVEGRQ | 242- DGAmiNYVEGdm |
| 7. LLAELFT | 334- ewsELFT |
| 8. DLFTKAGFSEYKILKEFGA | 337- eLFTKAGFSEYKILKEFGA |
Peptides nos. 1 and 7 were obtained with trypsin and the others with endoproteinase Asp-N. Parentheses indicate uncertain identification and X indicates unknown. Amino acids in lowercase indicate differences between the peptides and the deduced amino acid sequence of SBP1. Peptide no. 5 was obtained twice from two digestions.
A portion of the cDNA encoding SafBP was successfully amplified using primer 235 (5′-GAYAAYTTYGGNATHGA, based on the amino acid sequence DNFGIE in peptide no. 5 in Table I) and primer 240 (5′-GCNCCRAAYTCYTTNA, based on the amino acid sequence LKEFGA in peptide no. 8 in Table I). Y is C or T, N is any nucleotide, R is A or G, and H is A, T, or C. The PCR products were separated by gel electrophoresis (0.7% agarose) and blotted onto Nytran membranes (Schleicher & Schuell), and amplification of the correct DNA sequence was tested by hybridization at 46°C with radiolabeled primer 237 (5′-GARTAYAARATHCTNAA, based on the amino acid sequence EYKILK in peptide no. 8 in Table I).
For hybridization, the primer (50 pmol) was labeled in a 25-μL reaction at 37°C for 15 min using 66 pmol of [γ-32P]ATP (DuPont), 10 units of T4 polynucleotide kinase (New England Biolabs), and buffer supplied by the manufacturer. Hybridization was at 46°C. The approximately 500-bp PCR product was cloned into the EcoRV site of pBluescript (Stratagene), and was again confirmed by hybridization with probe 237 and by sequencing. The PCR product was used as a probe to isolate cDNAs from the cDNA library and the entire sequence of both strands of one was determined by automated fluorescence sequencing at the Michigan State University Plant Research Laboratory Biochemistry Facility. Sequences were analyzed by the BLAST (Altschul et al., 1990), DNASIS (Hitachi, San Bruno, CA), and PILEUP programs (Genetics Computer Group, 1994).
RNA Analysis
RNA was extracted from etiolated maize seedlings and tissues after freezing in liquid N2 by the method of DeVries et al. (1988). Total RNA (30 μg per lane) was fractionated by agarose gel electrophoresis with formaldehyde and blotted onto Nytran membranes (Schleicher & Schuell) (Sambrook et al., 1989). Hybridization was carried out using the method of Singh and Jones (1984) at 65°C, and filters were washed twice for 10 and 60 min at 65°C in 0.2× SSPE (1× SSPE is 150 mm NaCl, 10 mm NaH2PO4, and 1 mm EDTA, adjusted to pH 7.4 with NaOH) plus 0.2% SDS before autoradiography. The size of the SBP1 mRNA was estimated using a 0.24- to 9.5-kb RNA ladder (GIBCO-BRL). Equal loading of lanes was confirmed by staining of the rRNA bands with ethidium bromide.
Mapping
SBP1 was mapped using a set of recombinant inbred lines (Burr et al., 1994) provided by Dr. Benjamin Burr (Brookhaven National Laboratory, Upton, NY). Genomic DNA was extracted from 14-d-old plants of the parental maize lines CM37 and T232 and the 50 recombinant lines by the method of Hulbert and Bennetzen (1991). Eight micrograms of each DNA sample was digested with EcoRI, separated on a 0.7% agarose gel, and blotted onto Nytran membranes. Hybridization and washing were carried out as described above. The filters were then stripped of probe and rehybridized with control probe BNL17.18 to ensure that all samples were correctly labeled. The mapping data were analyzed using INBRED software (Burr et al., 1994).
Expression of SafBP in Escherichia coli
Vector pZL1 (GIBCO-BRL), containing the SafBP cDNA clone, was digested with EcoRI and NotI, the 1.5-kb insert was cloned into the corresponding sites in the E. coli expression vector pET21c (Novagen, Madison, WI), and the construct was transformed into E. coli strain BL21(DE3) (Novagen). A 50-mL culture of this strain was grown at 37°C to an A600 of 0.5, at which point expression of the cDNA was induced with 1 mm IPTG. As a control, E. coli BL21(DE3) transformed with pET21c with no insert was grown in parallel. Samples (12.5 mL) were removed 0, 1, 2, and 3 h after induction, centrifuged, extracted by the freeze-thaw method according to Novagen, and centrifuged again to remove cell debris. SafBA was in the supernatant and was not further purified. Each sample was in a final volume of 5 mL of 50 mm Tris-HCl, pH 8.0, and 150 μL of this was used in the [3H]Saf-binding assay according to the method of Walton and Casida (1995).
OMT Assays
OMT assays were done using substrates from Sigma in a total volume of 500 μL containing 50 mm Tris-HCl, pH 8.0, 5 mm MgCl2, and 0.5 μCi S-[methyl-3H]-S-adenosyl Met (New England Nuclear; specific activity, 14.4 Ci/mmol). The addition of 2 mm DTT did not influence the activity. Reactions were incubated for 30 min at 21°C and stopped by the addition of 50 μL of 2 m HCl. Organic scintillation cocktail (5 mL) was then added and nonpolar radioactivity was determined directly (Preisig et al., 1989).
RESULTS
Purification of SafBP
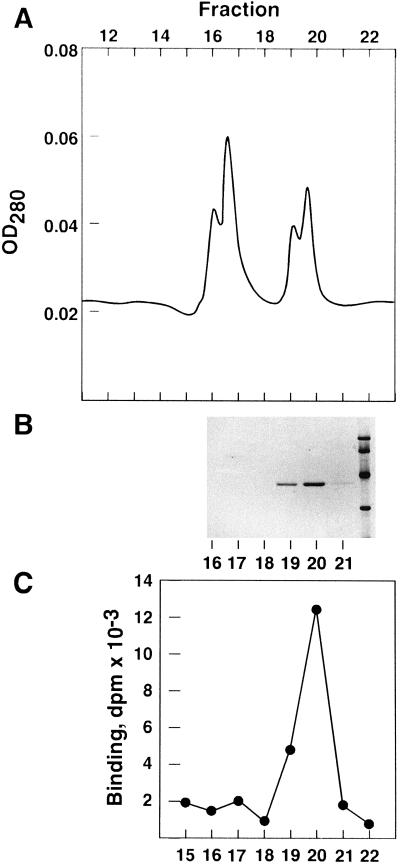
SafBA was purified by (NH4)2SO4 precipitation and three chromatographic steps to homogeneity, as judged by the presence of a single band on a one-dimensional SDS-polyacrylamide gel. SafBA is apparently the result of a protein (SafBP) with a molecular mass of 39 kD (Fig. 2). At no step in the purification was there any evidence of additional SafBPs. SafBP was shown by gel filtration to have a mass of 45 kD, and is therefore probably a monomer in its native state (data not shown). Based on Scatchard analysis showing that coleoptiles contain 55 pmol SafBA mg−1 protein (Walton and Casida, 1995), and assuming that the binding stoichiometry is 1, it is estimated that SafBP constitutes 0.2% of the total protein in the coleoptile. [3H]Saf binding to purified SafBP was competitively inhibited by thiocarbamate and chloroacetanilide herbicides (data not shown), consistent with our earlier results with SafBA in crude extracts (Walton and Casida, 1995).
Figure 2.
Final purification step of SafBP. A, A280 (OD280) of anion-exchange HPLC fractionation. B, SDS-PAGE of 50-μL aliquots of each HPLC fraction. C, [3H]Saf binding in 50 μL of each fraction. Equivalent HPLC fractions are aligned vertically in A, B, and C.
Immunological Analysis of SafBP
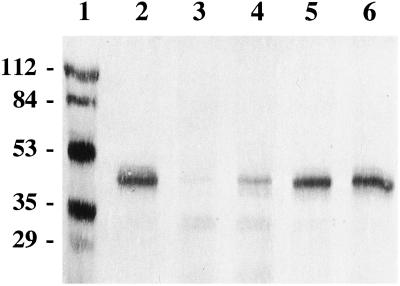
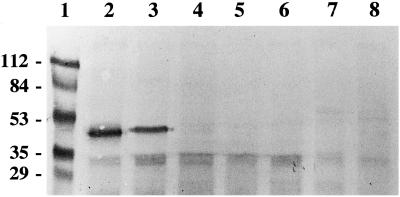
SafBP is present in the coleoptile, mesocotyl, and root, and to a lesser extent in the node and leaf (Fig. 3), consistent with earlier results showing that SafBA is most abundant in the coleoptile, present in the node, mesocotyl, and roots, and scarcest in the leaf (Walton and Casida, 1995). Antiserum raised against purified SafBP recognizes a major 39-kD protein in maize and sorghum (Sorghum bicolor L.), both of which contain SafBA, but not in wheat (Triticum aestivum L.), oats (Avena sativa L.), or barley (Hordeum vulgare L.), which do not contain SafBA (Fig. 4; Walton and Casida, 1995). Arabidopsis and tobacco, which also lack SafBP (J.D. Walton, unpublished results), also lack a protein recognized by anti-SafBP antiserum (Fig. 4). The nature of the cross-reacting protein(s) of approximately 30 kD is not known (Fig. 4).
Figure 3.
Immunodetection of SafBP in tissues of etiolated maize seedlings. Node extracts included approximately 0.5 mm of tissue on either side. Lane 1, Molecular mass markers (in kD); lane 2, coleoptile; lane 3, leaf; lane 4, node; lane 5, mesocotyl; and lane 6, root. Each lane of SDS-PAGE gel was loaded with 80 μg of protein. After transfer to nitrocellulose membranes, the blot was reacted with rabbit anti-SafBP antiserum and visualized with alkaline phosphatase coupled to goat anti-rabbit anti-IgG.
Figure 4.
Immunodetection of SafBP in different plant species. Blotting and visualization were as described in the legend to Figure 3 and Methods. Lane 1, Molecular mass markers (in kD); lane 2, maize; lane 3, sorghum; lane 4, wheat; lane 5, barley; lane 6, oats; lane 7, Arabidopsis; and lane 8, tobacco. Each lane was loaded with 80 μg of protein.
Cloning of a cDNA Encoding SafBP
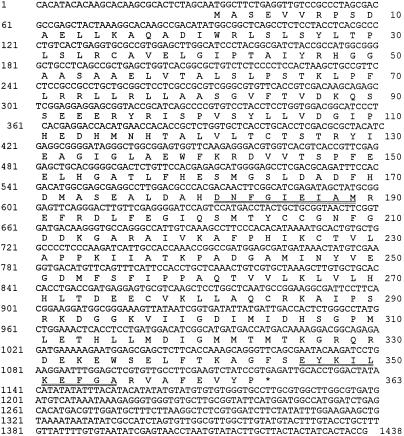
Based on the peptides obtained from proteolytic digests of SafBP shown in Table I, PCR was used to isolate three independent cDNAs encoding SafBP. All three cDNAs are approximately the same length and have the identical nucleotide sequences for at least 200 bp from either end in the regions where they overlap. The sequence of the longest cDNA, designated SBP1, has a single reading frame encoding a protein of 363 amino acids from the first in-frame Met to the first stop codon (Fig. 5). The predicted product is hydrophilic and has a Mr of 40,252 and a pI of 5.93, consistent with the measured biochemical properties of SafBP. All further experiments were done with SBP1.
Figure 5.
Sequence of SBP1, a cDNA encoding SafBP, and its deduced amino acid sequence. The two peptides used to design the PCR primers are indicated by underlining.
All of the peptides obtained from SafBP (Table I) were found in the predicted sequence of SBP1 (Table I; Fig. 5), although there are some discrepancies in individual amino acids. Based on peptide nos. 4, 7, and 8 (Table I), it appears that endoproteinase Asp-N sometimes cuts at Glu as well as Asp residues. Because only one SafBP was found during the purification of SafBA (Fig. 2), and because SBP1 encodes a protein with SafBA (see below), we conclude that the peptides in Table I are derived from the same gene as SBP1 and not from a related gene.
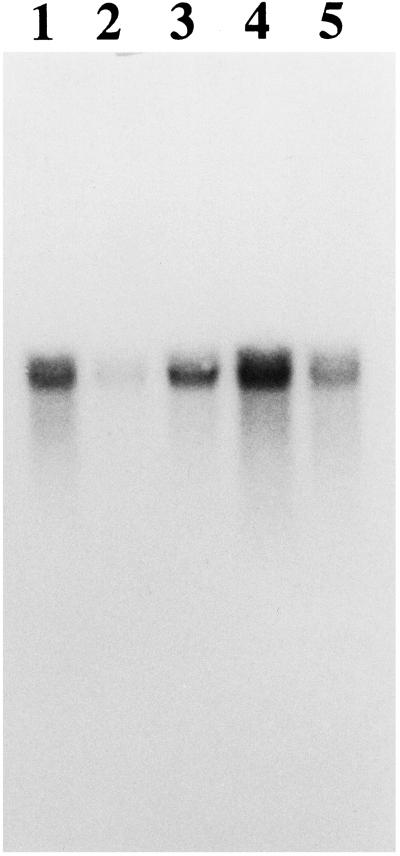
SBP1 hybridized strongly with one major band and weakly with at least one other band on Southern blots of total maize genomic DNA. SBP1 also hybridized with sorghum DNA but, even under low-stringency hybridization conditions, SBP1 did not hybridize with DNA from oats, wheat, or barley (data not shown). The SBP1 mRNA was present in all parts of the etiolated maize seedling but was present in the lowest levels in the leaves (Fig. 6). The low levels of SBP1 mRNA in the leaves is consistent with the low levels of SafBA (Walton and Casida, 1995) and SafBP (Fig. 3) in this tissue.
Figure 6.
RNA blot of etiolated maize tissues. Lane 1, Coleoptile; lane 2, leaf; lane 3, node; lane 4, mesocotyl; and lane 5, root. The node included about 0.5 mm of tissue on either side. Equal loading (30 μg of total RNA) in each lane was confirmed by staining of the rRNA bands with ethidium bromide. The SBP1 mRNA is approximately 1.5 kb.
The gene corresponding to SBP1 was mapped using the protocol of Burr et al. (1994) to a location near the centromere of chromosome 2, between markers npi242C and rbcS2. SBP1 has been entered on the maize map as marker msu1.
Expression of SafBP in E. coli
SBP1 was expressed in E. coli behind a promoter inducible by IPTG. Table II shows that extracts of E. coli expressing SBP1 have strong SafBA, whereas cells transformed with the empty expression vector have no binding. The results shown in Table III indicate that, like [3H]Saf binding to the native maize protein, [3H]Saf binding in E. coli extracts is inhibited by dichlormid, EPTC, metolachlor, acetochlor, and alachlor.
Table II.
[3H]Saf binding in 50-μL extracts of E. coli expressing SBP1
| Time | Control
|
SBP1
|
||
|---|---|---|---|---|
| Protein | Specific binding | Protein | Specific binding | |
| h | μg | dpm × 10−3 | μg | dpm × 10−3 |
| 0 | 3.8 | 0 | 6.6 | 0 |
| 1 | 10.1 | 0 | 17.1 | 17.9 |
| 2 | 9.5 | 0 | 20.9 | 51.1 |
| 3 | 13.3 | 0 | 12.3 | 24.6 |
After growing to an A600 of 0.5, cells were induced with IPTG and aliquots were harvested at the indicated times, lysed, and centrifuged to remove cell debris. The final volume of each extract was 5 mL. Control cells were transformed with the vector without insert. Values are the average of duplicates.
Table III.
Inhibition of [3H]Saf binding by safeners and herbicides in extracts of E. coli expressing SBP1
| Compounda | Concentration Tested | Total [3H]Saf Binding |
|---|---|---|
| μm | dpm × 10−3 | |
| None ([3H]Saf alone) | 67.3 | |
| [3H]Saf | ||
| + Saf (120 nm) | 2.5 | 3.0 |
| + Dichlormid (10 nm) | 1.4 | 0.7 |
| + EPTC (110 nm) | 1.6 | 1.7 |
| + Acetochlor (180 nm) | 1.1 | 11.6 |
| + Metolachlor (40 nm) | 1.1 | 3.3 |
| + Alachlor (70 nm) | 1.1 | 1.7 |
The E. coli/SBP1 2-h time-point preparation from Table I was used; protein content per sample was 20.9 μg. Values are the average of duplicates.
Concentrations that reduce specific binding of [3H]Saf by 50% (IC50) are shown in parentheses (from Walton and Casida, 1995).
Analysis of the Sequence of SafBP
The TBLASTN (Altschul et al., 1990) and TFASTA (Genetics Computer Group, 1994) programs showed that SafBP is related to a number of known or putative OMTs that methylate phenolic compounds such as catechol, lignin precursors, and flavonoids. Significant BLAST scores were also obtained against OMTs involved in methylation of inositol, acetylserotonin, and hydroxyindole. Like SafBP, most of the similar plant OMTs are soluble proteins of approximately 40 kD. The amino acid sequence of SafBP is most closely related to that of a putative OMT in barley that is induced by pathogens and UV light, with an overall amino acid identity and similarity of 38 and 63%, respectively (Gregersen et al., 1994; GenBank accession no. X77467). In contrast to the barley OMT, however, SafBP is constitutively expressed and is not induced further by safener or herbicide treatment (Walton and Casida, 1995; Fig. 6) or by infection with a fungal pathogen (data not shown). The sequence of SafBP is also related to that of a putative OMT, the mRNA of which, ZRP4, is abundantly expressed in maize roots (34% amino acid identity, 60% similarity; Held et al., 1993; GenBank accession no. L14063) and might be involved in the biosynthesis of suberin phenolics of the Casparian strip. Like SBP1, ZRP4 is poorly expressed in leaves (Held et al., 1993).
The closest match of SafBP to any protein of established function is to S-adenosyl-l-Met:6a-hydroxymaackiain 3-O-methyltransferase of pea, with 36% amino acid identity and 46% similarity (Preisig et al., 1989; GenBank accession no. U69554). SafBP is also related to a maize caffeic acid OMT (the product of the bm3 gene), with 30% amino acid identity and 54% similarity (Collazo et al., 1992; Vignols et al., 1995; GenBank accession no. M73235), and to caffeic acid OMTs from alfalfa and Monterey pine (Gowri et al., 1991; GenBank accession nos. M63853, U39301, and U70873). The sequence of SafBP shows no significant similarity to other classes of methyl transferases, including those that use CoA derivatives as substrates (e.g. Pakusch and Matern, 1991), protein methylases, and DNA methylases.
Three conserved motifs have been found in most OMTs (Kagan and Clarke, 1994). SafBP has a reasonable match to consensus motif II in the correct position (consensus: [G/P]- [T/Q][A/Y/F]DA[Y/V/I][I/F][L/V/C]; SafBP: PpAqtVvL, starting at amino acid 258; matches are shown in uppercase), but does not have the highly conserved central Asp residue. SafBP does not have a good consensus motif III in the proper position relative to motif II, but this motif is also not well conserved among known plant phenolic OMTs. In this region, the plant OMTs and SafBP share the sequence GKVI (starting at amino acid 295). However, SafBP lacks motif I, which is not only the most highly conserved of the motifs among all OMTs but is found in all of the plant phenolic OMTs.
Partially purified maize extracts have strong catechol and caffeic acid OMT activity, which are completely resolved from SafBA by chromatofocusing (data not shown). Therefore, despite its primary sequence similarity to phenolic OMTs, SafBP does not appear to catalyze methyl transfer to catechol or caffeic acid.
DISCUSSION
Based on its binding properties, SafBP is a good candidate for the initial site of action of Saf and perhaps other dichloroacetamide safeners. It binds active compounds with high affinity and is abundant in the coleoptile, an important tissue for the action of safeners and herbicides (Hickey and Krueger, 1974). SafBP is present in maize and sorghum, which respond to Saf, and absent in wheat, oat, and barley, which do not respond. SafBP also strongly binds chloroacetanilide and thiocarbamate herbicides in a competitive manner, suggesting that SafBP might also be involved in the response to the herbicides against which dichloroacetamide safeners protect.
Immunoblotting established that the absence of SafBA in plants other than maize and sorghum is due to lack of SafBP and is not due to an artifact of the binding assay. In addition, the species and tissue distribution of SafBP determined immunologically is consistent with the distribution based on SafBA (Walton and Casida, 1995).
Extracts of E. coli expressing a cDNA clone of SafBP bind [3H]Saf, which proves that SBP1 encodes SafBP. Like [3H]Saf binding to the native maize protein, [3H]Saf binding to SafBP from E. coli is inhibited by chloroacetanilide and thiocarbamate herbicides, indicating that the competitive binding of the herbicides and the safener to the same native maize protein as reported previously is not an artifact (Walton and Casida, 1995).
The predicted primary structure of SafBP suggests that it is an OMT. Ebert (1980) reported that metolachlor-treated sorghum seedlings had reduced lignification, consistent with the hypothesis that SafBP is involved in the production of lignin precursors such as caffeic acid and is the site of action of this herbicide, which binds SafBP with high affinity. Hickey and Krueger (1974) also found that corn coleoptiles treated with alachlor had reduced lignification. Insofar as lignification affects cell shape and tissue structure, perturbation of this process by the herbicides and/or safeners could account for some of the observed cytological and morphological effects of the herbicides (Ebert, 1980). However, SafBP does not catalyze methyl transfer from S-adenosyl Met to catechol or caffeic acid. SafBP might catalyze methyl transfer to an as-yet-unknown substrate such as a flavonoid, it might require special conditions (e.g. pH, cofactors, or metal ions) not present in our assays, or it might not actually be an OMT. The absence of an amino acid motif that is highly conserved among all known OMTs suggests that SafBP is not an enzymatically active OMT (Kagan and Clarke, 1994).
SafBP may not be the primary site of action of the dichloroacetamide safeners and/or herbicides, but might still be important in the pharmacokinetics of safener and/or herbicide action because of its relatively high abundance in the coleoptile. By temporarily sequestering the herbicides from their site of action, SafBP might allow detoxification mechanisms such as GSH conjugation time to be induced before phytotoxicity can occur.
The purification of SafBP and the isolation of its encoding gene will now permit a functional analysis of SafBP in the response of maize and sorghum to dichloroacetamide safeners and to chloroacetanilide and thiocarbamate herbicides by modification of its expression in maize and in heterologous plant hosts.
ACKNOWLEDGMENTS
We thank Benjamin Burr (Brookhaven National Laboratory, Upton, NY), Richard Kneusel (University of Freiburg, Germany), Maurice Snook (U.S. Department of Agriculture, Athens, GA), Scott Chilton (North Carolina State University, Raleigh), and Tim Helantjaris (Pioneer Hi-Bred International, Johnston, IA) for their generous gifts of biological and chemical materials. We thank Joe Leykam of the Michigan State University Macromolecular Structure Facility for proteolytic digestion, sequencing of SafBP peptides, and oligonucleotide synthesis.
Abbreviations:
- EPTC
S-ethyl dipropylthiocarbamate
- IC50
inhibitor concentration that reduces specific binding by 50%
- IPTG
isopropyl-β-d-thiogalactopyranoside
- OMT
O-methyltransferase
- Saf
the dichloroacetamide safener (R,S)-3-dichloroacetyl-2,2,5-trimethyloxazolidine (also known as R-29148)
- SafBA
safener-binding activity
- SafBP
safener-binding protein
Footnotes
This project was supported by Novartis, Inc., the Department of Energy, Division of Energy Biosciences (grant no. DEFG02-91ER20021 to J.D.W.), and the National Institute of Environmental Health Sciences (grant no. P01 ES00049 to J.E.C.).
The accession number for the SBP1 sequence described in this article is AF033496.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Burr B, Burr FA, Matz EC. Mapping genes with recombinant inbreds. In: Freeling M, Walbot V, editors. The Maize Handbook. New York: Springer-Verlag; 1994. pp. 249–254. [Google Scholar]
- Collazo P, Montoliu L, Puigdomènech P, Rigau J. Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol. 1992;20:857–867. doi: 10.1007/BF00027157. [DOI] [PubMed] [Google Scholar]
- DeVries S, Hoge H, Bisseling T. Isolation of total and polysomal RNA from plant tissues. Plant Mol Biol Manual. 1988;B6:1–13. [Google Scholar]
- Ebert E. Herbicidal effects of metolachlor (2-chloro-N-[ethyl-6-methyl-phenyl]-N-[2-methoxy-1-methylethyl]acetamide) at the cellular level in sorghum. Pestic Biochem Physiol. 1980;13:227–236. [Google Scholar]
- Fuerst EP. Understanding the mode of action of chloroacetamide and thiocarbamate herbicides. Weed Technol. 1987;1:270–277. [Google Scholar]
- Fuerst EP, Irzyk GP, Miller KD (1993) Partial characterization of glutathione S-transferase isozymes induced by the herbicide safener benoxacor in maize. Plant Physiol 102: 795–802 [DOI] [PMC free article] [PubMed]
- Genetics Computer Group . Program Manual for the Wisconsin Package. Madison, WI: Genetics Computer Group; 1994. [Google Scholar]
- Gowri G, Bugos RC, Campbell WH, Maxwell CA, Dixon RA. Stress responses in alfalfa (Medicago sativa L) Plant Physiol. 1991;97:7–14. doi: 10.1104/pp.97.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen PL, Christensen AB, Sommer-Knudsen J, Collinge DB. A putative O-methyltransferase from barley is induced by fungal pathogens and UV light. Plant Mol Biol. 1994;26:1797–1806. doi: 10.1007/BF00019493. [DOI] [PubMed] [Google Scholar]
- Hatzios KK. Herbicide antidotes: development, chemistry, and mode of action. Adv Agron. 1983;36:265–316. [Google Scholar]
- Hatzios KK. Mechanisms of action of herbicide safeners: an overview. In: Hatzios KK, Hoagland RE, editors. Crop Safeners for Herbicides. San Diego, CA: Academic Press; 1989. pp. 249–254. [Google Scholar]
- Held BM, Wang H, John I, Wurtele ES, Colbert JT. An mRNA putatively encoding for an O-methyltransferase accumulates preferentially in maize roots and is located predominantly in the region of the endodermis. Plant Physiol. 1993;102:1001–1008. doi: 10.1104/pp.102.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey JS, Krueger WA. Alachlor and 1,8-naphthalic anhydride effects on corn coleoptiles. Weed Sci. 1974;22:250–252. [Google Scholar]
- Hulbert SH, Bennetzen JL. Recombination at the Rp1 locus of maize. Mol Gen Genet. 1991;226:377–382. doi: 10.1007/BF00260649. [DOI] [PubMed] [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Latli B, Casida JE. Radiosynthesis of a chloroacetanilide herbicide ([phenyl-4-3H]acetochlor) and a dichloroacetamide safener for herbicides ([2,2-dimethyl-3H]R-29148) J Labelled Compd Radiopharm. 1995;36:147–155. [Google Scholar]
- Lay M-M, Hubbell JP, Casida JE. Dichloroacetamide antidotes for thiocarbamate herbicides: mode of action. Science. 1975;189:287–289. doi: 10.1126/science.1145201. [DOI] [PubMed] [Google Scholar]
- Pakusch A-E, Matern U. Kinetic characterization of caffeoyl-coenzyme A-specific 3-O-methyltransferase from elicited parsley cell suspensions. Plant Physiol. 1991;96:327–330. doi: 10.1104/pp.96.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig CL, Matthews DE, Van Etten HD. Purification and characterization of S-adenosyl-l-methionine:6a-hydroxymaack-iain 3-O-methyltransferase from Pisum sativum. Plant Physiol. 1989;91:559–566. doi: 10.1104/pp.91.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scott-Craig JS, Panaccione DG, Pocard J-A, Walton JD. The multifunctional cyclic peptide synthetase catalyzing HC-toxin production in the filamentous fungus Cochliobolus carbonum is encoded by a 15.7-kb open reading frame. J Biol Chem. 1992;67:26044–26049. [PubMed] [Google Scholar]
- Singh L, Jones KW. The use of heparin as a simple cost-effective means of controlling background in nucleic acid hybridization procedures. Nucleic Acids Res. 1984;12:5627–5638. doi: 10.1093/nar/12.14.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomènech P. The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell. 1995;7:407–416. doi: 10.1105/tpc.7.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD, Casida JE. Specific binding of a dichloroacetamide herbicide safener in maize at a site that also binds thiocarbamate and chloroacetanilide herbicides. Plant Physiol. 1995;109:213–219. doi: 10.1104/pp.109.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]